Abstract
shRNAs can trigger effective silencing of gene expression in mammalian cells, thereby providing powerful tools for genetic studies, as well as potential therapeutic strategies. Specific shRNAs can interfere with the replication of pathogenic viruses and are currently being tested as antiviral therapies in clinical trials. However, this effort is hindered by our inability to systematically and accurately identify potent shRNAs for viral genomes. Here we apply a recently developed highly parallel sensor assay to identify potent shRNAs for HIV, hepatitis C virus (HCV), and influenza. We observe known and previously unknown sequence features that dictate shRNAs efficiency. Validation using HIV and HCV cell culture models demonstrates very high potency of the top-scoring shRNAs. Comparing our data with the secondary structure of HIV shows that shRNA efficacy is strongly affected by the secondary structure at the target RNA site. Artificially introducing secondary structure to the target site markedly reduces shRNA silencing. In addition, we observe that HCV has distinct sequence features that bias HCV-targeting shRNAs toward lower efficacy. Our results facilitate further development of shRNA based antiviral therapies and improve our understanding and ability to predict efficient shRNAs.
Keywords: shRNA optimization, RNA secondary structure
The development of RNA interference (RNAi) technology to control gene expression has revolutionized biology and promises new therapeutic strategies for a variety of diseases (1). RNAi involves the endogenous production or artificial introduction of double-stranded RNA in cells, which can be recognized by the RNA-induced silencing complex (RISC). RISC selects one strand of the RNA (guide strand) and discards the other (passenger strand). Following strand selection, RISC uses the guide strand to recognize target mRNA substrates and directs the suppression of the mRNA by degradation and translational inhibition (2). Endogenous RNAi is triggered by microRNAs (miRNAs), ∼22 nucleotide (nt) RNAs derived from longer primary miRNAs (pri-miRNAs). Pri-RNAs are cleaved by the RNase III Drosha/DGCR8 complex to produce the ∼70-nt precursor miRNAs (pre-miRNAs) with hairpin-like stem loop structures. Pre-miRNAs are exported into the cytoplasm by exportin 5, where they are excised by the RNase III Dicer to produce the mature small double strand RNAs that are subsequently incorporated into RISC to suppress target mRNAs (3).
RNAi can be artificially triggered by introduction of synthetic duplex siRNAs into cells or expression of shRNAs. SiRNAs are synthetic double-stranded RNAs that mimic the endogenous Dicer excision product (4). SiRNAs are limited by their transient depletion and many cell types transfect poorly. ShRNAs are expressed from viral vectors in a form that mimics the stem-loop structure of the pre-miRNAs, and allows long-term, stable, and inheritable depletion of target mRNAs (5, 6). An improved expression approach is to express shRNAs in the context of endogenous miRNA transcripts that mimics pri-miRNAs. This approach allows expression from Pol II promoters and potent target depletion (7–10).
shRNAs have been applied to developing antiviral therapies in cell culture (11) and in clinical trials for HIV (12). A major challenge facing this strategy is that the high mutation rate of HIV makes it relatively easy for the virus to escape from suppression by mutating the shRNA target site (11). Consequently, multiple potent shRNAs will be required to prevent the occurrence of resistance, analogous to the highly active antiretroviral therapy (HAART) for HIV (11). In addition, a great concern for any RNAi application is off-target effects (i.e., the shRNA/siRNA can down-regulate not only the intended target but also unintended genes). Using lower doses of shRNAs with higher potency can reduce possible unwanted side effects without compromising their on-target effects. All things considered, the ability to accurately select highly potent shRNAs/siRNAs is key to the optimal application of RNAi and will improve our ability to use these molecules therapeutically.
We recently developed a multiplex high-throughput assay to identify potent shRNAs (13). This “sensor” assay uses a single vector to coexpress an shRNA and its target sequence fused to the 3′ UTR of a constitutively expressed fluorescence reporter. By monitoring the fluorescence intensity, we can measure the depletion efficiency of the shRNA and identify cells that express potent shRNAs. Combined with large-scale oligonucleotides synthesis and high-throughput sequencing technology or microarray hybridization, this single vector system allows multiplexed analysis of a large number of shRNA-target pairs. In this study, we applied the sensor assay to analyze ∼40,000 shRNAs from tiling the whole genomes of two HIV strains, one hepatitis C virus (HCV) strain, and one influenza A virus strain to identify strong antiviral shRNAs. This analysis uncovered sequence features of potent shRNAs and an overall dependence of silencing efficacy on the secondary structure of the target sequence. We also observe that distinct genome sequence features of HCV bias HCV-targeting shRNAs toward lower efficacy compared with HIV and influenza A virus.
Results
Sensor Assay Screen of ∼40,000 shRNAs from Tiling Four Viral Genomes.
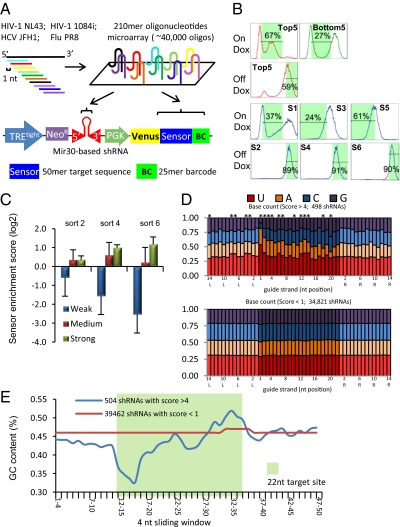
The sensor assay enables pooled screening for the discovery of potent shRNAs from a large number of candidates (13). Our original study validated the assay by evaluating 20,000 shRNAs from tiling of nine mammalian genes (13). In the present study we apply this platform to identify potent antiviral shRNAs from four viral genomes, including two HIV strains: NL43 of clade B and 1084i of clade C (14), one HCV strain: JFH1 of Genotype 2a (15), and one H1N1 influenza A strain: influenza A virus A/Puerto Rico/8/34 (PR8). We tiled the genomes to cover every possible target site in addition to a dozen previously validated shRNAs, also added as controls for the screen, which resulted in total 40,458 shRNAs. To construct the library, the stem-loop sequence of each shRNA was synthesized with a cognate 50-nt target sequence on the same 210-nt oligonucleotide. In addition, each 210-mer also includes a unique 25-mer barcode sequence (16) for microarray hybridization and restriction-enzyme sites for cloning. A two-step pooled cloning strategy was used to assemble the library into the pSensor vector, which is a retroviral vector with a Tet-responsive promoter (TREtight). The shRNA is under TRE promoter control, and the target sequence is constitutively expressed in the 3′ UTR of a fluorescence reporter (Venus). An engineered DF-1 chicken embryonic fibroblast cell line expressing ecotropic retroviral receptor and an improved reverse Tet-transactivator (rtTA3), termed the ERC (“Eco-rtTA-chicken”) cell line, was used for the sensor assay. To achieve 1,000-fold representation of each shRNA, 400 million ERC cells were infected with the retroviral library at a low multiplicity of infection (MOI = 0.1) to ensure single-copy integration of the retrovirus in the genome. After neomycin selection, cells went through three cycles of “Ping-Pong” sorting to select for highly potent shRNAs. Each cycle consisted of a doxycycline on (OnDox) sort for low fluorescence population and a doxycycline off (OffDox) sort for high-fluorescence population (Fig. 1B). In the OnDox sort, when shRNA expression is turned on, potent shRNAs would deplete Venus and shift their host cells to low fluorescence intensity. While the OffDox sort can eliminate cells with an intrinsically low fluorescence because of integration in a transcriptionally nonpermissive locus rather than because of depletion by shRNAs. Cells infected with five strong shRNAs (top 5) and five weak shRNAs (bottom 5) were sorted in parallel to determine the gating for library sorting (Fig. 1B). We extracted genomic DNA after sorts 2, 4, and 6 (OffDox) to measure shRNA abundance by microarray hybridization and deep sequencing (Fig. S1). Microarray hybridization and deep sequencing results show a strong correlation (Fig. S2) demonstrating the utility of both methods for the assay. We observed that the control shRNAs included in the library show clear separation after the sorts (Fig. 1C). Weak shRNAs dropped out starting from early sorts, medium-strength shRNAs show minimal enrichment after sort 6, and strong shRNAs controls are consistently enriched. We use the sort6 log2 enrichment ratio as the sensor score for ranking the efficacy of shRNAs.
Fig. 1.
Sensor assay screen of 40,000 shRNAs targeting four viral genomes. (A) Schematic for construction of the virus sensor library. Four viral genomes were tiled using ∼40,000 shRNAs that cover almost every possible target site. This library of 210-mer oligonucleotides that cover the shRNA sequence and cognate 50-nt target sites were synthesized on one microarray chip and cloned in two steps into the pSENSOR vector. (B) FACS sorting results of the sensor assay. First row: Top and bottom shRNA controls used for determining the gatings for OnDox conditions. Second row: Top shRNA controls used for determining the gatings for OffDox conditions. Third and Fourth rows: six sorts of the library. S1, S3, S5: sorts 1, 3, 5 are OnDox conditions and sorting for the low fluorescence population; S2, S4, S6: sorts 2, 4, 6 are OffDox conditions and sorting for the high fluorescence population. x axis is fluorescence intensity and y axis is normalized cell count. (C) Log2 Sensor enrichment scores of 13 control shRNAs in the library after sort 2, sort 4, and sort 6. Strong shRNAs are highly enriched compared with weak and medium shRNAs. (D) Comparison of nucleotide frequency of top shRNAs (sensor score >4) and low-scoring shRNAs (sensor <1). The 22-nt guide strand sequence is shown in dark colors; the 14-nt flanking sequences are shown in pastel colors. The flanking sequences of the reverse complements of the mRNA target region are shown: L corresponds to the 5′ flanking region and R to the 3′ flank of that reverse-complement strand covering the target sequence. Asterisks indicate positions that have a significant difference between top and low scoring shRNAs (P < 0.01). (E) Average GC content of the 4-nt sliding window of the 22-nt target site (green background) and the flanking regions, starting from 5′ to 3′ of the mRNA target.
Top-Scoring shRNAs Display Distinct Sequence Feature.
We analyzed the sequence patterns of the top-scoring shRNAs compared with the low-scoring shRNAs and observed distinct patterns of base compositions. The 498 top-scoring hairpins with sensor score >4 display a general enrichment for A/U in the guide strand and a thermodynamic asymmetry from 5′ to 3′ (Fig. 1E). The 5′ region of the guide strand is enriched for A/U (positions 1, 2, 3, 4, 5, 6, 10, 12, 13, and 14; P value <0.01) and the 3′ region tends to enrich for G/C (positions 18, 20; P value <0.01) (Fig. 1D). Guide strand position 1 is predominantly enriched for U (52% frequency), which can be explained by the preference for U to bind the RISC complex (17), but position 20 shows a significant depletion of A. Because this position pairs with position 1 of the passenger strand, depletion of A would make the passenger strand less likely have a U′ in the 5′ end, therefore less likely to compete with the guide strand for loading into RISC. These features confirm similar observations in our previous sensor screen (13). In addition, we also observe that the ∼8-nt region flanking the 3′ end of the 22-nt target site is also enriched for A, which has not been previously reported. Positions L3, L4, L7, and L8 in particular have ∼38% A, compared with an average of 31% with the low scoring shRNAs (P < 0.01) (Fig. 1D). Because this region is immediately adjacent to the shRNA seed-region binding site, the secondary structure in this region might affect the binding between shRNA and its target, which likely caused the sequence bias in this region.
Top-Scoring shRNAs Demonstrate Potent Antiviral Activity.
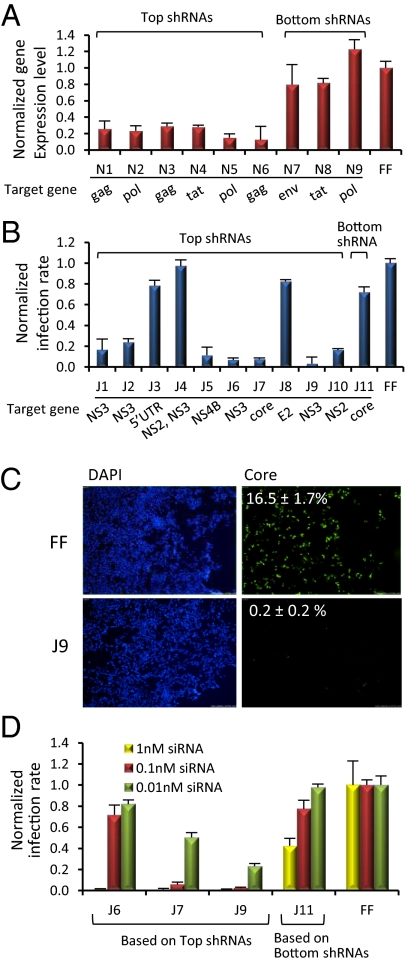
To test the efficacy of top-scoring HIV shRNAs, we generated HeLa-CD4 cell lines expressing shRNAs from a single-copy retroviral integration. We infected these cells with HIV-NL43 virus and examined the effect of different NL43-targeting shRNAs on HIV-NL43 virus gene expression. Six top-scoring NL43-targeting shRNAs (N1 to N6) depleted their target HIV genes by 70–90%, as measured by RT-PCR (Fig. 2A). In contrast, three low-scoring shRNAs (N7 to N9) displayed little or no depletion of their target genes. We also used an HCV cell-culture model (15) to validate top-scoring shRNAs targeting the HCV-JFH1 strain. Stable derivatives of Huh7.5.1 cells were generated to express individual shRNAs from single-copy retroviral integrations. Because the HCV genome is a single-stranded positive sense RNA that is also its mRNA, which is translated into a single polypeptide before further processing, the abundance of each HCV protein should correlate with the abundance of its mRNA. We used quantitative fluorescence imaging after immunostaining of the HCV core protein as a readout for shRNA-depletion efficiency. Of the top 10 scoring shRNAs, 7 had potent anti-HCV activity, reducing viral infection rates > 75% (Fig. 2B). Two low-scoring shRNAs were also tested: one (J11) has little effect on HCV infection, and another (J12) is highly cytotoxic, possibly explaining why it dropped out from the initial screen in the early sorts. Remarkably, four shRNAs achieved almost complete suppression of viral infection; J9 shRNA, in particular, brought down viral infection to background levels (Fig. 2C and Fig. S3). We then asked whether this potent shRNA antiviral effect could be translated to siRNAs. SiRNAs synthesized based on the sequences of shRNAs recapitulated the highly potent depletion efficiency. At 1 nM, siRNAs based on three top-scoring shRNAs completely suppressed HCV replication. Even at concentrations as low as 10 pM, two of the three siRNAs still have significant viral suppression activity, with J9 consistently demonstrating the highest potency (Fig. 2D). These results clearly demonstrate the ability of the sensor assay to identify very strong shRNAs. In addition, these shRNA sequences also convert into highly potent siRNAs.
Fig. 2.
Validation of top-scoring shRNAs in HIV and HCV cell-culture models. (A) Validation of six top-scoring shRNAs (N1–N6) targeting different HIV NL43 genes by rtPCR after NL43 infection of HeLa-CD4 cells in comparison with three low scoring shRNAs (N7–N9) and a control shRNA targeting firefly luciferase (FF). (B) Validation of the ten top scoring shRNAs (J1–J10) targeting different HCV-JFH1 genes by immunostaining of core protein of HCV in Huh 7.5.1 cells in comparison with a low scoring shRNA (J11) and a control shRNA targeting firefly luciferase (FF). (C) Immunostaining images of a top-scoring shRNAs J9 targeting HCV and shRNA targeting firefly luciferase were shown. (Left) DAPI staining showing nuclei of Huh 7.5.1 cells; (Right) staining of HCV core protein. The numbers are the percentages of cells infected by HCV. (Maginification: 20×.) (D) Three validated top shRNAs (J6, J7, J9) and a low-scoring shRNA (J11) were synthesized as siRNAs. Transfections of the siRNAs converted from top scoring shRNAs have strong anti-HCV effect compared with the low scoring J11 and a negative control siRNA targeting firefly luciferase.
RNA Secondary Structure Is a Major Determinant of shRNA Efficiency.
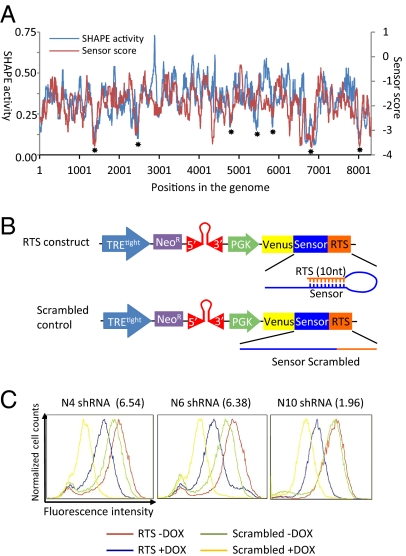
The secondary structure of the genome of HIV NL43 strain has been extensively studied using the SHAPE (selective 2'-hydroxyl acylation analyzed by primer extension) technology (18). We compared the sensor score and the SHAPE activity score of this strain and found they have a remarkable correlation. At regions of high secondary structure and thus low SHAPE reactivity, correspondingly low sensor scores were detected (asterisks shown in Fig. 3A). This correlation strongly suggests that shRNA efficacy is significantly reduced by secondary structures formed at the target site. This correlation also globally validates the results of the sensor screen. Although in regions with less secondary structure, and thus high SHAPE reactivity, sensor scores do not always show a strong degree of correlation. This finding suggests that favorable secondary structures of the target site are a necessary but not sufficient condition for shRNAs efficacy.
Fig. 3.
ShRNA efficacy is strongly influenced by target RNA secondary structure. (A) Correlation between the shRNA sensor score and SHAPE activity data for HIV NL43. Values are medians of a sliding window of 75 nt. The correlation coefficient is 0.3713 (P value < 10−100). Asterisks indicate regions with significant secondary structures. (B) RTS design scheme: 10-nt sequence reverse complementary to the seed-region binding site of the shRNA target site was inserted after the sensor sequence to form a duplex with the target site. The 10-nt scrambled sequences were inserted separately as negative controls. Three shRNAs targeting HIV-NL43 (N4, N6, N10) were tested using this method. (C) RTS experiments using the pSensor construct shows RTS can significantly reduce the depletion efficiency of shRNAs. Sensor scores are shown in the parentheses.
RNAi Target Shielding as a Method to Rescue RNAi.
Based on the result that secondary structure negatively correlates with shRNA efficacy, we reasoned that artificially introducing the formation of secondary structure should be able to inhibit the depletion by shRNAs. We named this strategy RNAi target shielding (RTS). We tested this idea using the pSensor construct by inserting a 10-nt sequence directly 3′ to the 50-nt sensor sequence (Fig. 3B). If the 10 nt is a randomly scrambled sequence, adding Dox induces shRNA expression and suppress fluorescence (compare Fig. 3C, green and yellow curves). However, if the 10 nt is reverse complementary to the 3′ of the 22-nt target site, when Dox is added, the degree of fluorescence intensity reduction is much less compared with the vector with a scrambled sequence (compare Fig. 3C, blue and yellow curves). We tested two top (N4 and N6) and one medium (N10) scoring shRNA using this method. All three cases demonstrate the capability of the reverse complementary sequence to inhibit depletion of the Venus fluorescent protein by the shRNAs. In addition, we observe that in the absence of Dox, the fluorescence intensity of the RTS construct-expressing cells is slightly higher than the scrambled construct-expressing cells. This result is likely because of RTS inhibition of the depletion by the shRNAs expressed due to low level Tet promoter activity in the absence of Dox, as the intensity differences (gaps between the red and green curves in Fig. 3C) correlates with the sensor scores of the three shRNAs.
HCV-Targeting shRNAs Demonstrate Lower Average Sensor Scores than HIV and Influenza A Virus.
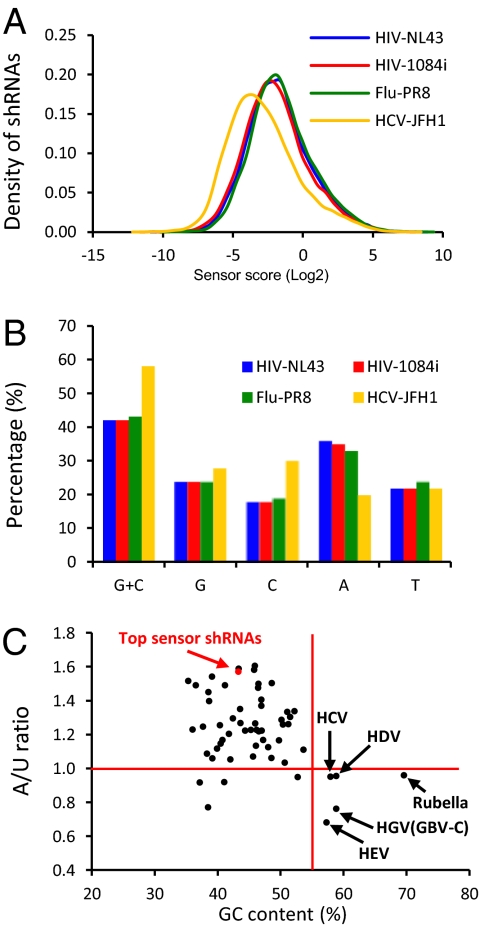
We analyzed the shRNA sensor scores of different viruses and found that HCV-targeting shRNAs have significantly lower sensor scores on average (P value < 0.001). As shown in Fig. 4A, although the sensor score curves of two HIV strains and influenza A PR8 are largely overlapping, the curve of HCV shRNAs shifts to the lower efficacy end, suggesting some general sequence features that distinguish HCV from HIV and influenza A virus. We examined extensively 28 sequence features that were known to affect the efficacy of siRNAs (19), and found that five features could distinguish HCV from HIV and influenza A virus, as shown in Table S1, which likely explain the decreased efficacy of HCV-targeting shRNAs. All five features directly correlate with the GC content of the viral genome. Indeed, the GC content of HCV is 58%, much higher than the other three viral genomes (42%, 42%, and 43% for HIV NL43, HIV 1084i, and influenza A PR8, respectively). One of the five features (UU at position 1 and 2) also directly correlates with the frequency of A in the viral genome. Indeed, the HCV genome contains 20% A, much lower than other three viral genomes (36%, 35%, and 33% for HIV-NL43, HIV-1084i, and influenza A-PR8, respectively). The significantly higher GC content and lower A content are likely the major reasons for HCV-targeting shRNAs’ lower efficacy, because we observe in our sensor screen that the top-scoring shRNAs have on average lower GC content and higher A content in their target sequences (Fig. 4C, red dot). We wondered how common are the two sequences features among RNA viruses. We analyzed 55 human RNA viruses and plotted their GC content against their A/U ratio (A% divided by U%) and found only five viruses share the pattern of high GC content and low A/U ratio, as HCV (Fig. 4C and Table S2). Four of the five viruses turned out to primarily infect human liver cells [HCV, HDV, HEV, and HGV (GBV-C)]. This connection might be explained by the highly active miRNA machinery in the liver (20), which may have exerted evolutionary pressure to bias the nucleotide compositions of the viruses infecting the liver.
Fig. 4.
HCV-targeting shRNAs have low sensor scores in average than HIV- and influenza A virus-targeting shRNAs. (A) Sensor score distribution of shRNAs by viral strains. (B) GC content and nucleotides frequencies of the four viral strains. (C) GC content and A/U ratio plot of 55 human RNA viruses and the average of 498 top shRNAs from the sensor screen.
Discussion
In this study, we applied an shRNA optimization assay to genomes of HIV, HCV, and influenza A virus and identified potent shRNAs targeting these pathogenic viruses from ∼40,000 shRNAs that cover the entire genomes of the four viral strains. The number of shRNAs used in this screen is twice the number we screened in our previous study (13), showing that large number of candidates can be examined in a single experiment. Using two cell-culture models for viral infection, we validated the top-scoring shRNAs and demonstrated their potent antiviral effect. These shRNAs target various genes of the viral genomes; therefore they could be tested for combinatorial treatment of viral infection (21, 22). Particularly, shRNAs targeting HCV demonstrate the ability to almost completely inhibit viral infectivity. This highly potent anti-HCV effect could be translated to synthetic siRNAs, which show strong antiviral activity at picomolar concentration. These data highlight the power of the sensor assay to identify strong shRNAs from a large number of candidates. Among the top 10 HCV-targeting shRNAs we tested, three had no anti-HCV effects. These false-positives are likely because of longer-range interaction of the RNA genome producing inhibitory structures or possibly because of the blocking of the target site by endogenous proteins that bind those sequences.
Our sensor assay screen uncovered several sequence features significantly enriched in top-scoring shRNAs. Several of these features confirm our previous observations, including thermodynamic asymmetry, high frequency of U at 5′ end of guide strand, bias for A/U at positions 2, 10, 13, 14, drop-out of A at position 20, and so forth. In addition, we also found sequence features not reported before. Notably, the ∼8-nt flanking region of the target site immediately adjacent to where the 5' end of the guide strand binds is enriched for A. This finding shows the importance of considering the sequence flanking the guide strand binding site for predicting strong shRNAs.
We observed a remarkable correlation of the sensor score with the SHAPE activity score along the whole genome of HIV-NL43. This finding clearly demonstrates that target RNA secondary structure is a major determinant of the efficacy of shRNA. We found SHAPE provides particularly useful information for excluding bad shRNAs because of highly structured target sites. Accordingly, SHAPE activity data, which can now be measured in a fast and high-throughput fashion (23), should be helpful for predicting shRNA efficacy. Furthermore, the ability of the sensor assay using only a 50-nt window to recapitulate experimentally derived secondary structure in native RNAs suggests that the vast majority of secondary structures are local in nature.
Inspired by these observations, we devised a unique way to rescue gene expression from the repressive effects of shRNAs by inserting a short stretch of nucleotide reverse complementary to the shRNA target site after the 3′ end of the reading frame. We named this method RNAi target shielding and showed that it can effectively inhibit depletion by shRNAs.
Comparing the sensor scores of four different viral genomes, we found that HCV shRNAs have on average lower sensor scores, indicating unique sequence features that make potent HCV-targeting shRNAs less frequent. This indication could be explained by the high GC content and low A content of the HCV genome. Remarkably, among 55 human ssRNA viruses analyzed, three of four viruses that share these sequence features with HCV also use liver cells as their natural hosts. These features are unlikely to be explained by codon usage as liver shows similar GC3 content (the GC content of the third nucleotide of codons) as other tissues (24). We hypothesize these unique sequence features are the outcome of viral evasion of the active miRNA machinery in the liver through evolution. Indeed, a large number of miRNAs are expressed in the liver. In addition, several miRNAs have been reported to be induced by IFN in liver cells to inhibit HCV infection (25). To survive the potential suppression by the abundant endogenous miRNAs in liver cells, the hepatitis viruses might have adapted to the high GC and low A content to reduce the chance of being inhibited by endogenous miRNAs. Given the facile delivery of RNAi reagent to the liver (26–28), we propose this susceptibility of HCV to RNAi could be exploited by RNAi-based therapy. Our results provide a rich database for selecting potent shRNAs to that end.
Experimental Procedures
See SI Experimental Procedures for additional experimental details. See Dataset S1 for the sequences and sensor scores by microarray hybridization and deep sequencing of the shRNAs used, and Dataset S2 for the genome sequences of the four viral strains used in this study.
Vectors, Library Construction, and Reporter Cell Lines.
We synthesized 40,458 210-mer oligonucleotides on a 55,000 features oligonucleotide array (Agilent Technology). Each 210-mer contains a 101-nt miR30-shRNA fragment, a 16-nt sequence containing EcoRI and MluI restriction enzyme sites for inserting the Venus-coding sequence, a 50-nt sensor cassette that is the target site for the shRNA, and a 25-mer unique barcode for microarray hybridization followed by an 18-nt primer binding site. A two-step pooled cloning strategy was used (13). In the first step, the 210-mer was cloned into the pSENSOR vector, the second step, the 3′miR30-PGK-Venus fragment was inserted between the shRNA and the sensor cassette. The ERC reporter cell line was as reported (13).
Sensor Ping-Pong Assay.
FACS sorting was performed on a BD FACSAria II sorting system (BD Biosciences). Four-hundred million cells were infected with the virus library to ensure 1,000-fold representation with single-copy integration of each virus. After selection by G418 (0.5 mg/mL), three cycles of Ping-Pong sorting were performed. In the “Ping” step, after treating the cells with Dox for 7 d, the cells were sorted to select for the low Venus population. In the “Pong” step, after 7 d of Dox withdrawal, cells were selected for high Venus expression. At least 1,000-fold representation was maintained after each sort and similar number of cells were frozen for extraction of genomic DNA. Five strong hairpins (top 5) and five weak hairpins (bottom 5) were used to determine the gating of the FACS sorting (13).
Supplementary Material
Acknowledgments
We thank members of the S.J.E. laboratory, D. Fusco and R. Chung for helpful discussion, and P. Yang for figure editing. X.T. is supported by a fellowship from the Damon Runyon Cancer Research Foundation (DRG-#[2008-09]); Z.J.L. is supported by grants from the National Natural Science Foundation of China (31100601) and the National Key Basic Research Program (2012CB316503); and Q.X. is supported by American Cancer Society Postdoctoral Fellowship 116410-PF-09-078-01-MGO. S.J.E., G.J.H., and S.W.L. are Howard Hughes Medical Institute investigators.
Footnotes
Conflict of interest statement: S.J.E., G.J.H., and S.W.L. are founders of Mirimus and serve on its scientific advisory board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119873109/-/DCSupplemental.
References
- 1.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 6.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 9.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 10.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkhout B. Toward a durable anti-HIV gene therapy based on RNA interference. Ann N Y Acad Sci. 2009;1175:3–14. doi: 10.1111/j.1749-6632.2009.04972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiGiusto DL, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2(36) doi: 10.1126/scitranslmed.3000931. 36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisson RD, et al. Infectious molecular clone of a recently transmitted pediatric human immunodeficiency virus clade C isolate from Africa: Evidence of intraclade recombination. J Virol. 2004;78:14066–14069. doi: 10.1128/JVI.78.24.14066-14069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc Natl Acad Sci USA. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 18.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 21.Liu YP, et al. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: A multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci USA. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen IM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrissey DV, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 28.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.