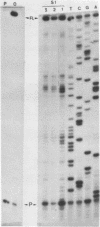
Abstract
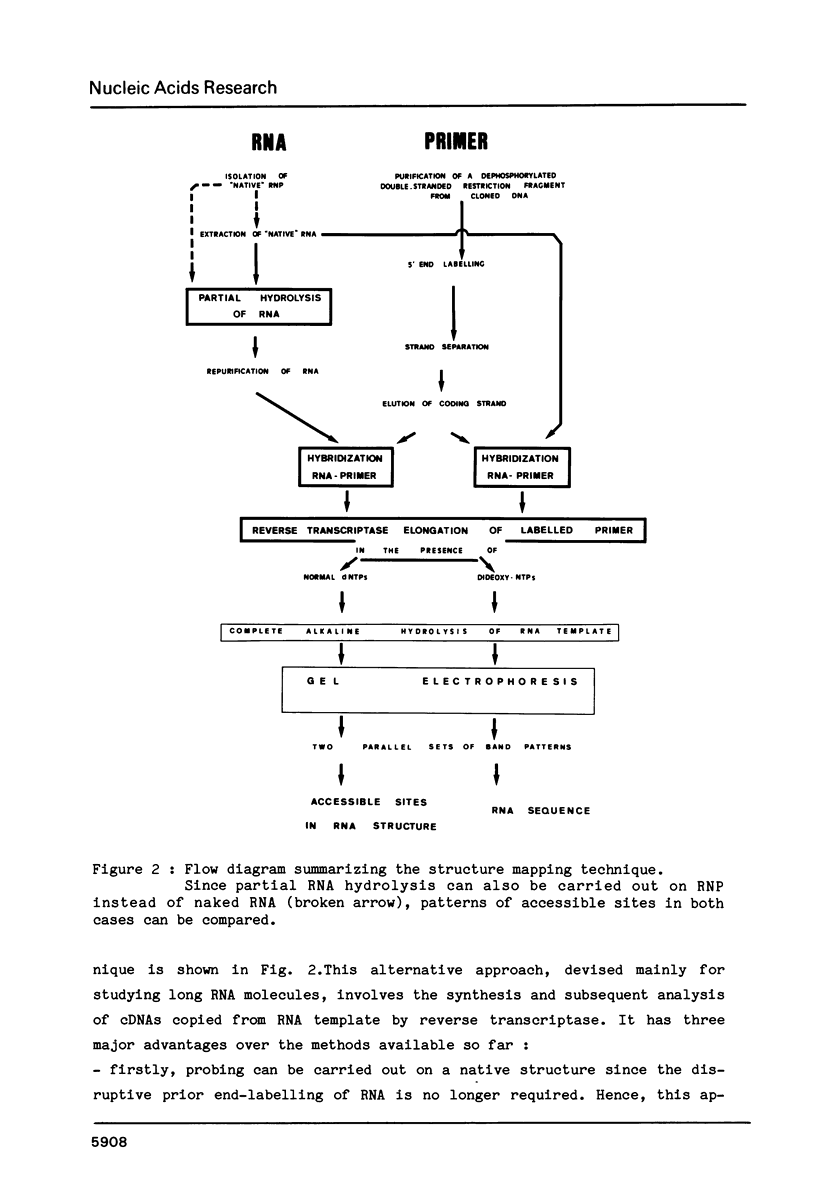
We have developed a combined approach for probing native structures in large RNAs. In the first method, after digestion with a structure specific nuclease, accessible sites are mapped at sequence resolution along the entire RNA molecule which is used as a template for the reverse transcriptase elongation of a 5' end labelled selected primer (coding strand of a small restriction fragment of the cloned gene). This method circumvents any prior end-labelling of RNA, a technique with major limitations for large RNAs. In the second approach, a rapid "heterologous" sequencing can be easily applied to definite domains of an RNA molecule in a variety of species (or individuals), without additional DNA cloning nor end-labelling of RNA. By taking advantage of the presence of evolutionary conserved tracts within an RNA sequence, it allows a rapid analysis of RNA folding patterns in terms of phylogenetic comparisons : when located within such a conserved tract, selected restriction fragments from a cloned gene can be used as heterologous primers for sequencing the upstream divergent region in RNAs of other species by currently available technology, i.e. reverse transcriptase elongation in the presence of chain terminator dideoxynucleotides.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Bachellerie J. P., Hearst J. E. Specificity of the photoreaction of 4'-(hydroxymethyl)-4,5',8-trimethylpsoralen with ribonucleic acid. Identification of reactive sites in Escherichia coli phenylalanine-accepting transfer ribonucleic acid. Biochemistry. 1982 Mar 16;21(6):1357–1363. doi: 10.1021/bi00535a039. [DOI] [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Thoren M., Salzman N., Thomspon J. A. Rapid sequence determination of late simian virus 40 16S mRNA leader by using inhibitors of reverse transcriptase. Proc Natl Acad Sci U S A. 1979 Feb;76(2):731–735. doi: 10.1073/pnas.76.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche M., Bachellerie J. P. RNA methylation and control of eukaryotic RNA biosynthesis. Effects of cycloleucine, a specific inhibitor of methylation, on ribosomal RNA maturation. Eur J Biochem. 1977 Mar 15;74(1):19–29. doi: 10.1111/j.1432-1033.1977.tb11362.x. [DOI] [PubMed] [Google Scholar]
- Dube D. K., Loeb L. A. On the association of reverse transcriptase with polynucleotide templates during catalysis. Biochemistry. 1976 Aug 10;15(16):3605–3611. doi: 10.1021/bi00661a031. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Kafatos F. C., Maniatis T. The primary structure of rabbit beta-globin mRNA as determined from cloned DNA. Cell. 1977 Apr;10(4):571–585. doi: 10.1016/0092-8674(77)90090-3. [DOI] [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. A slow tritium exchange study of the solution structure of Escherichia coli 5 S ribosomal RNA. J Mol Biol. 1981 Feb 25;146(2):223–239. doi: 10.1016/0022-2836(81)90433-2. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Friedman E. Y., Rosbash M. The syntheiss of high yields of full-length reverse transcripts of globin mRNA. Nucleic Acids Res. 1977 Oct;4(10):3455–3471. doi: 10.1093/nar/4.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Kawata Y., Ishikawa H. Primary and secondary structure of 5.8S rRNA from the silkgland of Bombyx mori. Nucleic Acids Res. 1982 Apr 10;10(7):2415–2418. doi: 10.1093/nar/10.7.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küntzel H., Heidrich M., Piechulla B. Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res. 1981 Mar 25;9(6):1451–1461. doi: 10.1093/nar/9.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMeur M., Glanville N., Mandel J. L., Gerlinger P., Palmiter R., Chambon P. The ovalbumin gene family: hormonal control of X and Y gene transcription and mRNA accumulation. Cell. 1981 Feb;23(2):561–571. doi: 10.1016/0092-8674(81)90152-5. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Connaughton J. F., Kumar A. Nucleotide sequence of the 5'- and 3'- domains for rabbit 18S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3445–3457. doi: 10.1093/nar/10.11.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta C. A., Wilson J. T., Forget B. G., Weissman S. M. Human beta-globin messenger RNA. III. Nucleotide sequences derived from complementary DNA. J Biol Chem. 1977 Jul 25;252(14):5040–5053. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F., Renalier M. H. Sequence of the 3'-terminal domain of mouse 18 S rRNA. Conservation of structural features with other pro- and eukaryotic homologs. FEBS Lett. 1982 Jun 7;142(2):260–266. doi: 10.1016/0014-5793(82)80148-8. [DOI] [PubMed] [Google Scholar]
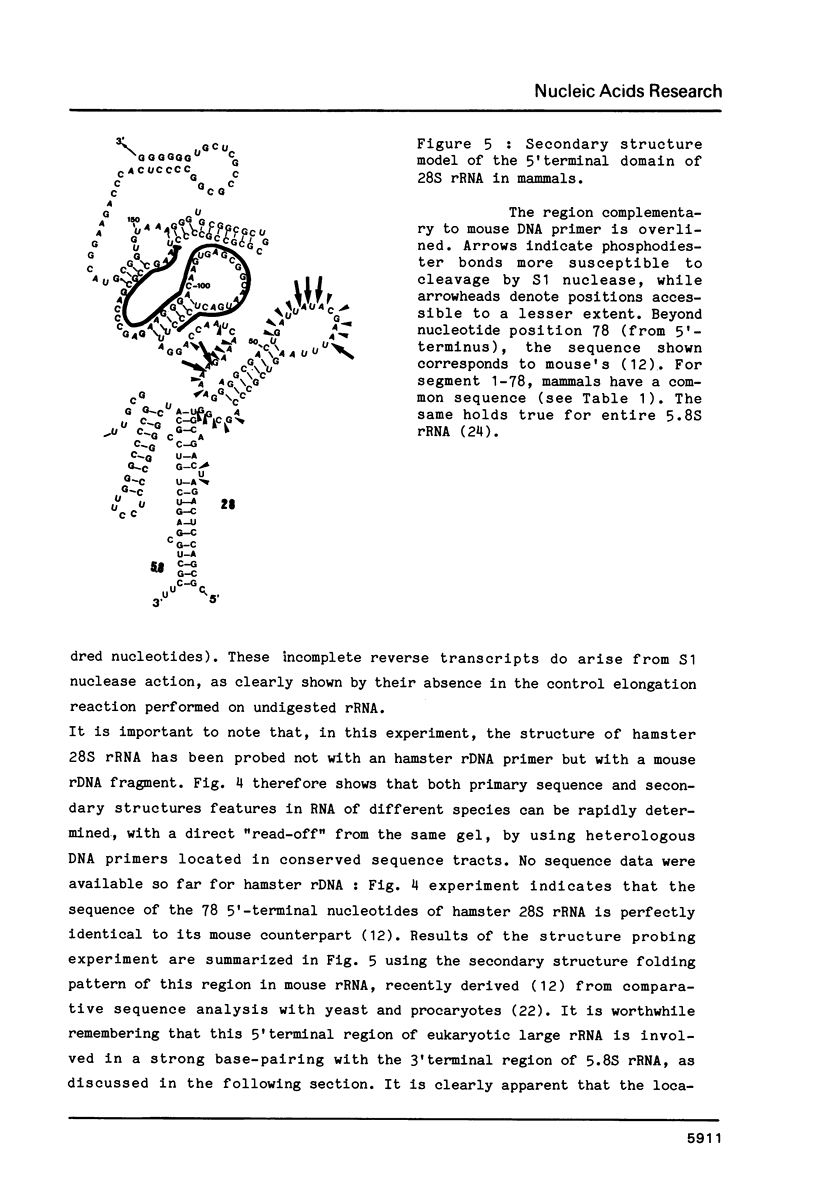
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Sogin M. L. Nucleotide sequence of Dictyostelium discoideum 5.8S ribosomal ribonucleic acid: evolutionary and secondary structural implications. Biochemistry. 1982 May 11;21(10):2335–2343. doi: 10.1021/bi00539a010. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979 Sep 27;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure of E. coli 16S RNA elucidated by psoralen crosslinking. Cell. 1983 Apr;32(4):1355–1365. doi: 10.1016/0092-8674(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Garrett R. A. A new RNA-RNA crosslinking reagent and its application to ribosomal 5S RNA. Nucleic Acids Res. 1978 Nov;5(11):4065–4075. doi: 10.1093/nar/5.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. A sequence from Drosophila melanogaster 18S rRNA bearing the conserved hypermodified nucleoside am psi: analysis by reverse transcription and high-performance liquid chromatography. Nucleic Acids Res. 1981 Apr 10;9(7):1723–1741. doi: 10.1093/nar/9.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. Reverse transcriptase pauses at N2-methylguanine during in vitro transcription of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3751–3754. doi: 10.1073/pnas.76.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965 Mar;8(2):357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]