Abstract
The scaffold protein for iron-sulfur cluster assembly, apo-IscU, populates two interconverting conformational states, one disordered (D) and one structured (S) as revealed by extensive NMR assignments. At pH 8 and 25 °C, approximately 70% of the protein is S, and the lifetimes of the states are 1.3 s (S) and 0.50 s (D). Zn(II) and Fe(II) each bind and stabilize structured (S-like) states. Single amino acid substitutions at conserved residues were found that shift the equilibrium toward either the S or the D state. Cluster assembly takes place in the complex between IscU and the cysteine desulfurase, IscS, and our NMR studies demonstrate that IscS binds preferentially the D form of apo-IscU. The addition of 10% IscS to IscU was found to greatly increase H/D exchange at protected amides of IscU, to increase the rate of the S → D reaction, and to decrease the rate of the D → S reaction. In the saturated IscU:IscS complex, IscU is largely disordered. In vitro cluster assembly reactions provided evidence for the functional importance of the S⇆D equilibrium. IscU variants that favor the S state were found to undergo a lag phase, not observed with the wild type, that delayed cluster assembly; variants that favor the D state were found to assemble less stable clusters at an intermediate rate without the lag. It appears that IscU has evolved to exist in a disordered conformational state that is the initial substrate for the desulfurase and to convert to a structured state that stabilizes the cluster once it is assembled.
Keywords: amino acid sequence effects on protein stability, protein order-disorder transition, two-dimensional exchange spectroscopy, biogenesis of Fe-S clusters
Iron-sulfur (Fe-S) clusters, which are among the most ancient and ubiquitous protein prosthetic groups, function in electron transport, enzymatic catalysis, and chemical sensing reactions or as structural units (1). Humans and other higher eukaryotes utilize the ISC (iron-sulfur cluster) system as the essential Fe-S cluster assembly mechanism in mitochondria, and defects in this system have been linked to a large number of human diseases (2). The prokaryotic ISC system has served as a useful model for understanding Fe-S cluster assembly and delivery. The bacterial system utilizes several proteins that have eukaryotic homologs: IscU (scaffold protein), IscS (cysteine desulfurase), HscB (cochaperone), HscA (chaperone), and CyaY (regulation or iron delivery, analog of human frataxin) (1). Interactions among these proteins have been shown to be critical for efficient Fe-S cluster biogenesis (3). The IscU protein acts as a scaffold on which the Fe-S clusters are assembled and from which the clusters are transferred to various apoproteins. IscS is a homodimeric pyridoxyl-5′-phosphate–dependent cysteine desulfurase (4). Each IscS subunit binds an IscU molecule and transfers sulfane sulfur generated from the conversion of cysteine to alanine to the cluster ligand cysteines of IscU (5). HscA and HscB, the DnaK-like chaperone and the DnaJ-like cochaperone proteins, respectively, facilitate the Fe-S cluster transfer mechanism in an ATP-dependent manner (6).
Although the steps leading to Fe-S cluster assembly on IscU have been under active examination, many key questions remain to be answered. We recently found that apo-IscU (the protein devoid of a metal ion or Fe-S cluster) from Escherichia coli exists in solution, under reducing conditions, as a mixture of two states: one that is structured (S) and one that is dynamically disordered (D) (7). We show here that substitutions of single amino acids at conserved sites can shift the equilibrium toward either the D or the S state.
By investigating the interaction between apo-IscU and IscS in solution by NMR spectroscopy, we found that IscS preferentially binds and stabilizes the D state of IscU. Iron-sulfur cluster assembly experiments conducted in vitro with a panel of IscU variants revealed the importance of the S⇆D equilibrium. Variants of apo-IscU predominantly in the S state exhibited a lag showing that the S → D reaction was the slow step in cluster assembly. Variants predominantly in the D state showed no lag but assembled clusters less rapidly than the wild type (WT) presumably because their S state, which supports the cluster, is less stable. The solution studies reported here suggest that the differential affinities of the D and S states of IscU play an important role in efficient iron-sulfur cluster assembly. Our finding that apo-IscU is largely disordered when bound to IscS differs from recent reports of X-ray (8), NMR (9), and small angle X-ray scattering (8, 10) investigations that found the IscU component to be structured within the IscU:IscS complex.
Results
Two Conformational States of Apo-IscU.
NMR spectra of apo-IscU (Fig. S1A) display two sets of signals reporting on two conformational states, a structured state and a dynamically disordered state. For a given residue, the signals from the two states can be linked on the basis of an exchange NMR spectrum, which show that the lifetimes of each state are on the order of 1 s (7). Our extensive assignments of the 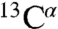 and
and 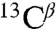 NMR signals from the S and D states of apo-IscU have enabled us to determine the secondary structural features of the two states (Fig. S1B). The D state exhibits little evidence of secondary structure, whereas the secondary structural elements of the S state correspond to those found in an NMR structure of the Zn2+ complex of IscU (11) and in the X-ray structure of the IscU:IscS complex (8). The assigned chemical shifts for the S (17837) and D (17836) forms of apo-IscU have been deposited at the BioMagResBank under the accession codes provided above.
NMR signals from the S and D states of apo-IscU have enabled us to determine the secondary structural features of the two states (Fig. S1B). The D state exhibits little evidence of secondary structure, whereas the secondary structural elements of the S state correspond to those found in an NMR structure of the Zn2+ complex of IscU (11) and in the X-ray structure of the IscU:IscS complex (8). The assigned chemical shifts for the S (17837) and D (17836) forms of apo-IscU have been deposited at the BioMagResBank under the accession codes provided above.
Single Amino Acid Substitutions in IscU Alter the Equilibrium Between the S and D States.
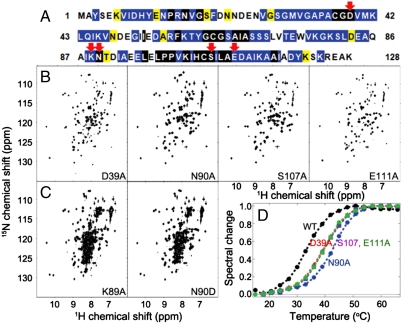
The D39A substitution (E. coli numbering system; Fig. 1A) of IscU is known to increase the stability of holo-IscU (12–14). In fact, the protein used in determining the X-ray structure of holo-IscU incorporated this stabilizing substitution (15). Our laboratory showed that E. coli apo-IscU(D39A) exists in solution primarily in the S state and adopts the same fold as the structured component of WT apo-IscU (7). This finding prompted us to search for other conserved residues in E. coli IscU (Fig. 1A) whose substitution perturbs the equilibrium between the S and D states. We report here three additional single-site substitutions that strongly favor the S state—N90A, S107A, and E111A (Fig. 1B)—and two that strongly favor the D state—K89A and N90D (Fig. 1C). Unlike D39A, these substitutions do not remove a potential cluster ligand. As expected, the substitutions that favor the S state increase the temperature at which the variants unfold (Fig. 1D). The structural differences among the variants were confirmed by circular dichroism (CD) spectroscopy (Fig. S2).
Fig. 1.
We have identified several single-site amino acid substitutions that shift the equilibrium between the S and D states of apo-IscU. Some substitutions favor the S state and some the D state. (A) Amino acid sequence of E. coli IscU: (black highlight) absolute conservation, (blue) conservative substitutions, and (yellow) semiconservative substitutions. Red arrows indicate positions substituted in the variants reported here. (B and C) Two-dimensional 1H-15N NMR spectra of E. coli IscU variant proteins labeled uniformly with nitrogen-15. The spectra in B show that IscU variants D39A, N90A, S107A, and E111A are largely structured, whereas those in C show that IscU variants K89A and N90D are largely disordered. (D) Thermal stabilities of wild-type apo-IscU (WT) and variants (D39A, S107, E111A, and N90A) that favor the S state. The variants that favor the D state, being largely unfolded, did not exhibit a thermal transition.
IscS Binds Preferentially to the D State of apo-IscU.
Given that apo-IscU exists in equilibrium between two states, S and D, it was of interest to determine whether one of these states interacts preferentially with IscS. We used three approaches to investigate this question: (i) We measured the effect of adding a substoichiometric quantity of IscS on the apo-IscU S → D and D → S transition rates as determined by two-dimensional exchange spectroscopy, (ii) we investigated the effect of added IscS on deuterium exchange at protected sites in the S state of apo-IscU, and (iii) we determined the conformational state of apo-IscU in the complex with IscS.
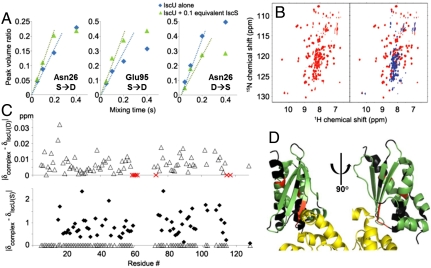
The S → D and D → S rates were determined by two-dimensional exchange spectroscopy by measuring the intensity of the exchange cross peak as a function of the exchange mixing time at short values of this delay before relaxation effects become dominant (16). We found that the addition of IscS increased the S → D rate and decreased the D → S rate for apo-IscU (Fig. 2A).
Fig. 2.
IscS binds preferentially to and stabilizes the D state of apo-IscU. (A) Volume of the S → D (Left and Center for Asn26 and Glu95, respectively) or D → S (Right for Asn26) exchange cross peak divided by the volume of the diagonal S or D peak, respectively, as a function of the mixing time in a two-dimensional NMR exchange experiment. Blue symbols are for IscU alone; green symbols are for IscU plus 0.1 equivalent of IscS. From the initial slopes, the S → D exchange rate increased from 0.77 s-1 for IscU alone to 1.1 s-1 for IscU in the presence of IscS; the D → S exchange rate decreased from 2.0 s-1 for IscU alone to 1.5 s-1 for IscU in the presence of IscS. The D⇆S equilibrium constant for apo-IscU from these measurements (0.24) is consistent with the relative populations of the S and D species under these conditions. (B) Two-dimensional 1H-15N NMR spectrum of apo-IscU labeled uniformly with nitrogen-15 (Left; red) and the spectrum of the same sample mixed with a stoichiometric amount of unlabeled IscS (Right; blue). (C) Extensive NMR assignments of signals from apo-IscU in the apo-IscU:IscS complex show that these IscU residues are disordered in the complex. The graphs show absolute values of the difference between the normalized backbone amide 1H-15N chemical shifts in the apo-IscU:IscS complex and those in the D (Upper) and S (Lower) state of free apo-IscU. For comparison, the data from the Upper panel (open triangle) are plotted on the same scale in the Lower panel. Red X’s in the Upper panel indicate residues whose signals were observed as apo-IscU(D) but broadened and disappeared upon addition of IscS. The normalized chemical shifts were calculated by the formula ΔδNH = [(ΔδN/6)2 + (ΔδH)2]1/2. The chemical shifts of the observed residues in the complex were similar to those of the D state and very different from those of the S state of apo-IscU. (D) X-ray crystal structure (Protein Data Bank ID code 3LVL) of the IscU:IscS complex (two views at right angles on one another) with IscU color coded to report information from NMR spectroscopy: Green are IscU residues determined to be disordered in the complex in solution on the basis of assigned NMR chemical shifts; red are residues observed in free apo-IscU that broadened and disappeared upon addition of IscS; and black are those residues whose NMR signals were not observed in either free apo-IscU or in the complex. Residues in yellow are from IscS.
Hydrogen exchange of protected amide groups in proteins can be measured by transferring 15N-labeled protein from H2O into D2O and following the decrease in intensity of two-dimensional (2D) NMR 1H-15N cross peaks as a function of time. The band-selective optimized flip-angle short transient NMR approach (17) enabled rapid recording of 2D spectra immediately after the solvent transfer. We found that the exchange rates of signals from WT apo-IscU increased in the presence of a small amount of added IscS; however, because protection factors for apo-IscU alone were low, the effects were difficult to quantify. By using N90A, a variant that exists primarily in the S state, the effects could be seen more clearly. In the absence of IscS, the exchange half-times for several residues were on the order of 1,000–2,000 s; however, in the presence of 0.11 equivalent of IscS, exchange was complete by the time (approximately 250 s) the first spectrum was collected (Fig. S3).
When we added unlabeled E. coli IscS to [U-15N]-apo-IscU, the 1H-15N heteronuclear single-quantum correlation (HSQC) spectrum changed to one indicative of the disordered state (Fig. 2B). By repeating this experiment with variants with differing stabilities of the D and S states, we found that higher concentrations of IscS were required to convert apo-IscU variants favoring the S state than those favoring the D state (Fig. S4), but with sufficient added IscS, conversion to the D state became nearly complete (Fig. S5).
Taken together, these results indicate that IscS binds preferentially to the D state of apo-IscU. Mass action serves to convert the remaining S state to a D-like state in the complex. The effect is reversible as shown by an experiment in which NMR spectra were taken of the complex formed between 15N-labeled apo-IscU and IscS before and after the addition of unlabeled apo-IscU; the spectrum of the initial complex showed signals only from disordered residues, whereas addition of unlabeled apo-IscU partially displaced 15N-apo-IscU from the complex, resulting in a spectrum showing signals from both the S and D states (Fig. S6).
Conformation of apo-IscU when Complexed with IscS.
We used 2D and 3D NMR methods to assign most of the observable backbone 1H-15N signals from WT apo-IscU in the complex with IscS (deposited at BioMagResBank under accession code 17844). The 1H-15N chemical shifts of apo-IscU in the IscU:IscS complex closely matched those of the D state of free apo-IscU but not those of the S state (Fig. 2C). These results identified which IscU residues are disordered in the complex in solution (residues colored green in the X-ray structure of the complex, Fig. 2D).
We used analytical gel filtration (Fig. S7) to estimate the mass of the complex in solution. The size determined (102 kDa) is in agreement with the (IscU)2∶(IscS)2 stoichiometry of the complex and the X-ray structure, which shows one IscU bound to each IscS subunit (8). Given the size of the complex, backbone signals from IscU residues in the binding site with IscS are expected to be exceedingly broad and not visible in the 1H-15N spectrum. Signals from residues (K59-G62, V73, and A117) in the D state of free apo-IscU broadened and disappeared upon addition of IscS (these residues are indicated by a red “X” in Fig 2C); they correspond to potential candidate residues in the interaction site with IscS (IscU residues colored red in the X-ray structure of the complex, Fig. 2D). IscU residues colored black in the X-ray structure of the complex (Fig. 2D) correspond to ones whose NMR signals were not observed in either free IscU or the complex.
Influence of Metal Binding on the Order-Disorder Processes.
It has been proposed that Zn2+ in the growth medium is required for the production of properly folded apo-IscU (9). To examine this claim, we produced WT [U-15N]-IscU in two M9 media, one supplemented with 8.4 μM ZnSO4 and the other with no added Zn2+; in both cases, we produced apo-IscU by EDTA treatment to remove metal ions followed by gel filtration to remove EDTA. The 2D 15N-HSQC spectra of apo-IscU produced from the two media showed the protein to be present as an identical mixture of S and D states (Fig. S8 A and B). We further observed that IscU adopts a single structured state upon the addition of either Zn2+ (Fig. S8C) or Fe2+ (Fig. S8D), with zinc forming a complex with higher thermal stability than iron (Fig. S8E). Thus, it is clear that the presence of metal ions can mask the interesting order-disorder equilibrium in apo-IscU by stabilizing an S-like state. We also examined whether IscU has similar properties under more physiological conditions of ionic composition or macromolecular crowding. The 2D 15N-HSQC spectrum of [U-15N]apo-IscU exhibited peaks from the S and D states under the following three conditions: (i) 150 mM KCl and 10 mM MgCl2 (Fig. S8F), (ii) 100 mg/mL Ficoll® PM 70 (Fig. S8G), and (iii) 100 mg/mL BSA (Fig. S8H). Furthermore, the addition of IscS to each of these solutions led to the appearance of disordered IscU peaks characteristic for the IscU:IscS complex (Fig. S8 I–M).
In Vitro Cluster Assembly.
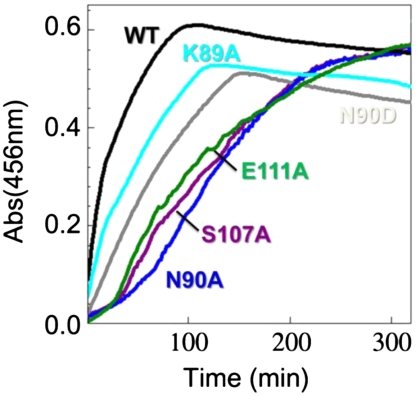
To examine the functional relevance of these findings, we carried out in vitro cluster assembly reactions (18) with a panel of IscU variants. We added each IscU variant anaerobically to the cluster assembly mixture (fivefold excess ammonium ferrous sulfate, fivefold excess l-cysteine, and 1/50-fold IscS) and monitored Fe-S cluster assembly by following the absorbance at 456 nm (Fig. 3). Under these conditions, IscU assembles a mixture of [2Fe-2S] and [4Fe-4S] clusters (18). We found that WT IscU assembled clusters most efficiently. The fully structured variants (N90A, S107A, and E111A) exhibited a biphasic reaction with an initial slow step and assembled clusters at an overall rate about seven times slower than WT IscU. By contrast, the partially disordered variants (K89A and N90D) exhibited no slow initial phase and assembled clusters about three times more slowly than WT IscU. The duration of the observed initial slow step in cluster assembly appears to correlate with the level of residual structured IscU in the mixture, as estimated from the relative intensities signals from the 15N-labeled side chain of tryptophan-76 of the apo-IscU variants, alone and in 1∶1 IscU:IscS mixtures (Fig. S4). Trp-76 is located in a mobile region in the complex, and its side-chain signal was observable in the complex.
Fig. 3.
Time course of iron-sulfur cluster assembly as monitored by absorbance at 456 nm. WT apo-IscU assembled clusters more efficiently than the other five variants studied. The more structured variants (E111A, S107A, and N90A) assembled clusters following an initial lag and at a slower rate than WT. Like WT, the less structured variants (K89A and N90D) did not exhibit an initial lag, but they assembled clusters at rates intermediate between WT and the more structured variants. The decay of intensity after its highest point arises from the instability of the cluster once formed.
In agreement with this model, we found that Fe-S cluster assembly in the presence of 250 μM ZnSO4, which stabilizes an S-like state of IscU, proceeded slowly with an initial lag (Fig. S9, scarlet). The rate of cluster assembly on WT IscU in the presence of Zn(II) was comparable to that of the most structured variant N90A in the absence of Zn(II), but the level of clusters assembled was lower. As expected, the reaction mixture without IscS did not show any cluster assembly (Fig. S9, orange).
Discussion
In most cases studied, protein-protein interactions are accompanied by increased order in the binding partners (19). However, several proteins have been identified that, like IscS, preferentially bind and stabilize a disordered target protein (20–22). Many of these are chaperone proteins that rescue misfolded proteins (23–25) or are proteins involved in protein degradation (26–32).
Apo-IscU and its interaction with IscS are governed by the equilibria shown in Eq. 1: 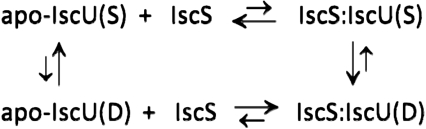 1 WT apo-IscU exists in equilibrium between its S and D states, and the relative levels were 70% S and 30% D under the experimental conditions of Fig. 1. In the presence of substoichiometric IscS, the S → D rate was found to increase and the D → S rate to decrease (Fig. 2A). The destabilization of apo-IscU by IscS was further confirmed by an increase in the H/D exchange rates at protected backbone amides (Fig. S3). Furthermore, apo-IscU became predominantly disordered in the IscS complex in solution (Fig. 2). Only a small number of apo-IscU residues interact with IscS; these may include some of the residues identified in the X-ray structure of the complex (8) as being in the contact region between the two proteins (Fig. 2D). However, the N-terminal residues of apo-IscU, which are disordered in free apo-IscU (Fig. S1) [also in the Zn2+∶IscU complex in solution (11)] but were found to become structured in the X-ray structure of the IscU:IscS complex (8), are not among those residues that interact with IscS, because we found that they remain disordered in the complex in solution (Fig. 2C). We interpret these results to indicate that IscS binds preferentially to the D form of apo-IscU over the S form (Eq. 1).
1 WT apo-IscU exists in equilibrium between its S and D states, and the relative levels were 70% S and 30% D under the experimental conditions of Fig. 1. In the presence of substoichiometric IscS, the S → D rate was found to increase and the D → S rate to decrease (Fig. 2A). The destabilization of apo-IscU by IscS was further confirmed by an increase in the H/D exchange rates at protected backbone amides (Fig. S3). Furthermore, apo-IscU became predominantly disordered in the IscS complex in solution (Fig. 2). Only a small number of apo-IscU residues interact with IscS; these may include some of the residues identified in the X-ray structure of the complex (8) as being in the contact region between the two proteins (Fig. 2D). However, the N-terminal residues of apo-IscU, which are disordered in free apo-IscU (Fig. S1) [also in the Zn2+∶IscU complex in solution (11)] but were found to become structured in the X-ray structure of the IscU:IscS complex (8), are not among those residues that interact with IscS, because we found that they remain disordered in the complex in solution (Fig. 2C). We interpret these results to indicate that IscS binds preferentially to the D form of apo-IscU over the S form (Eq. 1).
The importance of the S⇆D equilibrium to functional cluster assembly is revealed by studies of IscU variants that favor either the D or the S state. IscU variants that favor the S state (N90A, S107A, and E111A) exhibited an initial slow step in cluster assembly (Fig. 3); it appears that the S → D step is rate-limiting in these cases. WT IscU in the presence of Zn(II), which stabilizes the S state, also exhibited inhibited cluster assembly (Fig. S9). By contrast, apo-IscU variants predominantly in the D state (K89A and N90D) assembled clusters without a lag but at a slower rate than WT IscU, possibly because the intermediates in cluster assembly are less stable. WT apo-IscU accumulated clusters most efficiently, indicating that enough of the D form was present so as not to limit the rate of assembly and that the S form was sufficiently stable to support rapid cluster assembly.
The structure of the apo-IscU:IscS complex in solution differs profoundly from that observed by X-ray crystallography (8). It appears that the species crystallized was the small fraction of ordered apo-IscU in the complex stabilized by the crystallization cocktail. In the X-ray crystal structure of the complex, residues of IscU exhibited larger B factors (indicative of greater disorder) than those of IscS, particularly those residues of IscU far from the interaction site with IscS (8). In a previous NMR study of the interaction between [15N]-IscU and IscS, it was reported that nearly all signals became broadened in the complex (9). We have observed a similar effect with IscU∶Zn2+ in place of apo-IscU (Fig. S8I) and presume that the earlier study (8) involved the metal complex, which is resistant to the disorder transition.
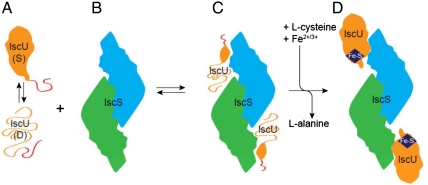
Our working model for iron-sulfur cluster assembly starts with apo-IscU existing in equilibrium between its D and S states (Fig. 4A). IscU bound to the IscS homodimer (Fig. 4B) is predominantly disordered (Fig. 4C) as the result of the much higher affinity of IscS for the D state over the S state. Once sulfur and iron have been recruited to the scaffold protein, the cluster assembles, and IscU switches to the S state, which stabilizes the cluster (Fig. 4D). The fact that IscS has a lower affinity for the S state helps enable dissociation of holo-IscU, which then transfers its cluster to an acceptor protein.
Fig. 4.
Working model for iron-sulfur cluster assembly. (A) The scaffold protein, IscU, is monomeric and populates two conformational states in solution, one S and one D, that yield separate sets of NMR signals. In both states, the N-terminal residues (represented in red) are disordered, and a single set of NMR signals is observed. (B) The cysteine desulfurase, IscS, is a homodimer. (C) Upon binding IscS, IscU becomes largely disordered. (D) The cysteine desulfurase converts l-cysteine to l-alanine and in doing so generates sulfur, which is picked up by mobile cysteine residues of IscU and, along with iron ions, is used in cluster assembly.
A number of advantages may accrue from the IscU S⇆D equilibrium and differential affinity of IscS for the D and S states shown by this model. Preferential binding to the disordered form of apo-IscU ensures minimal competition from holo-IscU. The disordering of IscU in the complex also ensures that metal ions are released. The model is consistent with the sulfur-first model for cluster assembly (33), and the mobility of the metal-free cysteines of IscU in the complex may assist in their interaction with active site residues of IscS that transfer the sulfur atoms. The dynamic nature of IscU should also make the cluster assembly site more accessible to proteins that deliver iron and electrons required for cluster assembly. The persulfated cysteines of IscU then accumulate iron, and the dynamics may assist in atom rearrangements leading to cluster assembly. Concurrent with, or subsequent to, cluster assembly, IscU switches from the D state to the S state to produce holo-IscU. The lower affinity of the S state may minimize product inhibition. We previously reported that the cochaperone HscB, which is involved in recruiting holo-IscU to the chaperone HscA, binds preferentially to the S form of apo-IscU (7). These factors may account for the exceedingly high efficiency of the ISC system, which must support high-throughput production of iron-sulfur proteins.
The panel of IscU variants presented here will be useful in determining the structural and dynamic features of the S⇆D transition of apo-IscU. The variants also can aid in uncovering the detailed steps in Fe-S cluster assembly and cluster transfer to acceptor proteins. The report that cluster transfer to apo-ferredoxin is slower with IscU variant D39A than with WT IscU (34) suggests that the S → D transition likely plays a role also in this process.
Methods
Production and Purification of Proteins.
[U-15N]-labeled samples of IscU variants were produced and purified in the apo form as described previously (35). The expression plasmids of IscU variants were created by applying the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) to the pTrcIscU expression vector (35). IscS was prepared as described in SI Methods. To check whether the presence of ZnSO4 in the expression media served to remove the conformational heterogeneity of apo-IscU as previously proposed (9), two separate [U-15N]-apo-IscU samples were prepared; one of them was expressed in a regular M9 medium without a zinc supplement, whereas the other was expressed in a M9 medium containing 8.4 μM ZnSO4. The two protein samples were purified by the same procedure as described previously (35).
NMR Spectroscopy.
All NMR spectra were acquired at 25 °C with 600- or 800-MHz Varian NMR System spectrometers equipped with a z-gradient cryogenic probe, and all samples contained 7% D2O, 0.7 mM 2,2-dimethyl-2-silapentane-5-sulfonate, and 0.02% sodium azide. NMRPipe (36) was used to process the raw NMR data, and Sparky (University of California, San Francisco) was used for data analysis. The samples for the signal assignments of the S and D states of WT apo-IscU and the exchange rate determination between the S and D states of WT apo-IscU were prepared in 50 mM Tris·HCl buffer at pH 7.5 containing 0.5 mM EDTA and 5 mM DTT. The samples for the hydrogen exchange rate measurements of apo-IscU(N90A) were prepared in 50 mM Tris·HCl buffer at pH 7.5 containing 150 mM NaCl, 0.5 mM EDTA, and 5 mM DTT. Salt was added to the samples used for the H/D exchange measurements because it stabilizes the S state of apo-IscU and made it easier to measure the exchange rates. We used fast maximum-likelihood reconstruction as implemented in the Newton software package (37) in analyzing the hydrogen exchange and 2D NMR exchange data. More experimental details are provided in SI Methods.
CD Spectroscopy.
The buffer used for CD spectroscopy contained 20 mM Tris·H2SO4 (pH 8.0) and 0.5 mM Tris(2-carboxyethyl)phosphine. The 1-mm path-length cuvettes were sealed with a rubber septum as needed. The concentration of apo-IscU was 10–20 μM. To prepare the metal complexes, IscU∶Zn2+ and IscU∶Fe2+, ZnSO4, or (NH4)2Fe(SO4)2 were added to a protein sample to achieve a metal ion concentration of 100 μM. Far-UV CD spectra of IscU variants and complexes were recorded with an Aviv 202SF CD spectrophotometer (Aviv Biomedical) at 25 °C. Thermal denaturation curves of IscU variants were obtained by following CD signals at 222 nm while increasing the temperature in 3 °C increments from 9 to 69 °C. The protein samples were incubated for 5 min at each temperature. Because of the variable secondary structure content of IscU variants, the final thermal denaturation curves were normalized so that “0” represented the structured state whereas “1” represented the unfolded state (Fig. 1D).
Reconstitution of [Fe-S]-IscU.
The approach used was adapted from a published protocol (18). All the experiments involving [Fe-S]-IscU were conducted in an anaerobic chamber (Coy Laboratory) filled with 90% N2 and 10% H2 at room temperature. The O2 level was maintained at less than 5 ppm. The buffer used to reconstitute [Fe-S]-IscU was 0.1 M Tris∙HCl (pH 7.5) and 5 mM DTT. The reconstitution mixture in this buffer contained 50 μM apo-IscU, 250 μM ferrous ammonium sulfate, and 1 μM IscS. The reaction was initiated by adding l-cysteine to achieve a concentration of 250 μM. We used 1-cm path-length cuvettes sealed with a rubber septum to measure the absorbance at 456 nm, which positively correlates with the increasing amount of the reconstituted [Fe-S]-IscU. A UV-1700 UV-visible spectrophotometer (Shimadzu) equipped with a temperature-controlled cell positioner was used for the absorbance measurements, and the raw data were processed with UVProbe 2.21 software (Shimadzu). All reconstitution experiments were repeated at least twice to confirm reproducibility.
Supplementary Material
Acknowledgments.
We thank University of Wisconsin–Madison colleagues Patricia Kiley for providing the expression plasmid for IscS, Taewook Kim for helping to prepare an expression plasmid for IscU variants, Roger Chylla for assistance with spectral reconstruction, and M. Thomas Record for suggesting the ion composition mimicking E. coli cytoplasm. Huanyao Gao and Michael K. Johnson (University of Georgia) trained J.H.K. in the use of their protocol for in vitro assembly of holo-IscU. This work was supported by U.S. National Institutes of Health grants R01 GM58667 and U01 GM94622. This study was in collaboration with the National Magnetic Resonance Facility at Madison, which is supported by US National Institute of Health Grant P41 RR02301.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 17837, 17836, and 17844).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114372109/-/DCSupplemental.
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 4.Cupp-Vickery JR, Urbina H, Vickery LE. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J Mol Biol. 2003;330:1049–1059. doi: 10.1016/s0022-2836(03)00690-9. [DOI] [PubMed] [Google Scholar]
- 5.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 6.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JH, et al. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry. 2009;48:6062–6071. doi: 10.1021/bi9002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi R, et al. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prischi F, et al. Of the vulnerability of orphan complex proteins: The case study of the E. coli IscU and IscS proteins. Protein Expr Purif. 2010;73:161–166. doi: 10.1016/j.pep.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Prischi F, et al. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat Commun. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramelot TA, et al. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 12.Unciuleac MC, et al. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, et al. Characterization of an iron-sulfur cluster assembly protein (ISU1) from Schizosaccharomyces pombe. Biochemistry. 2002;41:5024–5032. doi: 10.1021/bi016073s. [DOI] [PubMed] [Google Scholar]
- 14.Foster MW, et al. A mutant human IscU protein contains a stable [2Fe-2S](2+) center of possible functional significance. J Am Chem Soc. 2000;122:6805–6806. [Google Scholar]
- 15.Shimomura Y, Wada K, Fukuyama K, Takahashi Y. The asymmetric trimeric architecture of [2Fe-2S] IscU: Implications for its scaffolding during iron-sulfur cluster biosynthesis. J Mol Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Montelione GT, Wagner G. 2D chemical exchange NMR spectroscopy by proton-detected heteronuclear correlation. J Am Chem Soc. 1989;111:3096–3098. [Google Scholar]
- 17.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 18.Agar JN, et al. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 19.Tompa P, et al. Close encounters of the third kind: Disordered domains and the interactions of proteins. Bioessays. 2009;31:328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 20.Forster ML, Mahn JJ, Tsai B. Generating an unfoldase from thioredoxin-like domains. J Biol Chem. 2009;284:13045–13056. doi: 10.1074/jbc.M808352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothballer A, Tzvetkov N, Zwickl P. Mutations in p97/VCP induce unfolding activity. FEBS Lett. 2007;581:1197–1201. doi: 10.1016/j.febslet.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Gerega A, et al. VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J Biol Chem. 2005;280:42856–42862. doi: 10.1074/jbc.M510592200. [DOI] [PubMed] [Google Scholar]
- 23.Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. 2010;6:914–920. doi: 10.1038/nchembio.455. [DOI] [PubMed] [Google Scholar]
- 24.Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: An unfolding story. Curr Protein Pept Sci. 2009;10:432–446. doi: 10.2174/138920309789351930. [DOI] [PubMed] [Google Scholar]
- 25.Slepenkov SV, Witt SN. The unfolding story of theEscherichia coliHsp70 DnaK: Is DnaK a holdase or an unfoldase? Mol Microbiol. 2002;45:1197–1206. doi: 10.1046/j.1365-2958.2002.03093.x. [DOI] [PubMed] [Google Scholar]
- 26.Prakash S, Matouschek A. Protein unfolding in the cell. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Effantin G, Maurizi MR, Steven AC. Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase. J Biol Chem. 2010;285:14834–14840. doi: 10.1074/jbc.M109.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beskow A, et al. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 29.Farbman ME, Gershenson A, Licht S. Single-molecule analysis of nucleotide-dependent substrate binding by the protein unfoldase ClpA. J Am Chem Soc. 2007;129:12378–12379. doi: 10.1021/ja074168x. [DOI] [PubMed] [Google Scholar]
- 30.Hammarstrom P, Persson M, Owenius R, Lindgren M, Carlsson U. Protein substrate binding induces conformational changes in the chaperonin GroEL. A suggested mechanism for unfoldase activity. J Biol Chem. 2000;275:22832–22838. doi: 10.1074/jbc.M000649200. [DOI] [PubMed] [Google Scholar]
- 31.Benaroudj N, Tarcsa E, Cascio P, Goldberg AL. The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie. 2001;83:311–318. doi: 10.1016/s0300-9084(01)01244-5. [DOI] [PubMed] [Google Scholar]
- 32.Matouschek A. Protein unfolding—an important process in vivo? Curr Opin Struct Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 33.Smith AD, et al. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 34.Wu SP, Wu G, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: Role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 35.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, et al. NMRPIPE—A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Chylla RA, Hu K, Ellinger JJ, Markley JL. Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: Application to quantitative metabolomics. Anal Chem. 2011;83:4871–4880. doi: 10.1021/ac200536b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.