Abstract
Human cytomegalovirus infections involve the extensive modification of host cell pathways, including cell cycle control, the regulation of the DNA damage response, and averting promyelocytic leukemia (PML)-mediated antiviral responses. The UL35 gene from human cytomegalovirus is important for viral gene expression and efficient replication and encodes two proteins, UL35 and UL35a, whose mechanism of action is not well understood. Here, affinity purification coupled with mass spectrometry was used to identify previously unknown human cellular targets of UL35 and UL35a. We demonstrate that both viral proteins interact with the ubiquitin-specific protease USP7, and that UL35 expression can alter USP7 subcellular localization. In addition, UL35 (but not UL35a) was found to associate with three components of the Cul4DCAF1 E3 ubiquitin ligase complex (DCAF1, DDB1, and DDA1) previously shown to be targeted by the HIV-1 Vpr protein. The coimmunoprecipitation and immunofluorescence microscopy of DCAF1 mutants revealed that the C-terminal region of DCAF1 is required for association with UL35 and mediates the dramatic relocalization of DCAF1 to UL35 nuclear bodies, which also contain conjugated ubiquitin. As previously reported for the Vpr-DCAF1 interaction, UL35 (but not UL35a) expression resulted in the accumulation of cells in the G2 phase of the cell cycle, which is typical of a DNA damage response, and activated the G2 checkpoint in a DCAF1-dependent manner. In addition, UL35 (but not UL35a) induced γ-H2AX and 53BP1 foci, indicating the activation of DNA damage and repair responses. Therefore, the identified interactions suggest that UL35 can contribute to viral replication through the manipulation of host responses.
INTRODUCTION
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus subfamily and consists of an ∼230-kbp double-stranded DNA genome encased in an icosahedral capsid, surrounded by a proteinaceous matrix (tegument) layer and a host-derived lipid bilayer containing several viral glycoproteins. HCMV can establish both lytic and latent infections in human hosts yet causes little to no adverse effect in healthy adults. However, lytic HCMV replication is associated with significant disease and sometimes death in immunocompromised hosts, typically transplant recipients, neonates, and people with AIDS (16). HCMV encodes more than 200 viral proteins, although many remain poorly or completely uncharacterized (77). The expression of specific viral proteins is temporally controlled during the three general phases of the lytic replication cycle: the immediate-early (IE), early, and late phases (73). In addition, in the pre-IE phase, tegument-derived viral proteins are delivered to the host cell preformed and therefore can act before viral gene expression occurs to manipulate cells in ways that favor lytic replication (38).
Herpesvirus infections are associated with the extensive manipulation of host cell processes, including the control of the cell cycle, apoptosis, immune activation, and the DNA damage response (DDR) (2, 11, 64, 94). One of the first challenges to HCMV lytic replication in newly infected cells is overcoming the repressive effects of the promyelocytic leukemia (PML) protein (8, 90, 91). PML provides the molecular basis for the intrinsic immune response through the formation of PML nuclear bodies (NBs) that recruit, organize, and modify nuclear proteins that can silence viral gene expression (5, 17, 23, 74, 89). Soon after infection, HCMV genomes become associated with PML, and expression from the strong major immediate-early promoter (MIEP) is repressed, possibly through the histone modification of the MIEP promoter region (35, 67, 98). The tegument protein pp71 (UL82) contributes to host manipulation by alleviating the repressive effects of PML on the MIEP by displacing the transcriptional repressor ATRX and degrading Daxx (34, 59, 74). The activation of the MIEP results in the expression of the immediate-early protein IE1, which associates with, and mediates the dispersal of, PML NBs, further relieving PML-mediated repression and enhancing lytic replication (1, 44). In addition, the MIEP controls the expression of IE2 which, along with IE1, contributes to cell cycle arrest at the G1/S transition and viral gene expression (9, 73, 96).
Cell cycle control is essential for ensuring access to specific DNA replication machinery that the virus does not encode. To this end, herpesviruses, including HCMV, arrest cells at a G1/S transition in such a way that viral but not cellular DNA synthesis occurs (6, 10, 70). In the case of HCMV, the tegument proteins pp71 (UL82) (39, 40) and UL69 (58), as well as the immediate-early proteins IE1 and IE2, contribute to cell cycle control. Herpesvirus lytic replication also is associated with ATM (ataxia telangiectasia-mutated)-mediated DDR activation, and several proteins from this pathway are recruited to sites of viral replication (20, 45, 46, 84, 97). However, herpesviral proteins also interfere with some aspects of the ATM response such that apoptosis does not occur (11, 94). In addition, several individual herpesviral proteins have been shown to be sufficient to induce cell cycle arrest and/or ATM signaling (10, 52, 58, 60, 65, 66). Given the importance of controlling the cell cycle, apoptosis, and DDR pathways for HCMV replication, it is likely that other viral proteins contribute to the regulation of these processes to optimize the cellular environment for replication.
To better understand how the many uncharacterized HCMV proteins manipulate cellular processes, we previously conducted a screen of individual HCMV proteins for the ability to associate with and/or disrupt nuclear structures, including the nucleolus, Cajal bodies, nuclear speckles, and PML bodies (77). One interesting finding was that the UL35 protein formed ring-like nuclear bodies in transfected cells that recruited PML proteins and remodeled PML nuclear bodies, including the PML-associated proteins Sp100 and Daxx (76, 77, 82). The UL35 gene from HCMV encodes two proteins (UL35 and UL35a) which are expressed at different times during infection (55). The larger protein, UL35, consists of 640 amino acids and is produced late in infection (55). UL35 also is packaged into progeny virions as a minor tegument component (93) and therefore is delivered preformed to newly infected cells, where it is positioned to exert effects on the cell before viral gene expression. The shorter protein, UL35a, consists of amino acids 448 to 640 of UL35 and is produced both early and late in infection by the alternative transcription of the UL35 gene (55). Unlike UL35, UL35a does not appear to be a structural protein (55, 93). The deletion of the UL35 gene, and thus both UL35 and UL35a protein products, indicates that the gene is essential at low multiplicities of infection (MOI) and results in delayed replication and growth defects at higher MOI (19, 81). In addition, the deletion of the UL35 gene causes the improper subcellular trafficking of pp71 and other viral proteins (81).
Despite sharing identical C-terminal sequences, UL35 and UL35a do not appear to have similar biological functions. Reporter-based assays suggest that UL35 can activate the MIEP, whereas UL35a has no effect or is inhibitory (55, 82). In addition, the ability to form PML-altering nuclear bodies is limited to UL35, as UL35a adopts a strictly pan-nuclear localization pattern and has no apparent effect on the organization of PML (76, 77, 82). Both UL35 and UL35a can interact with the tegument protein pp71 (82). However, UL35a affects the nuclear shuttling of pp71 (76), while pp71 enhances the formation of UL35 nuclear bodies (76, 82). Finally, UL35 and UL35a can interact with each other (76), adding another level of complexity and regulation to the activity of these proteins. These divergent activities of UL35 and UL35a support the need for the temporal control of their expression throughout the replication cycle. While current data suggest that UL35 and UL35a have important but diverse effects, little is known about how these proteins affect cells or about the cellular proteins that are targeted by these proteins. Here, we use an affinity purification approach coupled to mass spectrometry (AP-MS) to identify host proteins that interact with UL35 and UL35a in human cells, leading to the discovery of a previously unrecognized role for UL35 in the manipulation of the host cell cycle and DDR.
MATERIALS AND METHODS
Cell lines.
U2OS human osteosarcoma and human embryonic kidney 293A cells were maintained in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human nasopharyngeal carcinoma cells (CNE-2Z) were maintained in α-minimal essential medium (MEM) supplemented with 10% FBS. CNE-2Z cells with the stable silencing of PML (CNE-2Z shPML) were previously described (80) and maintained in α-MEM supplemented with 10% FBS and 0.4 μg/ml puromycin (Bioshop). HeLa-Fucci cells are HeLa cells stably transformed with fluorescently tagged cdt1 (mKO2-hCDT1) and geminin (mAG-hGem) as described previously (75) and were cultured in DMEM supplemented with 10% fetal bovine serum, GlutaMax (Gibco), and penicillin-streptomycin.
Plasmids.
Plasmids encoding UL35 and UL35a (HCMV strain AD169) with a C-terminal sequential peptide affinity (SPA) tag (UL35-S) in the pMZS3F vector (101) or a triple FLAG epitope tag (UL35-F) in pCMV-3FC have been described previously (77). The SPA tag consists of a calmodulin binding peptide and a triple-FLAG epitope tag separated by a tobacco etch virus (TEV) protease cleavage site (101). The control vector (LacZ) encodes SPA-tagged β-galactosidase expressed from pMZS3F. Untagged wild-type UL35 (UL35-wt), UL35a, and UL35N (UL35 amino acids 1 to 447) in pCMV-3FC have been described previously (76). A Myc-tagged construct containing the WD40 motif and C terminus of DCAF1 was a gift from Yue Xiong (62). Full-length (FL) DCAF1 and a C-terminal deletion containing amino acids 1 to 1417 (DCAF1 1-1417), containing N-terminal tandem FLAG and hemagglutinin (HA) epitope tags, was a gift from Filippo Giancotti (51).
Primary antibodies.
Rabbit serum raised against UL35 was a gift from Bonita Biegalke (55). Mouse anti-FLAG (M2 clone) was from Sigma-Aldrich. Rabbit primary antibodies against FLAG, PML, DDB1, USP7, γ-H2Ax (A300-081A), and the DCAF1 N terminus (amino acids 25 to 75; A301-887A) and C terminus (amino acids 1457 to 1507; A301-888A) all were from Bethyl Laboratories. Sheep anti-PML was a gift from David Bazett-Jones (University of Toronto). Rabbit anti-myc was from AbCam. Antibodies against phospho-histone H3 Ser10 (8656-R) and Cdc2 (954) were from Santa Cruz, while the phospho-Cdc2 Tyr15 antibody (9111S) was from Cell Signaling. Rabbit anti-lysine 48-conjugated ubiquitin was from Millipore. 53BP1 antibody was from BD Biosciences (612522).
Immunofluorescence microscopy.
Cells were seeded into 6-well plates on glass coverslips (∼700,000 cells/well) and transfected with expression plasmids using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions, using a DNA/Lipofectamine 2000 ratio of 2 μg to 2 μl for 293A cells and 2 μg to 4 μl for U2OS and CNE2Z cells. Transfected cells were fixed at 48 h posttransfection (or at the indicated times) with 3.7% formaldehyde in phosphate-buffered saline (PBS) (20 min), permeabilized with 0.5% Triton X-100 in PBS (10 min), and blocked with 4% bovine serum albumin (BSA) in PBS (20 min) prior to incubation with primary (1 h) and secondary (45 min) antibodies in 4% BSA in PBS. Primary antibodies were detected using either goat anti-mouse or anti-rabbit FAb fragments conjugated with Alexa fluor 488 or Alexa fluor 555 (Invitrogen). In experiments using sheep anti-PML primary antibodies, donkey secondary antibodies raised against sheep, rabbit, and mouse (conjugated with Cy2, Cy3, and Cy5, respectively) were used to prevent cross-reaction between goat secondary antibodies and sheep primary antibody. Coverslips were mounted onto slides using ProLong Gold antifade fluorescent mounting medium (Invitrogen) containing 4′,6′-diamidino-2-phenylindole (DAPI) for the visualization of nuclear DNA. Images were acquired using the 63× oil objective (numeric aperture, 1.4) on a Leica DM IRE2 inverted fluorescence microscope. Images were processed using OpenLAB (ver.4.0.2) and Adobe Photoshop CS5 using only linear adjustments. For the quantification of USP7 nuclear bodies or γ-H2AX or 53BP1 foci, the foci were counted in each of 100 randomly transfected or untransfected cells in 3 to 6 separate experiments as indicated in the figure legends. Statistical analyses (Student's t test) were conducted using Microsoft Excel 2007 software.
Western blotting.
Cells were lysed in cell lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40) plus protease inhibitor cocktail (P8340; Sigma) and clarified by centrifugation at 13,000 × g for 15 min at 4°C, and the protein concentration was determined using Bradford reagent (Bio-Rad). Equal amounts of cell lysates were subjected to SDS-PAGE, and proteins were transferred to nitrocellulose membranes, blocked with 4% milk, and incubated with the indicated primary antibodies (1 h) and goat anti-mouse or goat anti-rabbit HRP-conjugated secondary antibodies (45 min; Santa Cruz). Antibodies were detected by chemiluminescence using Western Lighting chemiluminescent reagent (PerkinElmer) and exposure to photographic film (Amersham). For the detection of phosphorylated H3 and Cdc2 by Western blotting, cells were lysed in 9 M urea buffered by 10 mM Tris-HCl, pH 6.8, and 50 μg of each lysate was analyzed by SDS-PAGE and Western blotting as described above.
Coimmunoprecipitation.
293A cells in 10-cm plates were cotransfected with the indicated plasmids and, 40 h later, were harvested and lysed in cell lysis buffer as described above for Western blotting. One mg of cleared lysates was used for immunoprecipitation (IP) and diluted in cell lysis buffer to give a final concentration of 4 mg/ml. For anti-FLAG IP, cell lysates were incubated with anti-FLAG resin (20-μl bed volume; Sigma) for 2 h at 4°C while rotating. Beads were washed four times for 10 min in 1 ml of lysis buffer, and protein was eluted with 50 μl of protein sample buffer (5% SDS, 20 mM Tris, pH 8, 10% dithiothreitol [DTT], 20% glycerol). Western blotting then was performed as described above. For DCAF1 immunoprecipitations, cell lysates prepared as described above were precleared with a 20-μl bed volume of protein A/G beads (Santa Cruz) by incubation for 45 min at 4°C. Protein A/G beads were removed by centrifugation, and cell lysates then were incubated with 2 μg DCAF1 antibody overnight at 4°C with rotating. Antibody-DCAF1 complexes then were recovered by incubation with a 20-μl bed volume of protein A/G beads for 45 min at 4°C. Beads were washed four times with 1 ml cell lysis buffer and eluted in 50 μl of protein sample buffer.
DCAF1 silencing experiments.
293A cells (250,000 to 500,000 cells) seeded in 10-cm dishes were transfected with 100 pmol short interfering RNA (siRNA) against DCAF1 (sc-76898; Santa Cruz) or Allstars negative-control siRNA (1027281; Qiagen) using Lipofectamine 2000, followed by two more rounds of siRNA transfection 24 h apart. Twenty-four h after the final round of siRNA transfection, cells were transfected with 6 μg of plasmid expressing FLAG-tagged proteins using Lipofectamine 2000. Seventy-two h later, cells were lysed and Western blotting (using 30 μg of total cell lysate) and immunoprecipitations were performed as described above.
DNA content analysis by flow cytometry.
For DNA content analysis, cells were fixed overnight at −20°C in 70% ethanol, washed in PBS with 0.5% BSA, immunostained with rabbit anti-FLAG primary antibody (Bethyl) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology), treated with 100 μg/ml RNase A for 1 h at 37°C, and stained with 50 μg/ml propidium iodide. All samples were analyzed using a FACSCalibur flow cytometer (BD Biosciences), and data were collected using CellQuest software. Cell cycle analysis was performed using FlowJo software (Treestar Inc.).
Cell cycle analysis in HeLa-Fucci cells.
For microscopy, HeLa-Fucci cells were seeded into 6-well cluster plates containing coverslips and were transfected 24 h later using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and using a DNA/Lipofectamine 2000 ratio of 2 μg to 4 μl. Cells were fixed (3.7% formaldehyde) and permeabilized (0.1% Triton X-100) 24 h after transfection. Coverslips were blocked for 1 h in PBS containing 4% BSA and then incubated for 1 h each with rabbit anti-FLAG primary antibody (Bethyl) and goat anti-rabbit Alexa Fluor 350 (Molecular Probes) secondary antibody. Coverslips were mounted onto slides with ProLong Gold antifade mounting medium (Invitrogen), and images were acquired using a Leica DM IRE2 inverted fluorescence microscope. mKO2-Cdt1 and mAG-geminin fluorescent protein expression was detected using Tx2 (BP560/40nm) and yellow fluorescent protein (YFP) (BP500/20 nm) filter cubes, respectively. Images were processed with OpenLab software (version 4.0.2). For the quantification of cell cycle phase by fluorescence-activated cell sorting (FACS), HeLa-Fucci cells were seeded into 10-cm tissue culture plates and transfected as described above with a DNA/Lipofectamine 2000 ratio of 8 μg to 16 μl. Forty-eight h posttransfection, cells were collected, fixed, and permeabilized as described above. FLAG expression was detected with rabbit anti-FLAG followed by goat anti-rabbit Alexa fluor 647 (Molecular Probes) secondary antibody. Cells were filtered to remove aggregates before being examined using a FACSCalibur flow cytometer (Becton Dickinson). FLAG expression was detected following excitation at 633 nm and collection at 661 nm (661/16 nm BP filter, FL4 channel). Cdt1 (mKO2-hCDT1) and geminin (mAG-hGem) were excited using a 488-nm laser, and fluorescent signals were collected at 585 (585/42 nm BP filter, FL2 channel) and 530 nm (530/30 nm BP filter, FL1 channel), respectively. The data were analyzed using FloJo software (Tree Star, version 9.0.1).
Mass spectrometry.
For the mass-spectrometric analysis of UL35- and UL35a-interacting proteins, five 150-cm2 dishes of subconfluent (75 to 85%) transiently transfected FLAG- or SPA-tagged UL35 or UL35a-expressing cells were scraped into PBS, pooled, washed with PBS, and collected by centrifugation at 1,000 × g for 5 min at 4°C. Cell pellets were stored at −80°C. Cell pellets were weighed and resuspended with lysis buffer (50 mM HEPES-NaOH, pH 8.0, 100 mM KCl, 2 mM EDTA, 0.1% NP-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM DTT, and 1:500 protease inhibitor cocktail; Sigma, St. Louis, MO) in a 1:4 (wt/vol) ratio. Resuspended cells were incubated on ice for 10 min, subjected to one additional freeze-thaw cycle, and then centrifuged at 27,000 × g for 20 min at 4°C. The supernatant was transferred to a fresh 15-ml conical tube, and 1:1,000 benzonase nuclease (25 U/ml; Novagen, San Diego, CA) plus 30 μl packed, preequilibrated FLAG-M2 agarose beads (Sigma, St. Louis, MO) were added. The mixture was incubated for 2 h at 4°C with end-over-end rotation. Beads were pelleted by centrifugation at 1,000 × g for 1 min and transferred to a clean Eppendorf tube. The beads then were washed once with 1 ml lysis buffer and two times with 1 ml ammonium bicarbonate rinsing buffer (50 mM ammonium bicarbonate, pH 8.0, 75 mM KCl). Elution was performed by incubation with 150 μl of 125 mM ammonium hydroxide (pH 11.0). The elution step was repeated twice. The eluate was centrifuged at 1,000 × g for 1 min, transferred to a fresh centrifuge tube, and lyophilized. One μg of mass spectrometry-grade tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin (Promega, Madison, WI) dissolved in 70 μl of 50 mM ammonium bicarbonate (pH 8.3) was added to the eluate and incubated at 37°C overnight. The resulting peptides were lyophilized and resuspended in buffer A (0.1% formic acid). Liquid chromatography (LC) analytical columns (75-μm inner diameter) and precolumns (100 μm) were prepared in house from fused silica capillary tubing from InnovaQuartz (Phoenix, AZ) and packed with 100-Å C18-coated silica particles (Magic, Michrom Bioresources, Auburn, CA). Peptides were subjected to LC-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) using a 120-min reversed-phase LC (RPLC; 95% water-acetonitrile, 0.1% formic acid) buffer gradient running at 250 nl/min on a Proxeon EASY-nLC pump in line with a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A parent ion scan was performed in the Orbitrap using a resolving power of 30,000, and then the six most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation) using standard collision-induced dissociation (CID) fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS findings of the same m/z (within a −0.1 and +2.1 Thompson window; exclusion list size, 500) detected 3 times within 45 s were excluded from analysis for 60 s. For protein identification, Thermo.RAW files were converted to the .mzXML format using Proteowizard (41) and then searched using X!Tandem (15) against the human RefSeq database (version 37). X!Tandem search parameters were the following: complete modifications, none; cysteine modifications, none; potential modifications, +16@M and W, +32@M and W, +42@N terminus, +1@N, and Q. Data were analyzed using the ProHits (54) and SAINT software tools (14).
RESULTS
Identification of UL35 and UL35a host protein interactions by AP-MS.
To identify host proteins that interact with UL35 and UL35a, FLAG-tagged UL35 and UL35a were expressed in 293A cells by transfection, and cell lysates were subjected to affinity purification with an anti-FLAG resin. Eluates were subjected to trypsin digestion, and the resulting peptides were identified using nanospray liquid chromatography electrospray ionization-tandem mass spectrometry (nLC-ESI-MS/MS). For UL35, we conducted five FLAG immunoprecipitations (IPs; two with the SPA tag and three with the triple FLAG tag), and for UL35a four FLAG IPs (two with the SPA tag and two with the triple FLAG tag) were performed. Four control affinity purifications also were conducted using 293A cells transfected with the same plasmid containing the LacZ open reading frame. MS data were subjected to SAINT (for significance analysis of interactome) analysis (14), and putative UL35/UL35a interactors with an average SAINT probability score of greater than 0.80 (AvgP) are listed in Fig. S1 in the supplemental material. All of the hits also were cross-referenced against a large in-house AP-MS database using ProHits (54). Proteins observed in more than 20% of the AP-MS analyses in our database (i.e., associated with a project frequency of >0.2) were removed, as these proteins are likely to interact with the antibody or solid-phase resin material in a nonspecific manner. Proteins that met both the SAINT and the ProHits project frequency criteria are included in Table 1 and represent those polypeptides with a high probability of interacting specifically with UL35 and/or UL35a. Despite displaying a project frequency of greater than 20%, we also included DDB1 in Table 1, as it is known to associate with DCAF1 and DDA1 (68) and did not appear in the control samples. Also, the high number of peptides recovered for DDB1 (spectral counts) in each experiment is considerably above that seen for nonspecific interactions, showing that DDB1 was efficiently recovered with UL35. A complete list of the recovered cellular proteins and SAINT analysis is provided in Table S1 in the supplemental material.
Table 1.
Cellular protein interactions with UL35 and UL35a identified by AP-MS
| Protein and targeta | AvgPb | Specc | CtrlCountsd |
|---|---|---|---|
| UL35 | |||
| DCAF1 | 0.97 | 99|101|149|116|21 | 5|2|0|0|0 |
| DDB1e | 0.98 | 54|60|104|94|13 | 1|0|0|0|0 |
| DDA1 | 0.80 | 9|7|9|9|0 | 0|0|0|0|0 |
| SART3 | 1.00 | 28|28|42|39|20 | 0|0|0|0|0 |
| USP7 | 0.99 | 4|2|18|11|7 | 0|0|0|0|0 |
| OGT | 0.99 | 42|41|103|99|40 | 6|2|0|0|0 |
| IPO4 | 1.00 | 34|35|15|7|16 | 0|0|0|0|0 |
| UL35a | |||
| USP7 | 1.00 | 8|10|41|27 | 0|0|0|0|0 |
| OGT | 1.00 | 48|96|45|34 | 6|2|0|0|0 |
| IPO4 | 1.00 | 39|114|37|46 | 0|0|0|0|0 |
| TNPO1 | 0.99 | 5|33|8|8 | 0|0|0|0|0 |
| G3BP2 | 0.97 | 5|3|5|2 | 0|0|0|0|0 |
| DDX18 | 0.98 | 2|4|10|5 | 0|0|0|0|0 |
Target indicates the identity of cellular proteins copurified with UL35 and/or UL35a.
The average SAINT probability score assigned to each Bait-Prey interaction.
The number of spectral counts assigned to each prey protein for each of 4 (UL35a) or 5 (UL35) mass spectrometric analyses.
The number of spectral counts for each prey protein observed in each of 5 control experiments with LacZ.
DDB1 has a project frequency of >20% (46.7%) but is included here because of the high Spec and AvgP score (indicating that this is not a nonspecific interaction) and its ability to form complexes with DDA1 and DCAF1.
Since UL35 and UL35a share sequence identity, it is not surprising that some cellular proteins associated with both viral proteins. These include the ubiquitin-specific protease USP7, O-linked N-acetylglucosaminyl (O-GlcNAc) transferase (OGT), and the nuclear import factor importin 4 (IPO4). USP7 is an important regulator of p53 stability (49) and also is targeted by other herpesviruses, namely, the herpes simplex virus protein ICP0 (21) and the Epstein-Barr virus (EBV) protein EBNA1 (31). O-GlcNAc is an important posttranslational modification catalyzed by OGT that regulates several important nuclear events, including the regulation of p53 and several other transcription factors that help control expression from the CMV MIEP, including NF-κB, Ying-yang 1 (YY1), and CREB (69, 85).
Four proteins interacted specifically with UL35, of which three (DCAF1, DDB1, and DDA1) are known to form an E3-ubiquitin ligase complex with Cullin 4A (Cul4A) and Roc1 that may regulate cell cycle and/or DNA repair processes (32, 68). DCAF1 (DDB1-Cul4 associated factor 1) also is known as Vpr binding protein (VprBP) and was identified as a target of the human immunodeficiency virus type 1 (HIV-1) Vpr protein (48). DDB1 (DNA damage binding protein 1) connects DCAF1 and DDA1 (DET1 and DDB1 associated 1) to Cul4A (36, 68). The fourth UL35-specific interaction identified was with SART3 (squamous cell carcinoma antigen recognized by T cells 3), an RNA binding protein involved in tumor immunology and mRNA splicing (28, 63).
Three proteins (TNPO1, G3BP2, and DDX18) were recovered with UL35a only. Transportin 1 (TNPO1) is a nuclear import factor, and G3BP2 (RasGAP SH3 binding protein 2) has been implicated in regulating NF-κB activity (72) as well as p53 and mdm2 (43). DDX18 is a DEAD-box RNA helicase that may have a role in regulating cell proliferation (18), but it is largely uncharacterized.
UL35 inhibits formation of USP7 NBs in a PML-independent manner.
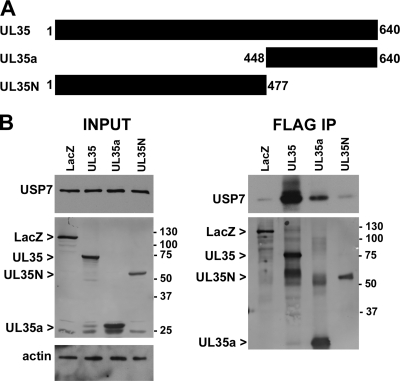
We used coimmunoprecipitation to validate some of the protein interactions identified by mass spectrometry. The USP7 interaction was further explored for several reasons. First, both UL35 and UL35a were found to associate with this ubiquitin-specific protease. Second, although other herpesvirus proteins are known to interact with USP7, this is the first example of a betaherpesvirus interacting with this cellular target. Finally, USP7 is known to associate with PML (22, 80), and UL35 has dramatic effects on the organization of PML bodies in transfected cells (76, 77, 82). 293A cells were transfected with plasmids expressing FLAG-tagged β-galactosidase (LacZ), UL35, UL35a, or UL35N (76) (Fig. 1). UL35N consists of the first 447 amino acids of UL35, which do not contain the UL35a sequence (Fig. 1A). We analyzed the levels of USP7 before FLAG IP and did not observe a change in the overall levels of USP7 in the presence of UL35 or UL35a compared to those in LacZ control transfections and actin loading controls (Fig. 1) despite good expression and more than 90% transfection efficiency (data not shown). Transfected proteins were recovered by anti-FLAG immunoprecipitation and analyzed for the recovery of USP7 by Western blotting (Fig. 1B). USP7 copurified with UL35 very strongly, while UL35a was recovered less efficiently despite similar levels of expression and recovery by IP (Fig. 1B). Unlike results with UL35 and UL35a, USP7 was not recovered by UL35N to a greater degree than the β-galactosidase negative control (Fig. 1B), suggesting that USP7 interacts with the C terminus of UL35 (i.e., UL35a) but that additional UL35 sequences also contribute to the affinity or stability of the UL35-USP7 interaction.
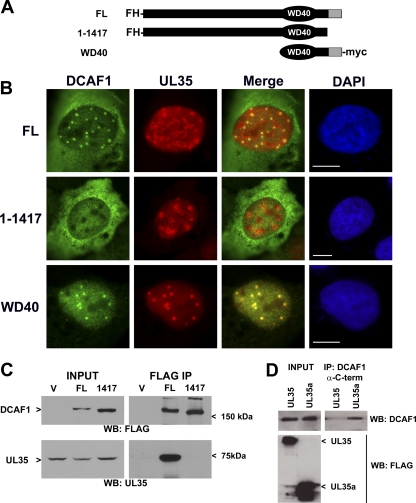
Fig 1.
UL35 and UL35a interact with USP7. (A) Schematic representation of UL35, UL35a, and UL35N showing amino acid numbers. (B) Western blot analysis of 293A cells transfected with FLAG-tagged LacZ, UL35, UL35a, or UL35N plasmid and subjected to anti-FLAG immunoprecipitation (IP) at 48 h posttransfection. Cell lysates (input) and post-IP elutions (FLAG IP) were probed for USP7 and/or actin as indicated. Transfected proteins were detected with anti-FLAG antibody, and their positions are indicated to the left of the images. Molecular mass markers (in kDa) are indicated to the right of the images.
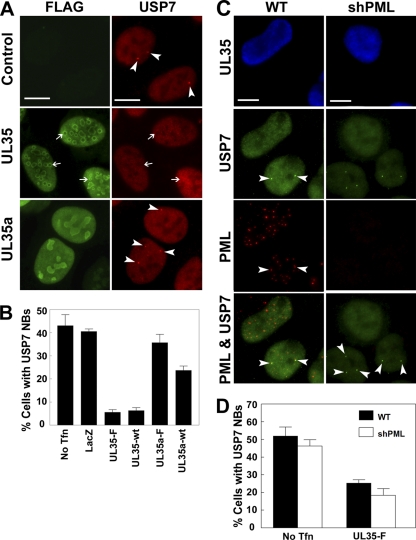
USP7 has been shown previously to associate with PML NBs, and we have recently demonstrated that USP7 is a negative regulator of PML (80). We also have recently described the ability of UL35 to alter PML NBs by recruiting PML to UL35 NBs (76). Since USP7 associates with both PML and UL35, we tested if USP7 localization was affected by the expression of UL35. U2OS cells expressing FLAG-tagged UL35 or UL35a were immunostained for FLAG and USP7 (Fig. 2A). In UL35-transfected cells, there was no significant recruitment of USP7 to UL35 NBs (Fig. 2A, arrows). In untransfected control cells, USP7 had a pan-nuclear staining, with some cells containing a small number of NBs (Fig. 2A, arrowheads). The formation of USP7 NBs has been reported previously and in some cases reflects the association of USP7 with PML NBs (22, 80, 86). Similarly to untransfected cells, USP7 also formed NBs in UL35a-transfected cells; however, the presence of USP7 NBs was noticeably reduced in UL35-transfected cells. Thus, although USP7 did not associate with UL35 structures, UL35 did appear to affect the ability of USP7 to form NBs. We quantified the inhibition of USP7 NB formation by UL35 by determining the percentage of transfected cells containing USP7 NB (Fig. 2B). Cells transfected with the LacZ negative control showed amounts of USP7 NBs similar to those of untransfected cells (∼40%), indicating that the transfection process did not alter USP7 NB formation. Compared to untransfected control cells, both FLAG-tagged UL35 (UL35-F) and untagged UL35 (UL35-wt) caused a significant decrease (P < 0.01) in USP7 NBs, resulting in only ∼5% of transfected cells containing USP7 NBs. In contrast, untagged UL35a (P = 0.02) and FLAG-tagged UL35a (P = 0.27) had a much smaller effect on the level of USP7 NBs, while the LacZ control vector had no appreciable effect on USP7 NB formation (P = 0.57).
Fig 2.
UL35 inhibits formation of USP7 nuclear bodies. (A) U2OS cells were transfected with plasmids expressing FLAG-tagged UL35 (UL35-F) or UL35a (UL35a-F) and immunostained at 48 h posttransfection for FLAG and USP7 as indicated. The positions of selected UL35 NBs are indicated with arrows (UL35 panels). Arrowheads (control and UL35a panels) indicate USP7 NBs. (B) Cells from panel A were quantified for USP7 NBs. The percentage of transfected cells containing at least one USP7 NB is presented. No Tfn, untransfected control cells; wt, untagged UL35 or UL35a; LacZ, transfection control plasmid expressing β-galactosidase. (C) Wild-type CNE-2Z (WT) and CNE-2Z shPML cells were transfected with FLAG-tagged UL35, fixed at 48 h posttransfection, and immunostained for USP7, PML, and UL35 (anti-FLAG) as indicated. Arrowheads in the WT panels indicate USP7 NBs that are associated with PML. Arrowheads in the shPML panels show USP7 NBs formed in the absence of PML. (D) Quantification of USP7 NBs from panel C. The percentage of transfected cells containing at least one USP7 NB is presented for WT and shPML cells. No Tfn, untransfected control cells. Values in panels B and D represent the means ± standard errors; n = 3 to 6. Scale bars in panels A and C are 10 μm.
Since some USP7 NBs are associated with PML NBs and since UL35 alters PML NBs, we investigated whether the apparent loss of USP7 NBs is a consequence of UL35-mediated PML alterations that could prevent USP7 association with PML. To this end, we expressed UL35 in CNE-2Z cells in which all PML isoforms were silenced with shRNA (Fig. 2C) (23, 80). As expected, compared to wild-type PML-expressing CNE-2Z cells, UL35 expression inhibited USP7 NB formation (Fig. 2D) (P < 0.01) and PML association (Fig. 2C). Neighboring untransfected cells still formed USP7 NBs, which associated with PML (Fig. 2C, arrowheads). Interestingly, in untransfected cells in which PML was silenced and PML bodies therefore were absent, USP7 still formed NBs (Fig. 2C, arrowheads), indicating that PML is not required for USP7 NB formation. As with wild-type CNE-2Z cells, the expression of UL35 inhibited USP7 NB formation in the absence of PML (P < 0.01) relative to untransfected control cells, indicating that the effect of UL35 on USP7 NB formation is independent of PML. The quantification of the results in CNE-2Z cells showed that although UL35 did not inhibit USP7 NB formation as efficiently as it did in U2OS cells, the inhibition was very similar in wild-type and PML-silenced cells (P > 0.05) (Fig. 2D). Thus, the data provide evidence that UL35 interacts with USP7 in cells and can affect its subnuclear localization.
UL35 associates with components of the CUL4A-DDB1-DCAF1 E3 ubiquitin ligase complex.
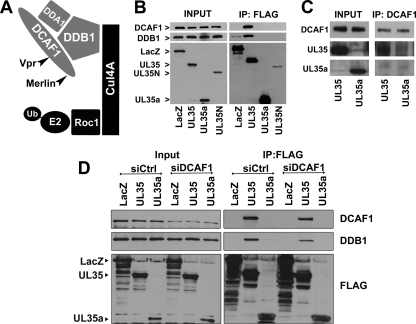
In addition to USP7, UL35 was found to associate with DCAF1, DDB1, and DDA1. These three proteins all are components of a Cullin 4-based E3 ubiquitin ligase complex (Cul4DCAF1) consisting of the scaffold protein Cullin4A, the E3 ligase Roc1, the substrate recognition protein DCAF1, and the adaptor protein components DDB1 and DDA1 (Fig. 3A) (36, 68). Interestingly, Cul4-DDB1 complexes are targeted by several different viruses that hijack the ubiquitin ligase activity of these complexes to direct specific cellular proteins for degradation that may not normally be targeted by this E3 ligase (94). In addition, CUL4-DDB1-based complexes are associated with DNA damage responses (DDR) and cell cycle control (68). Given the complex interactions between CMV and host cells, which often involve the manipulation of DDR and the cell cycle, we chose to further explore the interaction of UL35 with this important cellular complex.
Fig 3.
UL35 interacts with DCAF1. (A) Cartoon of the Cul4DCAF1 complex showing previously defined interactions. The proteins isolated with UL35 are indicated in gray. (B) Western blot analysis of 293A cells transfected with FLAG-tagged LacZ, UL35, UL35a, or UL35N expression plasmid and subjected to anti-FLAG immunoprecipitation (IP) at 48 h posttransfection. Cell lysates (Input) and post-IP elutions (IP: FLAG) were probed for DCAF1 and DDB1, while transfected proteins were detected with anti-FLAG antibody. Their positions are indicated to the left of the images. (C) Western blot analysis of 293A cells transfected with FLAG-tagged UL35 or UL35a and subjected to immunoprecipitation with anti-DCAF1 antibody directed against the N terminus of DCAF1. Cell lysates (Input) and post-IP elutions (DCAF1 IP) were probed for DCAF1 and anti-FLAG (UL35 and UL35a). (D) Western blot analysis of 293A cells treated with siRNA against DCAF1 (siDCAF1) or negative-control Allstars siRNA (siCtrl) and transfected with FLAG-tagged LacZ, UL35, or UL35a expression plasmid. Cell lysates were subjected to anti-FLAG immunoprecipitation 72 h posttransfection. Whole-cell extracts (Input) and post-IP elutions (IP: FLAG) were probed for DCAF1 and DDB1, while transfected proteins were detected with anti-FLAG antibody. Their positions are indicated to the left of the images.
First, we confirmed the association between UL35 and DCAF1 and DDB1 by coimmunoprecipitation and Western blotting (Fig. 3B). 293A cells were transfected with LacZ, UL35N, UL35, and UL35a plasmids with FLAG tags. UL35 did not affect the levels of DCAF1 or DDB1 (Fig. 3A, input lanes). Consistently with our mass spectrometry analysis, DCAF1 and DDB1 were recovered with UL35 following FLAG immunoprecipitation but not with UL35a. Similarly, UL35N did not recover detectable levels of DDB1 or DCAF1, indicating that full-length UL35 is required for association with these two cellular proteins (Fig. 3B). In addition, the immunoprecipitation of endogenous DCAF1 using an antibody directed against the N terminus of DCAF1 recovered UL35 but not UL35a (Fig. 3C).
We next examined whether the association of UL35 with DDB1 was dependent on DCAF1, as would be expected if UL35 interacted with the Cul4DCAF1 complex through DCAF1. To this end, we depleted DCAF1 with siRNA and then expressed FLAG-tagged UL35, UL35a, or β-galactosidase (LacZ) followed by immunoprecipitation with anti-FLAG antibody (Fig. 3D). In cells treated with siRNA directed against DCAF1, the expression of this protein was decreased while there was no change in the level of DDB1. In cells receiving the control siRNA treatment, DCAF1 and DDB1 both were recovered with UL35 but not with UL35a or LacZ (Fig. 3D). However, the recovery of both DCAF1 and DDB1 with UL35 was reduced in DCAF1-depleted cells, in keeping with the degree of decrease in DCAF1 levels. The decreased association of DDB1 with UL35 after DCAF1 downregulation supports the hypothesis that UL35 associates with DDB1 through DCAF1 as part of a complex, such as the Cul4DCAF1 complex shown in Fig. 3A.
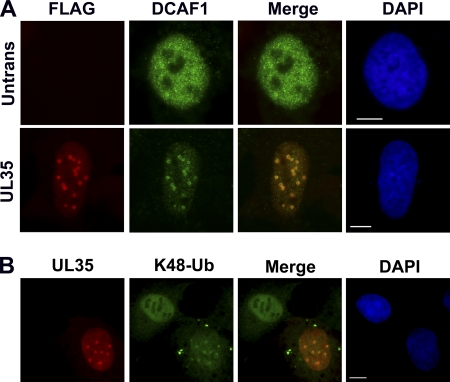
We also used immunofluorescence microscopy to determine if DCAF1 localization was affected by UL35. U2OS cells were transfected with FLAG-tagged UL35 and stained with anti-FLAG and anti-DCAF1 antibodies (Fig. 4A). In untransfected cells, DCAF1 adopts a diffuse nuclear localization pattern; however, when UL35 is expressed, most of the visible DCAF1 becomes associated with UL35 NBs, indicating the ability of UL35 to relocalize DCAF1. Since DCAF1 forms part of an E3 ubiquitin ligase complex, we wanted to determine if the recruitment of DCAF1 to UL35 NBs was associated with an increase in polyubiquitinated substrates. Since the addition of polyubiquitin chains containing the lysine 48 (K48-Ub) linkage targets proteins for proteasomal degradation, we analyzed UL35-transfected cells for the accumulation of K48-Ub by immunofluorescence microscopy (Fig. 4B). In the presence of UL35, a portion of K48-Ub was found to localize to the UL35 NBs in a pattern similar to that observed for DCAF1, while adjacent untransfected cells contained more diffuse K48-Ub staining. These results indicate that UL35 NBs can act as sites for protein ubiquitination and are consistent with UL35 recruitment of Cul4DCAF1 E3 ligase complexes to UL35 NBs.
Fig 4.
DCAF1 and conjugated ubiquitin localize to UL35 NBs. (A) U2OS cells with (UL35) and without (Untrans) transfection with FLAG-tagged UL35 plasmid were immunostained for FLAG and DCAF1. (B) Cells transfected as described for panel A were immunostained for UL35 and lysine 48-conjugated polyubiquitin chains (K48-Ub). Cellular DNA is stained with DAPI. Scale bar, 10 μm.
UL35 interacts with the C terminus of DCAF1.
The C-terminal third of DCAF1 (amino acids 1000 to 1501) contains the WD40 region (amino acids 1041 to 1377), which is important for association with both DDB1 and Vpr (48). In addition, the cellular protein Merlin negatively regulates the Cul4-DCAF1 complex (51) through interactions with the C-terminal region between amino acids 1417 and 1501 (33). Thus, we wanted to determine if UL35 functions in a similar manner by associating with the C-terminal region of DCAF1. To this end, U2OS cells were cotransfected with a plasmid expressing untagged UL35 (UL35-wt) and a second plasmid expressing either full-length DCAF1 (FL) or DCAF1 lacking the part of the C terminus (1-1417) (Fig. 5A) that contains the Merlin binding site. Both FL and 1-1417 contain N-terminal FLAG and HA (FH) tags (Fig. 5A). In addition, U2OS cells were cotransfected with FLAG-tagged UL35 and a plasmid encoding the DCAF1 C-terminal region that includes the WD40 motif (important for DDB1 and Vpr interactions) and the Merlin binding region (62). Full-length DCAF1 showed colocalization with UL35 NBs (Fig. 5B) similar to that observed with endogenous DCAF1 (Fig. 4A). However, DCAF1 1-1417 did not colocalize with UL35 in NBs (Fig. 5B), suggesting that the extreme C terminus of DCAF1 is needed for association with UL35. Consistently with this result, the WD40 DCAF1 fragment, which contains the entire C terminus, was strongly associated with UL35 NBs (Fig. 5B), indicating that the C terminus of DCAF1 is required for association with UL35.
Fig 5.
C terminus of DCAF1 is required for UL35 association. (A) Schematic of various DCAF1 mutants used in this figure showing the positions of the FLAG-HA (FH) and myc tags. The gray region indicates the portion of DCAF1 required for association with Merlin. WD40 indicates the location of the WD40 motif required for association with DDB1. (B) U2OS cells were cotransfected with the indicated DCAF1 and either UL35-wt (no tags; top two rows) or UL35-F (bottom row) construct and fixed 48 h later for immunofluorescence microscopy. FL and 1-1417 DCAF1 proteins were detected with anti-FLAG, while UL35-wt was detected with anti-UL35 antiserum (top two rows). In the bottom row, the WD40 construct was detected with anti-myc antibodies, while UL35-F was detected with anti-FLAG antibodies (bottom row). (C) 293A cells were cotransfected with the plasmid expressing UL35-wt and a plasmid expressing either full-length DCAF1 (FL), DCAF1 1-1417 (1417), or nothing (V) and harvested 48 h later. Cell lysates before (Input) and after FLAG immunoprecipitation (FLAG IP) were analyzed by Western blotting for DCAF1 (WB: FLAG) and UL35 (WB: UL35). (D) Western blot analysis of 293A cells transfected with FLAG-tagged UL35 or UL35a plasmids and subjected to immunoprecipitation with anti-DCAF1 antibody directed against the C terminus of DCAF1. Cell lysates (Input) and post-IP elutions (IP: DCAF1) were probed for DCAF1 and FLAG (UL35 and UL35a).
We also used Western blot analysis to examine the importance of the DCAF1 C terminus for association with UL35. First, 293A cells were cotransfected with plasmids expressing UL35-wt and FLAG-tagged FL or 1-1417 versions of DCAF1 (Fig. 5C). UL35 was recovered only with full-length DCAF1 and not with 1-1417 or the empty vector negative control (Fig. 5C). Second, we used an antibody directed against the C terminus of DCAF1 (amino acids 1457 to 1507), which spans the Merlin binding site, and attempted to immunoprecipitate endogenous DCAF1 in the presence of UL35 or UL35a (Fig. 5D). Contrary to results obtained with antibody directed against the N terminus of DCAF1 (Fig. 3B), when UL35 was present the recovery of DCAF1 by the C-terminal DCAF1 antibody was substantially reduced relative to that of UL35a-expressing cells. These results suggest that the UL35 interaction with the DCAF1 C terminus prevents the DCAF1 antibody from recognizing its epitope, whereas UL35a, which does not interact with DCAF1, does not affect this interaction. Taken together, these results indicate that residues 1418 to 1501 of DCAF1 are important for interaction with UL35.
UL35 causes cells to accumulate in the G2 phase of the cell cycle.
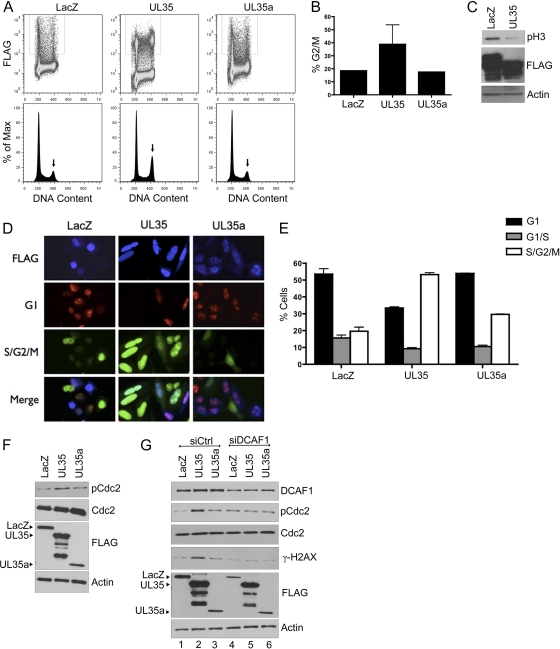
The interaction of HIV-1 Vpr with DCAF1 has been shown to induce the accumulation of cells in the G2 phase of the cell cycle (32, 48, 88, 95). Therefore, we assessed whether UL35 caused an increase in G2 cells as an indicator of altered DCAF1 activity. To this end, 293A cells were transfected with plasmids expressing FLAG-tagged UL35, UL35a, or β-galactosidase (LacZ), and cell cycle profiles were determined by flow-cytometric analyses for DNA content. UL35-transfected cells consistently showed an accumulation of cells with the DNA content of the G2 or M phase of the cell cycle (39% on average) compared to the level for the LacZ negative control (19% on average), as shown in the representative experiment in Fig. 6A and the composite quantification of the G2/M peaks in Fig. 6B. In contrast, UL35a expression did not noticeably affect the cell cycle profile (Fig. 6A and B). To determine whether the increased G2/M peak seen with UL35 was due to increased levels of G2 or M cells, Western blotting for phosphorylated histone H3 (pH 3), a marker of M phase, was performed on samples from the same experiment. As shown in Fig. 6C, UL35 did not increase pH 3 levels but rather decreased this cell population, indicating that fewer cells had entered mitosis, as expected if progression through G2 is slowed or arrested.
Fig 6.
UL35 causes the accumulation of cells in the G2 phase of the cell cycle. (A) 293A cells were transfected with plasmids expressing FLAG-tagged β-galactosidase (LacZ), UL35, or UL35a, and 72 h later they were stained with anti-FLAG antibody and propidium iodide and analyzed by flow cytometry for FLAG and DNA content (top panels). The DNA content of the gated FLAG-positive cells is shown in the bottom panels, and the G2/M peak is indicated by the arrows. (B) Quantification of the G2/M peak from two independent experiments as described for panel A. Average values with standard deviations are shown. Standard deviations for UL35a samples were too small to be seen on the histogram. (C) Western blot of whole-cell extracts of 293A cells transfected with FLAG-tagged LacZ and UL35 expression plasmids and probed with the antibodies against FLAG, phosphorylated histone H3 (pH 3), and actin (loading control). (D) HeLa-Fucci cells were transfected with the same expression plasmids as those for panel A and then were fixed 24 h later and stained for anti-FLAG primary antibody and an Alexa fluor 350-coupled secondary antibody to detect transfected cells. Cells on coverslips were imaged for Alexfluor 350 (FLAG; blue) as well as for the fluorescent proteins mKO2-Cdt1 (G1; red) and mAG-geminin (S/G2/M; green). A representative image is shown for each plasmid, where images were captured with the same exposure time. (E) HeLa-Fucci cells transfected as described for panel D were fixed 48 h posttransfection and stained with anti-FLAG antibody followed by Alexa fluor 647-coupled secondary antibody. Cells then were analyzed for FLAG, mKO2-hCDT1 (G1), and mAG-hGem (S/G2/M) using FACSCalibur flow cytometry, and the data were analyzed using FloJo software. Average results for FLAG-containing cells from two experiments are shown. G1/S cells are those expressing both mKO2-hCDT1 and mAG-hGem. (F) Western blot of 293A whole-cell extracts transfected as described for panel A and probed with antibodies against total (Cdc2) or phosphorylated (pCdc2) Cdc2, FLAG, or actin as indicated. Positions of full-length FLAG-tagged proteins are indicated on the left. (G) Western blot of 293A cells treated with siRNA against DCAF1 (siDCAF1) or Allstars negative-control siRNA (siCtrl), transfected with plasmid expressing FLAG-tagged UL35, UL35a, or β-galactosidase (LacZ), and probed for the specific antibodies indicated on the right.
We analyzed effects of UL35 and UL35a on cell cycle progression in another cell line, HeLa-Fucci, which is engineered to change colors at different stages of the cell cycle due to the expression of two fluorescently tagged proteins, Cdt1 (mKO2-hCDT1) and geminin (mAG-hGem) (75). mKO2-hCDT1 is expressed in G1, resulting in red fluorescence, while mAG-hGem is expressed in S, G2, and M, resulting in green fluorescence. Both proteins are expressed at the G1/S interphase, resulting in yellow cells. These cells were transfected with the LacZ, UL35, or UL35a plasmid, and FLAG-expressing cells were visualized by microscopy and assessed for green and red fluorescence. As shown in Fig. 6D, UL35 expression resulted in an increase in the proportion of cells with green fluorescence relative to that of LacZ- and UL35a-expressing cells, indicating that UL35 increased the size of the S/G2/M population. These effects were quantified by FACS for cells with red, green, and yellow fluorescence (Fig. 6E). UL35-expressing cells were consistently seen to increase the S/G2/M population approximately 2-fold over that seen with the LacZ control (P = 0.01), whereas UL35a had a much smaller (and not statistically significant) effect on this population (P = 0.1). Therefore, these results also are consistent with an increased accumulation of G2 cells caused by UL35 but not UL35a.
We further examined the cell cycle perturbation of UL35 by determining if UL35 activated a G2 checkpoint response, as can be detected by the phosphorylation of cdc2 (at Tyr15). To this end, we compared levels of total and Tyr15-phosphorylated cdc2 (pcdc2) in UL35-, UL35a-, and LacZ-transfected samples (Fig. 6F). UL35, but not UL35a, resulted in an increase in pcdc2 without affecting total cdc2 levels, which is consistent with the activation of a G2 checkpoint. This suggests that the activation of the G2 checkpoint by UL35 results in the observed accumulation of G2 cells. We then tested the hypothesis that G2 checkpoint activation involves the interaction of UL35 with DCAF1 by determining if this effect of UL35 was dependent on DCAF1. Therefore, we treated cells with siRNA against DCAF1 (or negative-control siRNA) and then compared the abilities of UL35, UL35a, and LacZ to induce cdc2 phosphorylation (Fig. 6G). As expected, UL35, but not UL35a, induced cdc2 phosphorylation with control siRNA treatment (Fig. 6G, lanes 1 to 3). However, this induction of pcdc2 was abrogated by the downregulation of DCAF1, such that the level of pcdc2 was identical to that seen in the UL35a or LacZ sample (Fig. 6G, lanes 4 to 6). We conclude that the UL35-mediated G2 checkpoint activation is dependent on DCAF1.
UL35 induces a DNA damage response.
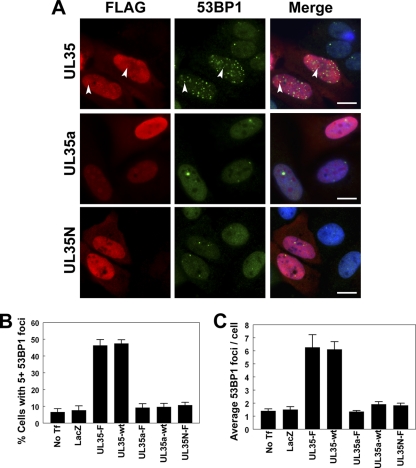
In addition to inducing G2 arrest, Vpr has been shown to induce repair foci (containing γ-H2AX and 53BP1), which are indicative of DNA damage, in a DCAF1-dependent manner (3). Therefore, we examined UL35-transfected cells for DNA damage markers. First, we transfected cells with FLAG-tagged UL35, UL35a, or UL35N and immunostained them for FLAG and 53BP1 (Fig. 7). By immunofluorescence microscopy, cells expressing UL35 but not UL35a or UL35N showed an increase in 53BP1 foci in many cells (Fig. 7A). Occasionally, some 53BP1 foci appeared to be associated with UL35 NBs (Fig. 7A, arrows). We quantified the ability of UL35 to induce 53BP1 foci by determining the percentage of cells with five or more 53BP1 foci (Fig. 7B) and determining the overall average number of 53BP1 foci in transfected cells (Fig. 7C). Only about 10% of untransfected control cells had five or more 53BP1 foci with an average of about 1.5 foci per cell, which was similar to the level for LacZ-transfected cells. Compared to untransfected control cells (No Tf), the expression of FLAG-tagged (UL35-F) and untagged UL35 (UL35-wt) caused similar significant increases in 53BP1 foci (P < 0.01 for UL35-wt and UL35-F in Fig. 7B and C), resulting in about 45% of transfected cells with five or more foci and an average of more than six foci per cell. In contrast, neither UL35a (tagged or untagged) nor UL35N induced 53BP1 foci.
Fig 7.
UL35 increases 53BP1 foci. (A) U2OS cells transfected with plasmid expressing FLAG-tagged UL35, UL35a, or UL35N were immunostained for FLAG and 53BP1 as indicated. Nuclei were visualized by DAPI staining (blue) in the merged panels. Arrows indicate the colocalization of UL35 NBs and 53BP1 foci. (B and C) U2OS cells were transfected with plasmids expressing the indicated wild-type (wt) or FLAG-tagged (-F) proteins and immunostained for FLAG and 53BP1. No Tf, untransfected control cells; LacZ, transfection control plasmid expressing β-galactosidase. The percentage of transfected cells with five or more 53BP1 foci (B) and the average number of 53BP1 foci per transfected cell (C) was determined. Values in panels B and C represent the means ± standard errors; n = 3 to 4. Scale bar, 10 μm.
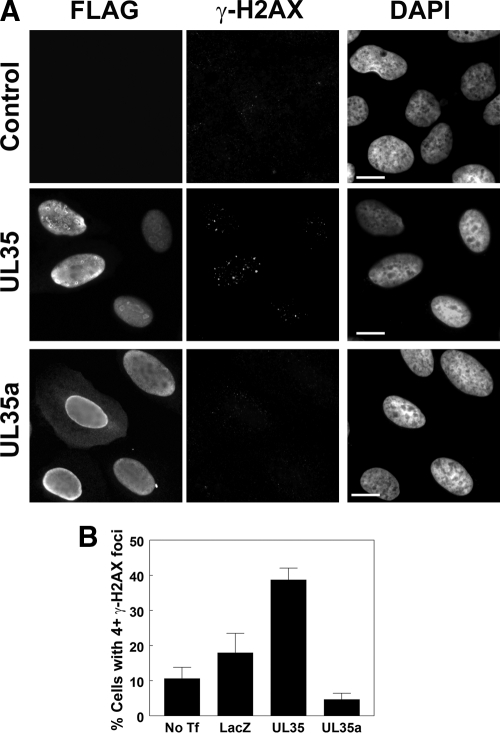
We also examined γ-H2AX as a marker for DNA damage by comparing cells expressing FLAG-tagged UL35 or UL35a to untransfected (control) cells (Fig. 8A). Control cells and UL35a-transfected cells looked similar, with very few to no γ-H2AX foci per cell, whereas UL35 induced an obvious increase in γ-H2AX foci in 20 to 30% of the transfected cells (Fig. 8A). There was no obvious correlation between the number of UL35 NBs and the number of γ-H2AX foci, and γ-H2AX foci did not obviously localize to UL35 NBs, suggesting that UL35 NB formation is not the cause of these effects. We also quantified these effects by determining the percentage of cells with four or more γ-H2AX foci per cell (Fig. 8B). This population of cells was found to increase from 10.7% in untransfected cells and 18.0% in LacZ-transfected cells to 40.3% in UL35-transfected cells (P < 0.05 for UL35 versus LacZ; P < 0.01 for UL35 versus No Tf). In contrast, only 4.7% of UL35a-transfected cells had four or more γ-H2AX foci (P > 0.05 for UL35a versus LacZ or No Tf), indicating that only full-length UL35 caused the activation of this DNA damage response. Therefore, the results from both γ-H2AX and 53BP1 foci show that UL35, but not UL35a, induces DNA damage.
Fig 8.
UL35 causes an increase in γ-H2AX foci. (A) U2OS cells transfected with FLAG-tagged UL35 or UL35a were fixed and immunostained for FLAG and γ-H2AX as indicated. Control cells were untransfected. Nuclei were visualized by DAPI staining. (B) U2OS cells were transfected with plasmids expressing the indicated FLAG-tagged proteins (No Tf, no transfection; LacZ, control plasmid expressing β-galactosidase) and immunostained for FLAG and γ-H2AX. γ-H2AX foci were counted, and the percentages of cells with four or more γ-H2AX foci are shown. Values represent the means ± standard errors; n = 3. Scale bar, 10 μm.
Given that UL35 interacts with components of the Cul4DCAF1 complex and this complex is known to function in DNA damage responses, the results described above are consistent with the hypothesis that UL35 interacts with and disrupts the normal function of DCAF1, resulting in increased DNA damage. We further investigated the involvement of DCAF1 in DNA damage induction by UL35 by examining the induction of γ-H2AX protein levels by UL35 with and without DCAF1 depletion (Fig. 6G). In keeping with the effect on γ-H2AX foci, UL35 but not UL35a expression increased the levels of γ-H2AX (Fig. 6G, lanes 1 to 3) in control siRNA-treated cells. However, the UL35-mediated induction of γ-H2AX was abrogated in cells with decreased levels of DCAF1 (Fig. 6G, lanes, 4 to 6). Therefore, the results as a whole suggest that UL35 induces DNA damage, leading to G2 checkpoint activation through interactions with DCAF1.
DISCUSSION
Our analysis of the HCMV UL35 and UL35a proteins by AP-MS revealed several unique and shared cellular targets, suggesting that UL35 and UL35a both have distinct and overlapping roles during the viral replication cycle. This notion is further supported by the controlled differential expression of UL35 and UL35a throughout the virus replication cycle, providing times when UL35 and UL35a are expressed alone (UL35 at the pre-immediate-early stage and UL35a at the early stage) or together (late in infection) (55). UL35 and UL35a also can interact with themselves and each other, which could further regulate their interaction with their cellular targets (76). Thus, the UL35 gene products may be involved in coordinating the complex regulation of host cell manipulation during the virus replication cycle.
To our knowledge, UL35 and UL35a are the first HCMV proteins shown to associate specifically with OGT; however, at least one CMV protein (UL32, a basic phosphoprotein) is modified by this enzyme (27). O-GlcNAc modification is akin to phosphorylation and is a reversible posttranslational modification of serine and threonine residues that can occur on cytoplasmic or nuclear proteins (69). Thus, O-GlcNAc modification can compete for and regulate phosphorylation sites. Many important cellular proteins are modified by OGT, including p53 (99), as are several transcription factors involved in the regulation of the MIEP (e.g., NF-κB [100], YY1 [29], and CREB [47]), making this enzyme an attractive target for subversion (69, 85). Alternatively (or in addition), UL35 and/or UL35a may associate with OGT because they are substrates for this enzyme and are modified by O-GlcNAc. Both proteins contain multiple serine and threonine residues and are known to be phosphorylated (55), thus O-GclNAc modification could contribute to the regulation of UL35 and/or UL35a activity.
In addition to associating with OGT, UL35 and UL35a also both associate with components of the nuclear import machinery (TNPO1 and IPO4). This is perhaps not surprising, considering that both proteins are nuclearly localized; however, UL35 and/or UL35a may have other effects on nuclear-cytoplasmic shuttling, since the deletion of the UL35 gene, and thus both UL35 and UL35a, affects the trafficking of at least two viral proteins (pp65 and pp71) (81). Interestingly, only UL35a interacted with TNPO1, and UL35a appears to be sufficient to affect pp71 nucleocytoplasmic transport (76), thus UL35a might affect pp71 localization through alterations to TNPO1 rather than direct interaction with pp71. Such effects on TNPO1 also could result in alterations to other cases of viral or cellular protein localization. TNPO1 typically is associated with the transport of mRNA binding proteins and ribosomal proteins (26); however, it has been implicated in the nuclear transport of other cellular proteins. For example, the chemokine receptor CCR2 has been shown to interact with TNPO1, a process enhanced by ligand binding (24). HCMV encodes chemokine receptor homologues and immune modulation is a prominent feature of HCMV infections, thus HCMV might target nuclear transport machinery to gain finer control over chemokine receptor signaling or other important cellular processes (83).
UL35a also copurified with Ras-GTPase-activating protein-SH3-domain-binding protein 2 (G3BP2). G3BP2 acts as a Ras inhibitor by interacting with the SH3 domain of GTPase activating protein (GAP) (see the references in reference 43). G3BP2 also interacts with p53 and mdm2 and can modulate p53 activity (43). Interestingly, USP7 (described below) is also a regulator of p53 and mdm2 and is a target of UL35a. In addition, G3BP2 contributes to the retention of the transcription factor NF-κB in the cytoplasm in its inactive state (72), and NF-κB is an important transcriptional activator for the CMV MIEP.
Both UL35 and UL35a interact with the ubiquitin-specific protease USP7, as determined by AP-MS (Table 1) and coimmunoprecipitation (Fig. 2). USP7 is a deubiquitinating enzyme that was first characterized as a target for the herpes simplex virus protein ICP0 (22). ICP0 has E3 ubiquitin ligase activity and is susceptible to autoubiquitination (7). Association with USP7 promotes the deubiquitinated state of ICP0, thereby enhancing ICP0 stability (7). USP7 also is targeted by EBNA1 of EBV, a gamma herpesvirus (31, 78). Unlike ICP0, EBNA1 interacts with the N-terminal TRAF domain of USP7 and acts as a competitive inhibitor of USP7 association with the tumor suppressor p53 (30). This leads to p53 stabilization by blocking p53 deubiquitination by USP7 (78). In addition, EBNA1 recruits USP7 to the viral regulatory sequences, where it may alter the chromatin structure by removing monoubiquitin from histone H2B (79).
Here, we describe UL35 and UL35a as the first examples of betaherpesvirus proteins that interact with USP7, further implicating USP7 as an important and common target of herpesviruses. UL35, but not UL35a, significantly inhibited the ability of USP7 to form NBs, demonstrating that the interaction between UL35 and USP7 is a functional one, but also suggesting that UL35 and UL35a have divergent functions with respect to USP7. While the functions of USP7 NBs are unknown, they often are associated with PML NBs (22, 80). Interestingly, and perhaps not coincidentally, ICP0, EBNA1, and UL35 all associate with PML and cause its degradation or alteration (12, 76, 87). In the case of EBNA1, the disruption of PML is dependent on its ability to interact with USP7 (87), and accordingly, USP7 recently has been shown to act as a negative regulator of PML in the absence of EBNA1 (80). We have shown here that although UL35 forms NBs that recruit PML, USP7 is not associated with these structures, possibly due to the general inhibition of USP7 to form NBs. The lack of USP7 at UL35 NBs also may be due to the fact that the same region of UL35 that mediates the multimerization that would occur in UL35 NBs also mediates the USP7 interaction (our unpublished data), hence UL35 may not be able to interact with USP7 and form NB as the same time. In addition, the UL35-mediated inhibition of USP7 NB formation is independent of PML, which, combined with the interaction data presented here, suggests that UL35 mediates specific and direct effects on USP7. Thus, although proteins from all three herpesvirus subfamilies target USP7, their purposes in doing so seem diverse, suggesting that USP7 is a regulatory hub for cellular pathways relevant to successful virus infection.
In addition to interacting with the deubiquitinase USP7, UL35 also interacts with DCAF1, DDB1, and DDA1, all of which are components of the Cul4ADCAF1 E3 ubiquitin ligase complex. Although the precise biological functions of DCAF1 and the Cul4ADCAF1 complex are unclear, DCAF1 is involved in DNA replication and cell proliferation (32, 62), and this progrowth activity is inhibited by the tumor suppresser Merlin (51). Since we did not recover Cul4A in immunoprecipitations with UL35, we do not know if UL35 interacts with an intact Cul4ADCAF1 complex. However, our results are consistent with similar affinity purifications of Vpr or Merlin from human cells coupled to mass spectrometry, which also failed to identify Cul4A in the DCAF1-DDB1 complex (32, 33, 95). Rather, the importance of Cul4A in the Vpr-targeted complex was determined from more directed assays (32, 88, 95). However, the enrichment of lysine 48-conjugated polyubiquitin at DCAF1-containing UL35 NBs suggests that, like Vpr and Merlin, UL35 interacts with DCAF1 in such a way that it allows association with the entire Cul4ADCAF1 complex (48, 51, 95). The normal cellular targets of Cul4ADCAF1 are unknown, making the functional analysis of the direct effects of the UL35-DCAF1 interaction difficult. However, studies on the Merlin-DCAF1 interaction showed that the inhibition of this complex can affect cell cycle regulation and tumorigenesis (51), raising the possibility that UL35 also affects these processes.
There is substantial precedence for viral proteins usurping and subverting Cullin-based E3 ligase complexes (50, 94). Several diverse viruses encode proteins that target DDB1-based complexes, including the paramyxovirus SV5 V protein, which binds directly to DDB1 in the absence of DCAF1and promotes the ubiquitination and degradation of STAT (71). Similarly, the murine cytomegalovirus protein M27 binds to DDB1, which facilitates the degradation of STAT2 to promote cell survival in interferon-treated cells (92). This study is particularly relevant for our findings with UL35, as it shows the importance of DDB1 for CMV infection. Interestingly, in addition to binding DDB1, the SV5 V protein causes cell cycle alterations, including G2/M arrest, as a consequence of its association with DDB1, which is similar to our observations with UL35 (53). In addition, the hepatitis B virus and woodchuck hepatitis virus X proteins both bind to DDB1, and expression is associated with altered progression through S phase, genomic instability, and apoptosis (4, 13, 50, 61). Another herpesvirus, murine gamma herpesvirus 68 (MHV68), also targets DDB1 through its M2 latency protein, which interacts with DDB1 as well as ATM and inhibits DNA damage-induced apoptosis (52). Thus, like USP7, DDB1-based E3 ligase complexes appear to be a convergence point for the virus manipulation of important host cell pathways.
Of particular relevance to our findings is the association of HIV Vpr with DCAF1. Like UL35, Vpr associates with DCAF1 (48, 95), causes G2 checkpoint activation (3, 32, 88), and increases γ-H2AX and 53BP1 foci (3). For Vpr, these effects are dependent on the interaction between Vpr and the assembled Cul4ADCAF1 complex (3, 32, 48, 88). Some Vpr mutants can bind DCAF1 but do not cause G2 arrest, suggesting that other cellular factors also are required for this effect, and it is hypothesized that these factors represent atypical targets for ubiquitination (48). Vpr forms chromatin-associated nuclear foci, and the recruitment of DCAF1 to these foci is required for G2 arrest and DDR activation (3). UL35 also forms NBs that recruit DCAF1 and are associated with polyubiquitin, although we did not see a strong correlation between the extent of UL35 NB formation and the ability to cause γ-H2AX or 53BP1 foci (Fig. 7 and 8 and data not shown), suggesting that the formation of NBs by UL35 is not a requirement for the DDR response. In keeping with this conclusion, most of the DNA damage foci induced by UL35 did not colocalize with UL35 NBs (Fig. 7 and 8). However, given the similarities between the cellular effects of Vpr and UL35, it is likely that the UL35-mediated cell cycle and DDR effects are due to the ability of UL35 to interact with DCAF1 and impair its normal functions. In support of this model, the induction of γ-H2AX by UL35 was found to be DCAF1 dependent. In addition, since HCMV infection is known to promote G1/S arrest as a requirement for viral replication, the G2 arrest that we observe with UL35 is likely a consequence of DDR activation rather than the specific function of UL35 expression during replication. However, we note that there is evidence that, in some situations, HCMV infections result in some accumulation of infected cells in the G2/M phase of the cell cycle (25, 37, 57), and it is possible that UL35 contributes to these effects. It also is worth noting that the disruption of either Cul4, DDA1, or DDB1 is associated with increased DNA damage and cell cycle alterations (56, 68), further implicating UL35 as a potential regulator of Cul4DCAF1 activity. Further, since UL35a can interact with UL35 (76), it is possible that UL35a, which is expressed with UL35 late in infection, could regulate or attenuate UL35 effects on DCAF1 by competing with DCAF1 for association with UL35.
The manipulation of DNA damage and repair pathways is a common feature of herpesvirus and DNA viruses in general. Considerable evidence indicates that the manipulation of these pathways is necessary for ensuring viral DNA replication while limiting apoptotic and other responses unfavorable to viral infection, and that such manipulations are carried out by specific viral proteins. We have now shown that UL35 is one such protein that stimulates the DDR and suggest that, like Vpr, this effect is due to binding the Cul4DCAF1 complex. UL35 may disrupt normal Cul4-DCAF1 complex function by hijacking the complete complex or by sequestering some of the components (DCAF1, DDB1, and DDA1) and disrupting the complex. In either case, the expression of UL35 contributes to the activation of the DDR, likely through the alteration of normal DCAF1 functions. Interestingly, among its many roles, USP7 also has been shown to play a role in the DDR (42), so the ability of UL35 to activate the DDR and induce G2 arrest may be multifactorial. Taken together, our results suggest that UL35 has important roles in the manipulation and fine-tuning of the host cell to support efficient replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kathy Shire, June Tan, and Tobias Flecken for technical assistance and Yue Xiong (University of North Carolina) and Filippo Giancotti (Sloan-Kettering Institute) for DCAF1 constructs.
This work was supported by Canadian Institutes of Health Research operating grants awarded to L.F. (grant number 12477), G.D. (MOP-84260), and B.R. (MOP-812268). J.S. is supported by a Terry Fox Foundation Research Fellowship awarded by the National Cancer Institute of Canada. G.D. is a Canadian Institutes of Health Research (CIHR) New Investigator, a Senior Scientist of the Beatrice Hunter Cancer Research Institute, and the Cameron Research Scientist of the Dalhousie University Cancer Research Program. B.R. holds the Canada Research Chair (Tier 2) in Proteomics and Molecular Medicine. L.F. is a tier 1 Canada Research Chair in Molecular Virology and therefore receives salary support from the Canada Research Chairs program.
Footnotes
Published ahead of print 9 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ahn JH, Hayward GS. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39–55 [DOI] [PubMed] [Google Scholar]
- 2. Andoniou CE, Degli-Esposti MA. 2006. Insights into the mechanisms of CMV-mediated interference with cellular apoptosis. Immunol. Cell Biol. 84:99–106 [DOI] [PubMed] [Google Scholar]
- 3. Belzile JP, Abrahamyan LG, Gerard FC, Rougeau N, Cohen EA. 2010. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6:e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergametti F, Sitterlin D, Transy C. 2002. Turnover of hepatitis B virus X protein is regulated by damaged DNA-binding complex. J. Virol. 76:6495–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernardi R, Pandolfi PP. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8:1006–1016 [DOI] [PubMed] [Google Scholar]
- 6. Bresnahan WA, Boldogh I, Thompson EA, Albrecht T. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150–160 [DOI] [PubMed] [Google Scholar]
- 7. Canning M, Boutell C, Parkinson J, Everett RD. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279:38160–38168 [DOI] [PubMed] [Google Scholar]
- 8. Cantrell SR, Bresnahan WA. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castillo JP, Kowalik TF. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19–34 [DOI] [PubMed] [Google Scholar]
- 10. Cayrol C, Flemington EK. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748–2759 [PMC free article] [PubMed] [Google Scholar]
- 11. Chaurushiya MS, Weitzman MD. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amsterdam) 8:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chelbi-Alix MK, de Thé H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 13. Chen HY, Tang NH, Lin N, Chen ZX, Wang XZ. 2008. Hepatitis B virus X protein induces apoptosis and cell cycle deregulation through interfering with DNA repair and checkpoint responses. Hepatol. Res. 38:174–182 [DOI] [PubMed] [Google Scholar]
- 14. Choi H, et al. 2011. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8:70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467 [DOI] [PubMed] [Google Scholar]
- 16. Crough T, Khanna R. 2009. Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol. Rev. 22:76–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dellaire G, Bazett-Jones DP. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963–977 [DOI] [PubMed] [Google Scholar]
- 18. Dubaele S, Chene P. 2007. Cellular studies of MrDb (DDX18). Oncol. Res. 16:549–556 [DOI] [PubMed] [Google Scholar]
- 19. Dunn W, et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everett RD. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol. 8:365–374 [DOI] [PubMed] [Google Scholar]
- 21. Everett RD, Meredith M, Orr A. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett RD, et al. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Everett RD, et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Favre N, et al. 2008. Chemokine receptor CCR2 undergoes transportin1-dependent nuclear translocation. Proteomics 8:4560–4576 [DOI] [PubMed] [Google Scholar]
- 25. Fortunato EA, Sanchez V, Yen JY, Spector DH. 2002. Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 76:5369–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fried H, Kutay U. 2003. Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 60:1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greis KD, Gibson W, Hart GW. 1994. Site-specific glycosylation of the human cytomegalovirus tegument basic phosphoprotein (UL32) at serine 921 and serine 952. J. Virol. 68:8339–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harada K, Yamada A, Yang D, Itoh K, Shichijo S. 2001. Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int. J. Cancer 93:623–628 [DOI] [PubMed] [Google Scholar]
- 29. Hiromura M, et al. 2003. YY1 is regulated by O-linked N-acetyl-glucosaminylation (O-glcNAcylation). J. Biol. Chem. 278:14046–14052 [DOI] [PubMed] [Google Scholar]
- 30. Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. 2003. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 278:47753–47761 [DOI] [PubMed] [Google Scholar]
- 31. Holowaty MN, et al. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987–29994 [DOI] [PubMed] [Google Scholar]
- 32. Hrecka K, et al. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J, Chen J. 2008. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 27:4056–4064 [DOI] [PubMed] [Google Scholar]
- 34. Hwang J, Kalejta RF. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334–338 [DOI] [PubMed] [Google Scholar]
- 35. Ishov AM, Stenberg RM, Maul GG. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jackson S, Xiong Y. 2009. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34:562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jault FM, et al. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalejta RF. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalejta RF, Bechtel JT, Shenk T. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kalejta RF, Shenk T. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virol. 77:3451–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kessner D, Chambers M, Burke R, Agus D, Mallick P. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khoronenkova SV, Dianova II, Parsons JL, Dianov GL. 2011. USP7/HAUSP stimulates repair of oxidative DNA lesions. Nucleic Acids Res. 39:2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. 2007. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP). Oncogene 26:4209–4215 [DOI] [PubMed] [Google Scholar]
- 44. Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158 [DOI] [PubMed] [Google Scholar]
- 45. Kudoh A, et al. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 46. Kudoh A, et al. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lamarre-Vincent N, Hsieh-Wilson LC. 2003. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125:6612–6613 [DOI] [PubMed] [Google Scholar]
- 48. Le Rouzic E, et al. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188 [DOI] [PubMed] [Google Scholar]
- 49. Li M, et al. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653 [DOI] [PubMed] [Google Scholar]
- 50. Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. 2010. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 17:105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li W, et al. 2010. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell 140:477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liang X, et al. 2006. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J. Virol. 80:5862–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin GY, Lamb RA. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu G, et al. 2010. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28:1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y, Biegalke BJ. 2002. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J. Virol. 76:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lovejoy CA, Lock K, Yenamandra A, Cortez D. 2006. DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 26:7977–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu M, Shenk T. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850–8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lu M, Shenk T. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lukashchuk V, McFarlane S, Everett RD, Preston CM. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin-Lluesma S, et al. 2008. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology 48:1467–1476 [DOI] [PubMed] [Google Scholar]
- 62. McCall CM, et al. 2008. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 28:5621–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Medenbach J, Schreiner S, Liu S, Luhrmann R, Bindereif A. 2004. Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol. Cell. Biol. 24:7392–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miller-Kittrell M, Sparer TE. 2009. Feeling manipulated: cytomegalovirus immune manipulation. Virol. J. 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Murphy EA, Streblow DN, Nelson JA, Stinski MF. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nikitin PA, et al. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nitzsche A, Paulus C, Nevels M. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Olma MH, et al. 2009. An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J. Cell Sci. 122:1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ozcan S, Andrali SS, Cantrell JE. 2010. Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799:353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Petrik DT, Schmitt KP, Stinski MF. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 80:3872–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE. 2005. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 79:13434–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Prigent M, Barlat I, Langen H, Dargemont C. 2000. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 275:36441–36449 [DOI] [PubMed] [Google Scholar]
- 73. Reeves MB. 2011. Chromatin-mediated regulation of cytomegalovirus gene expression. Virus Res. 157:134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sakaue-Sawano A, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132:487–498 [DOI] [PubMed] [Google Scholar]
- 76. Salsman J, Wang X, Frappier L. 2011. Nuclear body formation and PML body remodeling by the human cytomegalovirus protein UL35. Virology 414:119–129 [DOI] [PubMed] [Google Scholar]
- 77. Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4:e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saridakis V, et al. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18:25–36 [DOI] [PubMed] [Google Scholar]
- 79. Sarkari F, et al. 2009. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 5:e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sarkari F, Wang X, Nguyen T, Frappier L. 2011. The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS One 6:e16598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schierling K, Buser C, Mertens T, Winkler M. 2005. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J. Virol. 79:3084–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schierling K, Stamminger T, Mertens T, Winkler M. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J. Virol. 78:9512–9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sherrill JD, Miller WE. 2008. Desensitization of herpesvirus-encoded G protein-coupled receptors. Life Sci. 82:125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shirata N, et al. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336–30341 [DOI] [PubMed] [Google Scholar]
- 85. Sinclair J. 2010. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta 1799:286–295 [DOI] [PubMed] [Google Scholar]
- 86. Sivachandran N, Cao JY, Frappier L. 2010. Epstein-Barr virus nuclear antigen 1 hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 84:11113–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sivachandran N, Sarkari F, Frappier L. 2008. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 4:e1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tan L, Ehrlich E, Yu XF. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81:10822–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 157:128–133 [DOI] [PubMed] [Google Scholar]
- 92. Trilling M, et al. 2011. Identification of DNA-damage DNA-binding protein 1 as a conditional essential factor for cytomegalovirus replication in interferon-gamma-stimulated cells. PLoS Pathog. 7:e1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Varnum SM, et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Weitzman MD, Lilley CE, Chaurushiya MS. 2011. Changing the ubiquitin landscape during viral manipulation of the DNA damage response. FEBS Lett. 585:2897–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wen X, Duus KM, Friedrich TD, de Noronha CM. 2007. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282:27046–27057 [DOI] [PubMed] [Google Scholar]
- 96. Wiebusch L, Hagemeier C. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wilkinson DE, Weller SK. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660 [DOI] [PubMed] [Google Scholar]
- 99. Yang WH, et al. 2006. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8:1074–1083 [DOI] [PubMed] [Google Scholar]
- 100. Yang WH, et al. 2008. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. U. S. A. 105:17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zeghouf M, et al. 2004. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.