Abstract
Estrogen receptor (ER) and NF-κB are transcription factors with profound effects on breast cancer cell proliferation and survival. While many studies demonstrate that ER and NF-κB can repress each other, we previously identified a gene signature that is synergistically upregulated by these two factors in more aggressive luminal B breast tumors. Herein, we examine a novel mechanism of cross talk between ER and NF-κB that results in the upregulation of the antiapoptotic gene BIRC3 (also known as cIAP2). We demonstrate that NF-κB, acting through two response elements, is required for ER recruitment to an adjacent estrogen response element (ERE) in the BIRC3 promoter. This effect is accompanied by a major increase in NF-κB-dependent histone acetylation around the ERE. Interestingly, CBP, a histone acetyltransferase previously implicated in repressive interactions between ER and NF-κB, plays a permissive role by promoting histone acetylation and ER recruitment, as well as enhanced expression of BIRC3. These findings suggest a new gene regulatory mechanism by which inflammation and NF-κB activation can influence ER recruitment to inherently inactive ER binding sites. This fine-tuning mechanism may explain how two factors that generally repress each other's activity may work together on certain genes to promote breast cancer cell survival and tumor progression.
INTRODUCTION
The estrogen receptor (ER) is expressed in approximately 75% of breast cancers, and women with such tumors are generally treated with endocrine therapies, such as tamoxifen or aromatase inhibitors. However, not all ER-positive tumors respond to these therapies. Through gene expression profiling studies, ER-positive tumors have been delineated into two intrinsic subtypes, luminal A and luminal B (48, 49). Women with the luminal A subtype of breast tumors respond well to therapy and have a good prognosis, whereas the outcome is poor for women with the luminal B subtype of tumors, nearly as poor as that seen in the case of ER-negative tumors. Our lab recently identified a gene signature synergistically upregulated by cross talk between ER and NF-κB that is highly associated with luminal B but not luminal A-type tumors (16). This signature is enriched for cell survival genes, including the cellular inhibitor of apoptosis gene, cIAP2, which is also known as BIRC3. We have previously shown that BIRC3 is upregulated by estradiol (E2) and the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) in a number of ER-positive but not ER-negative cell lines. Using chemical inhibitors and a small interfering RNA (siRNA) approach, our lab has further demonstrated that BIRC3 plays an important role in promoting estrogen-dependent breast cancer cell survival and protecting against TNF-α-induced cell death (51). Understanding the mechanism by which BIRC3 is upregulated by cross talk between ER and NF-κB is therefore of clinical relevance.
The ER subtypes, ERα and ERβ, are ligand-dependent transcription factors that interact with DNA and control transcription of ER target genes in response to estradiol (E2). In breast cancer cells, thousands of ER target genes and binding sites have been identified through genome-wide approaches (6, 15, 19, 31, 35). In the classical mechanism of ER action, binding to DNA occurs at palindromic estrogen response elements (EREs). However, many ER binding sites do not contain recognizable EREs (5, 35, 38); hence, in addition to direct binding to DNA sequences, ER tethering to other transcription factors appears to play a significant role in mediating estrogen action (21, 27, 33, 39, 47). The NF-κB pathway is activated by a variety of extracellular stimuli, such as proinflammatory cytokines and growth factors. In the canonical pathway, activation of the upstream IκB kinase (IKK) kinase complex leads to downstream phosphorylation and subsequent proteasomal degradation of the IκB inhibitory proteins. This liberates NF-κB family members p65 and p50, which then translocate to the nucleus, where they bind to their response elements (NF-κB-REs) and transactivate gene expression. Like ER, NF-κB can interact with many other transcription factors to regulate target gene expression.
ER and NF-κB family members have been shown to influence each other's transcriptional activity. Much work has been done to delineate the multiple mechanisms by which ER can repress NF-κB action to exert an anti-inflammatory effect (7, 13, 28, 41). Similarly, there are several pieces of evidence indicating that NF-κB can repress ER expression and transcriptional activity (2, 10, 14, 23, 36). However, there are relatively few examples of ER and NF-κB working together to increase transcription (1, 30, 45, 53). Recently, we found that these factors can work cooperatively on a number of genes, including PTGES, which codes for prostaglandin E synthase, and ABCG2, which codes for a drug efflux pump (17, 43). For these genes, we demonstrated that NF-κB may potentiate ER action by stabilizing ER occupancy on DNA at functional EREs. For the PTGES gene, cross talk occurs at the ERE itself, whereas for the ABCG2 gene, a weak NF-κB-RE located downstream of the ERE is required. This suggests that many gene-specific mechanisms of positive cross talk can occur between ER and NF-κB and that the specificity may be based on the nature and arrangement of the regulatory elements in the gene.
However, the mechanism by which ER and NF-κB work together to increase expression of BIRC3 to promote cell survival is not known. In the present study, we show that ER can potentiate TNF-α-dependent BIRC3 expression by binding to an ERE directly upstream of two NF-κB-REs. The ability of ER to access its binding site, however, is NF-κB dependent and is accompanied by CBP-mediated changes in histone acetylation around the ERE. This transcriptional mechanism represents a novel interaction between two potent transcription factors that are known to be important regulators of breast cancer growth and progression. An important implication of our findings is that inflammatory factors can alter where ER binds in the genome and that ER can, in turn, enhance the effect of inflammatory factors on the regulation of their target genes.
MATERIALS AND METHODS
Materials.
17β-Estradiol (E2) and 4-hydroxytamoxifen (TAM) were obtained from Sigma. Cytokines were obtained from R&D Systems. ICI 182,780 (ICI) was purchased from Tocris. The small molecule theophylline, 8-[(benzylthio) methyl]-(7CI, 8CI) (TPBM) was a generous gift from David Shapiro (University of Illinois at Urbana-Champaign [UIUC]). Adenoviral vectors for green fluorescent protein (GFP) and IκBα-DN were kindly provided to us by Michael O'Donnell (University of Illinois at Chicago [UIC]) and Ruxana Sadikot (UIC), respectively, and used as previously described (17, 43). Antibodies for chromatin immunoprecipitation (ChIP) assays, ERα (sc-543), p65 (sc-372), and CBP (sc-369) were obtained from Santa Cruz Biotechnology; anti-histone H3 (ab1791-100) was obtained from Abcam, and anti-histone H4 (17-10047), anti-acetyl-H3 (06-599), and anti-acetyl-H4 (06-866) were obtained from Millipore.
Cell culture.
MCF-7 cells were kindly provided by Benita Katzenellenbogen (UIUC). These cells express ERα but not ERβ (8). Cells were cultured in minimal essential medium (MEM) containing 5% calf serum with antibiotics. Prior to E2 treatment, cells were cultured in phenol-red free media containing 5% charcoal-dextran stripped calf serum for at least 3 days.
Regulation of BIRC3 mRNA expression and promoter activity.
RNA was isolated and analyzed by reverse transcription-quantitative PCR (RT-qPCR) using primers specific for BIRC3 and for 36B4, which served as an internal control, as previously described (16, 51). Fragments of the BIRC3 promoter (from −527 to +55 and −247 to +55) subcloned into the PGL2-basic reporter plasmid (24) were kindly provided to us by T. H. Lee (Yonsei University, South Korea). Mutations to the ERE and NF-κB-REs were made using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). Wild-type and mutated sequences are given in Table S1 in the supplemental material. MCF-7 cells were transiently transfected with reporter constructs along with the Renilla luciferase construct pGL4.70 (Promega), using Lipofectamine 2000 (Invitrogen) in antibiotic-free Opti-MEM (Invitrogen). Dual-luciferase assays (Promega) were carried out after 4 h of treatment with E2 and/or TNF-α.
ChIP.
ChIP assays were carried out as previously described (43). For initial ChIP assays, chromatin was sonicated three times for 10 s to generate fragments of ∼500 bp in size. However, for ChIP studies using qPCR primers tiled along the BIRC3 promoter, sonication was increased to 10 s 30 times to generate fragments ∼200 bp in size. Inputs were serially diluted to generate standard curves and DNA enrichment was calculated as percent input for each sample. The percents input for individual experiments were converted to fold change relative to an untreated control. Average fold changes from a minimum of 3 independent experiments were then plotted. Primer sequences for ChIP qPCR are listed in Table S1 in the supplemental material.
siRNA transfections.
siRNA targeting p65, ER, or CBP or a nonspecific control (siNeg) was purchased from Ambion and transfected using the Dharmafect 1 transfection reagent as previously described (51). Treatments for RNA measurements or ChIP assays were carried out 48 h after siRNA transfection.
Statistical analysis.
qPCR and reporter data were analyzed by two-way analysis of variance (ANOVA) followed by a post hoc Bonferroni test. Significance for all statistical tests was set at P < 0.05. The data shown are the mean ± standard error of the mean (SEM) from at least three independent determinations.
RESULTS
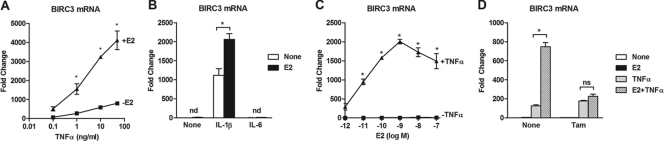
In MCF-7 breast cancer cells, BIRC3 expression is upregulated by TNF-α in a dose-dependent manner, and this upregulation is further enhanced by the combination of TNF-α and E2 (Fig. 1A). E2 in combination with interleukin-1β (IL-1β), but not IL-6, can also enhance BIRC3 expression over that seen with the respective cytokine alone (Fig. 1B). In the absence of cytokines, E2 has no effect on BIRC3 expression across a wide range of doses (Fig. 1C). However, in the presence of cytokines, such as TNF-α, a typical E2 dose-dependent increase in BIRC3 expression is observed. The effect of E2 is prevented by a number of ER antagonists, including TAM (Fig. 1D) and ICI 182,780 (16), indicating that ER is required for E2 activity. These data demonstrate that although E2 cannot regulate BIRC3 expression on its own, it is capable of potentiating cytokine action on this gene.
Fig 1.
Estradiol (E2) cannot induce expression of BIRC3 but potentiates regulation by proinflammatory cytokines. MCF-7 cells were treated for 2 h with increasing doses of TNF-α in the absence or presence of 10 nM E2 (A), 10 ng/ml IL-1β or IL-6 in the absence or presence of 10 nM E2 (B), increasing doses of E2 in the absence or presence of 10 ng/ml TNF-α (C), or 10 nM E2 and/or 10 ng/ml TNF-α in the absence or presence of 1 μM 4-hydroxytamoxifen (TAM) (D). BIRC3 mRNA was measured by qPCR, and fold change was calculated using the ΔΔCT method with 36B4 as an internal control. The data represent the means ± SEM for three independent replicates. *, P < 0.05 compared to treatment with either E2 or cytokine alone. nd, not detectable; ns, not significant.
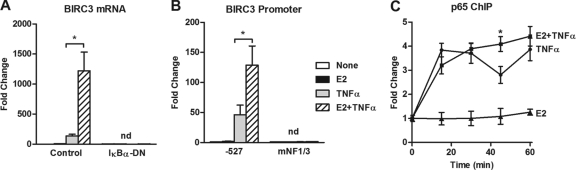
Previous studies have shown that cytokines regulate BIRC3 expression in an NF-κB-dependent manner through the first (NF1) and third (NF3) of three NF-κB-REs located in the promoter of the gene (24). We find that inhibition of the NF-κB pathway with IκBα-DN (Fig. 2A) or mutation of both NF-κB sites (Fig. 2B) not only prevents TNF-α from regulating expression of BIRC3, but also completely blocks E2 from potentiating TNF-α action, suggesting that both E2 and TNF-α require NF-κB activity to mediate their effects on the BIRC3 gene. We confirmed that IκBα-DN potently inhibits nuclear translocation of NF-κB family members, p65 and p50, in MCF-7 cells (see Fig. S1 in the supplemental material). The NF-κB family member p65 has been shown to bind to the promoter region containing NF1 and NF3 in response to cytokines (24). p65 recruitment to this region was assessed in breast cancer cells by ChIP assays, which demonstrated that the combination of E2 and TNF-α had little effect on p65 occupancy compared to that seen with TNF-α alone (Fig. 2C). It is therefore unlikely that E2 potentiates TNF-α action by influencing p65 recruitment to the promoter, which is in contrast to what we have previously shown for the ABCG2 gene (43).
Fig 2.
The NF-κB pathway is required for enhanced BIRC3 expression by the combination of E2 and TNF-α. (A) BIRC3 mRNA expression was examined in MCF-7 cells following 24 h of exposure to adenoviral vectors for GFP (control) or a dominant-negative form of IκBα (IκBα-DN) and an additional 2 h of treatment with E2, TNF-α, or both. (B) MCF-7 cells were transiently transfected with a BIRC3 promoter reporter construct spanning bp −527 to +55 of the promoter, in which the NF-κB response elements (NF1 and NF3) were intact (−527) or mutated (mNF1/3). Dual-luciferase assays were carried out following 4 h of treatment with E2, TNF-α, or both. (C) ChIP assays were carried out for p65 following treatment of MCF-7 cells with E2, TNF-α, or both for various lengths of time. The fold-increase in p65 occupancy at the BIRC3 promoter was calculated from the percent input of each pulldown and then comparing each treatment to untreated controls from four independent experiments. *, P < 0.05 compared to treatment with TNF-α alone. nd, not detectable.
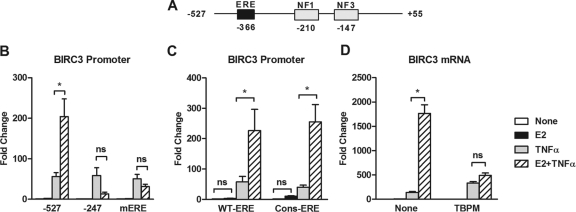
Further analysis of the BIRC3 promoter sequence revealed a putative ERE located 156 bp upstream of NF1 (Fig. 3A). Deletion (−247) or mutation of this ERE (mERE) completely prevented E2 from potentiating TNF-α action but had little effect on TNF-α alone (Fig. 3B). This finding suggests that although the ERE does not appear to be functional in response to E2 alone, it possesses a latent functionality that becomes apparent in response to both E2 and TNF-α—or, in other words, TNF-α allows this ERE to function. Although the sequence of the ERE is near consensus (GGGCA TAT TGACC), we mutated it to a full consensus site (GGTCA CCG TGACC) according to MatInspector (44) because the single-nucleotide difference is at an essential position in the sequence. A BIRC3 promoter construct containing the consensus ERE also failed to respond to E2, indicating that the lack of functionality of the ERE is not because of a weak binding sequence (Fig. 3C). In addition, TPBM, a small molecule that prevents ER binding to DNA (37), prevented potentiation of BIRC3 expression by E2 plus TNF-α over TNF-α alone, suggesting that ER binding to the BIRC3 ERE is essential for enhanced regulation of the gene (Fig. 3D).
Fig 3.
The ability of estrogen to enhance BIRC3 expression requires an intact estrogen response element (ERE) and ER binding to DNA. (A) Schematic representation of the BIRC3 promoter showing the relative position of a putative ERE upstream of two functional NF-κB response elements (NF1 and NF3) (44). (B) MCF-7 cells were transfected with the BIRC3 promoter reporter construct in which the ERE was intact (−527), a region of the promoter containing the ERE was deleted (−247), or the ERE was mutated (mERE). Reporter activity was measured by dual-luciferase assay following 4 h of treatment with E2, TNF-α, or both. (C) Luciferase activity of the −527 promoter fragment, in which the ERE was intact (wild type [WT-ERE]) or mutated to a consensus ERE (Cons-ERE), was measured after 4 h of treatment with E2, TNF-α, or both. (D) BIRC3 mRNA levels were measured in MCF-7 cells treated with E2, TNF-α, or both in the absence or presence of an ER-DNA binding inhibitor, TPBM (20 μM). *, P < 0.05 compared to treatment with TNF-α alone. ns, not significant.
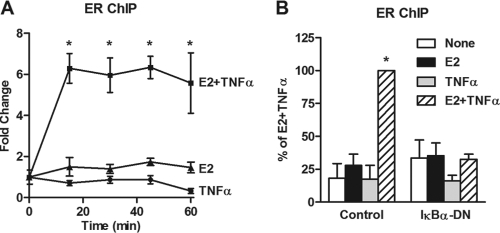
To examine recruitment of ER to the BIRC3 promoter, ChIP assays were carried out. In agreement with the lack of an E2 effect on BIRC3 expression, E2 had no effect on ER recruitment to the BIRC3 promoter region containing the ERE (Fig. 4A; see Fig. S2 in the supplemental material). However, the combination of E2 and TNF-α led to a rapid and robust recruitment of ER, which was sustained over 60 min and completely prevented by blockade of the NF-κB pathway with IκBα-DN (Fig. 4A and B). These results indicate that E2 alone cannot bring about ER recruitment to the BIRC3 gene and that activation of NF-κB is necessary to allow ER to be recruited by the combination of E2 and TNF-α. This mechanism appears to be gene specific since TNF-α can actually enhance (17, 43), repress, or have no effect on (see Fig. S3 in the supplemental material) ER recruitment to other ER target genes.
Fig 4.
ER recruitment to the BIRC3 promoter requires E2, TNF-α and the NF-κB pathway. (A) ChIP assays for ER were carried out following treatment of MCF-7 cells with E2, TNF-α, or both for up to 60 min, and recruitment to the BIRC3 promoter was examined. (B) ER recruitment to BIRC3 was carried out by ChIP assay following 24 h of exposure to IκBα-DN and 45 min of treatment with E2, TNF-α, or both. *, P < 0.05 compared to treatment with E2 or TNF-α alone.
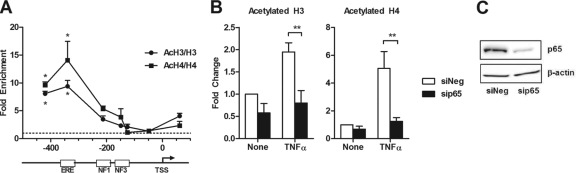
We next conducted a series of experiments to examine whether TNF-α influenced ER recruitment through chromatin remodeling around the ERE of the BIRC3 gene. Formaldehyde-assisted isolation of regulatory elements (FAIRE), micrococcal nuclease (MNase) sensitivity, and restriction enzyme accessibility assays all failed to detect any changes in chromatin architecture at the ERE, but chromatin remodeling was observed at the NF-κB-REs (see Fig. S4 in the supplemental material) (data not shown). However, TNF-α induced a robust enrichment of acetylated histones 3 and 4 (AcH3 and AcH4) around the ERE (Fig. 5A; see Fig. S5 and S6 in the supplemental material). A similar increase in acetylation throughout this region, as well as increased AcH4 nearer the transcription start site, was observed in response to E2 plus TNF-α (see Fig. S6). The effect of TNF-α on acetylation around the ERE was blocked by siRNA targeting p65 (Fig. 5B and C), indicating that TNF-α acting through NF-κB leads to an increase in histone acetylation at this site. This finding suggests that TNF-α-stimulated histone acetylation may be a potential mechanism by which NF-κB influences ER recruitment to the BIRC3 promoter.
Fig 5.
TNF-α and p65 cause enrichment of acetylated histones around the BIRC3 ERE. (A) AcH3 and AcH4 were examined by ChIP assay in MCF-7 cells treated with TNF-α for 15 min. Enrichment was calculated by first dividing the percentage of input for each AcH3 or AcH4 pulldown by the percentage of input for the total amount of H3 or H4, respectively, and second by comparing the ratio of acetylated to total histones in TNF-α-treated cells to that in vehicle-treated control cells. PCR was carried out using small amplicons that tile along the BIRC3 promoter. *, P < 0.05 compared to an untreated control. (B) Acetylation of H3 and H4 at the ERE was assessed following transfection with siNeg (control) or sip65 and treatment with or without TNF-α for 15 min. Fold change was calculated from the percentage of input for each pulldown and comparison to untreated siNeg controls for 3 independent experiments. **, P < 0.01 compared to untreated siNeg control. (C) p65 protein levels were examined by Western blotting in MCF-7 cells transfected with siNeg or sip65 for 48 h.
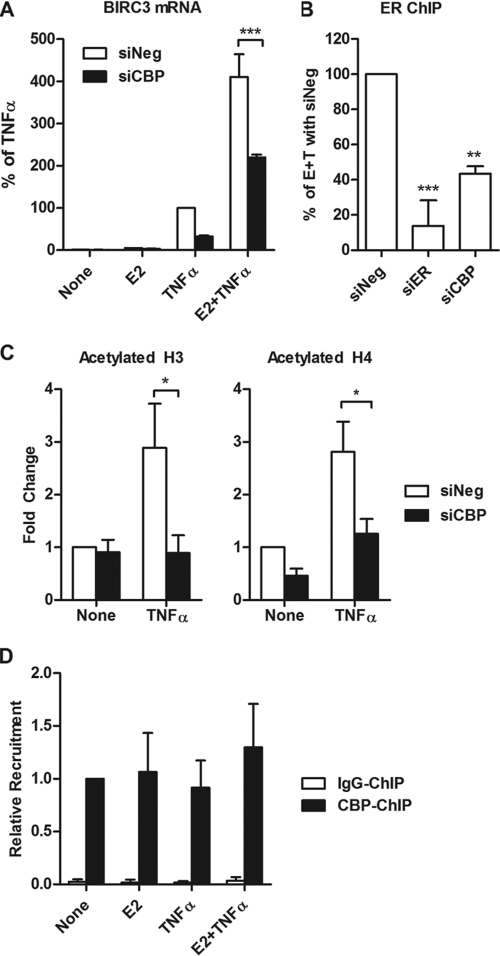
To determine which histone acetyltransferases (HATs) may be involved in histone acetylation around the ERE and subsequent ER recruitment, siRNAs for a number of coactivators and HATs known to interact with ER and p65 were used (see Fig. S7 in the supplemental material). As expected, knockdown of ER and p65 reduced regulation of BIRC3. Of the HATs, we found that CBP significantly reduced BIRC3 expression in response to E2 plus TNF-α treatment (Fig. 6A). CBP knockdown, as well as ER knockdown, significantly reduced ER recruitment to the BIRC3 promoter in the presence of E2 plus TNF-α (Fig. 6B). Furthermore, knockdown of CBP significantly reduced AcH3 and AcH4 enrichment around the ERE (Fig. 6C). The presence of CBP at the BIRC3 promoter was observed by ChIP assay but was not regulated by E2 and/or TNF-α treatment (Fig. 6D). Taken together, these data suggest that CBP is required for regulation of BIRC3, and this may be due to its ability to acetylate histones around the ERE and promote ER access to the gene, thereby leading to potentiation of BIRC3 gene expression.
Fig 6.
Role of CBP in BIRC3 expression, ER recruitment, and histone acetylation around the ERE. (A) MCF-7 cells were transfected with siRNA for CBP, and regulation of BIRC3 mRNA by E2, TNF-α, or both was assessed by qPCR. (B) ER recruitment to the BIRC3 promoter was assessed by ChIP assays following transfection with siNeg, siER, or siCBP and treatment with E2 plus TNF-α for 15 min. Data are represented as a percentage of ER recruitment relative to the siNeg control. (C) Acetylated H3 and H4 at the ERE were examined following transfection with siNeg or CBP. *, P < 0.05, **, P < 0.01, and ***, P < 0.001, compared to the siNeg control. (D) ChIP assays for CBP or IgG were carried out after 15 min of treatment with E2, TNF-α, or both.
DISCUSSION
In this study, we find that BIRC3, an important antiapoptotic factor in breast cancer cells (51), is upregulated by TNF-α and that this regulation is greatly potentiated by E2, despite the fact that E2 cannot regulate the gene on its own. This occurs through a novel mechanism of cross talk between ER and NF-κB not previously observed. We find that E2 potentiates TNF-α action through a near-consensus ERE that is located upstream of two functional NF-κB-REs. The endogenous ERE appears to be inaccessible to ER in the presence of E2 alone, but NF-κB activation leads to a high degree of histone acetylation around the ERE. This is mediated by CBP, which allows ER to access its binding site and work synergistically with NF-κB to potently and rapidly increase transcription of the BIRC3 gene.
At first we expected that the lack of functionality of the ERE could be the result of its sequence. It has been demonstrated that the sequence of the ERE can affect the binding affinity of ER for its response element and in turn influences transcription of the target gene (18, 32). Furthermore, we have shown that cooperativity between ER and NF-κB occurs between a nonconsensus NF-κB-RE and an ERE for another gene whose expression is increased by E2 and TNF-α (43). However, the nature of the BIRC3 ERE is nearly identical to a consensus sequence, and conversion to a perfect consensus sequence does not significantly increase ER activity, suggesting another factor may be preventing ER from interacting with this binding site. This is in accordance with the finding that far more high-confidence ER binding sites have been identified computationally than are actually detected by genome-wide ChIP-on-chip studies for ER recruitment with E2 alone (6, 52).
We also considered that the endogenous chromatin structure may be preventing ER from interacting with its binding site. The fact that many transcription factor binding sites are occluded during DNA packaging has been proposed as a common mechanism to confer a higher degree of regulation to gene expression in response to various stimuli (11). Furthermore, previous studies have demonstrated that other transcription factors, such as FOXA1, are necessary to facilitate ER recruitment to specific regions of chromatin (26). NF-κB has previously been shown to enhance DNA accessibility around the NF-κB-RE to which it binds. For example, at the granulocyte-macrophage colony-stimulating factor gene (GM-CSF) promoter, NF-κB causes increase in chromatin accessibility via remodeling through recruitment of the ATPase component of the SWI/SNF chromatin remodeling complex, BRG1 (4, 22). More recently, constitutive activation of NF-κB along with AP1 has been associated with increased accessibility of the IL-6 promoter in highly invasive breast cancer cells (40). However, in the case of the BIRC3 gene, NF-κB had little to no effect on the chromatin structure around the ERE, suggesting that NF-κB may be influencing ER recruitment through another mechanism. We did observe a significant enrichment of acetylated histones around the ERE in the presence of TNF-α or E2 plus TNF-α, which suggested that NF-κB interaction with HATs may increase accessibility of the ERE to ER. Previous studies have shown that histone acetylation can contribute to DNA accessibility to transcription factors by loosening histone-DNA contacts (3).
In our study, a number of HATs were identified that are required for E2 plus TNF-α-induced expression of BIRC3, several of which are known to interact independently with ER or NF-κB. CBP was of particular interest to us since it was required not only for enhanced BIRC3 expression, but also for ER recruitment as well as histone acetylation around the ERE. Previous studies have shown that ER recruitment to the CRH promoter is accompanied by an increase in H3 acetylation, as well as CBP recruitment (34). CBP has also been shown to be important for NF-κB action on the BIRC3 gene (25). In MCF-7 breast cancer cells, CBP appears to be present constitutively at the BIRC3 promoter. This has also been shown for other genes where CBP is thought to play a role in the rapid induction of transcription (42, 46). For the c-Fos gene, CBP is constitutively present at a serum response element where it interacts with the transcription factor, Elk-1 (42). Similarly, CBP is constitutively present at the mitogen-activated protein kinase phosphatase 1 gene (MKP-1) promoter, potentially through interactions with Sp3 and CREB transcription factors (46). The BIRC3 promoter contains a large number of putative response elements in close proximity to the ERE, including binding sites for AP-1, SP1, PPAR, C/EBP, and STAT transcription factors. All of these are known to interact with CBP, but determining which factors are responsible for maintaining CBP at the BIRC3 promoter requires further investigation.
Notably, CBP has been described to play a role in both synergy and repression between transcription factors. CBP appears to be required for synergy between NF-κB and STAT1 on the CXCL10 gene (12), while several studies describe a role for CBP, or the related factor p300, in mutual transrepression between ER and NF-κB (20, 41, 50). These studies suggest that transrepression may occur through (i) a competition between ER and NF-κB for the limited amounts of cellular CBP or p300 or (ii) ER causing displacement of CBP from NF-κB target gene promoters (41, 50). One study suggested that a repressive complex involving all three players may occur. However, this was not supported by a mammalian two-hybrid study that demonstrated a synergistic action between ER, NF-κB, and CBP (20). In the case of BIRC3 regulation, we propose that CBP plays a synergistic role between ER and NF-κB, potentially because of the nature and arrangement of the ER and NF-κB binding sites. Loss of CBP is correlated with reduced histone acetylation and ER binding, suggesting that the function of CBP is to assist ER in accessing the ERE, potentially through increased histone acetylation. Alternatively, CBP could function to acetylate nonhistones, such as ER and p65. These posttranslational modifications are associated with increased DNA binding and activity of both transcription factors (9, 29) and therefore require further investigation. A third possibility is that CBP may play a scaffolding role, allowing formation of a more stable complex, consisting of ER, p65, and other HATs, which cause acetylation of histones around the ERE.
In conclusion, we find that ER and NF-κB cooperate with each other via adjacent response elements in the BIRC3 promoter and cause enhanced regulation of the gene by a novel mechanism. Our findings indicate that inflammatory factors can confer functionality upon the ERE of BIRC3 by recruiting HATs and inducing histone acetylation at this site. These epigenetic modifications appear necessary to allow ER recruitment to the site, which permits ER to work together with NF-κB to increase expression of BIRC3. This scenario may be clinically important for those ER-positive breast tumors that have concomitant inflammation. Estrogen and proinflammatory cytokines may thus act together to cause enhanced transcription of prosurvival factors, leading to tumor promotion and more aggressive tumors.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the National Institutes of Health grant CA130932-2 (J.F.).
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Adamson AD, et al. 2008. Human prolactin gene promoter regulation by estrogen: convergence with tumor necrosis factor-alpha signaling. Endocrinology 149: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bodine PV, Harris HA, Komm BS. 1999. Suppression of ligand-dependent estrogen receptor activity by bone-resorbing cytokines in human osteoblasts. Endocrinology 140: 2439–2451 [DOI] [PubMed] [Google Scholar]
- 3. Brower-Toland B, et al. 2005. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346: 135–146 [DOI] [PubMed] [Google Scholar]
- 4. Cakouros D, et al. 2001. A NF-kappa B/Sp1 region is essential for chromatin remodeling and correct transcription of a human granulocyte-macrophage colony-stimulating factor transgene. J. Immunol. 167: 302–310 [DOI] [PubMed] [Google Scholar]
- 5. Carroll JS, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- 6. Carroll JS, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- 7. Cerciat M, Unkila M, Garcia-Segura LM, Arevalo MA. 2010. Selective estrogen receptor modulators decrease the production of interleukin-6 and interferon-gamma-inducible protein-10 by astrocytes exposed to inflammatory challenge in vitro. Glia 58: 93–102 [DOI] [PubMed] [Google Scholar]
- 8. Chang EC, Frasor J, Komm B, Katzenellenbogen BS. 2006. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology 147: 4831–4842 [DOI] [PubMed] [Google Scholar]
- 9. Chen LF, Mu Y, Greene WC. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21: 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu S, Nishi Y, Yanase T, Nawata H, Fuller PJ. 2004. Transrepression of estrogen receptor beta signaling by nuclear factor-kappab in ovarian granulosa cells. Mol. Endocrinol. 18: 1919–1928 [DOI] [PubMed] [Google Scholar]
- 11. Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- 12. Clarke DL, et al. 2010. TNFalpha and IFNgamma synergistically enhance transcriptional activation of CXCL10 in human airway smooth muscle cells via STAT-1, NF-kappaB, and the transcriptional coactivator CREB-binding protein. J. Biol. Chem. 285: 29101–29110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cvoro A, et al. 2006. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol. Cell 21: 555–564 [DOI] [PubMed] [Google Scholar]
- 14. Feldman I, Feldman GM, Mobarak C, Dunkelberg JC, Leslie KK. 2007. Identification of proteins within the nuclear factor-kappa B transcriptional complex including estrogen receptor-alpha. Am. J. Obstet. Gynecol. 196: 394.e1–394.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frasor J, et al. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144: 4562–4574 [DOI] [PubMed] [Google Scholar]
- 16. Frasor J, et al. 2009. Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer. Cancer Res. 69: 8918–8925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frasor J, Weaver AE, Pradhan M, Mehta K. 2008. Synergistic up-regulation of prostaglandin E synthase expression in breast cancer cells by 17beta-estradiol and proinflammatory cytokines. Endocrinology 149: 6272–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. 2004. Anatomy of the estrogen response element. Trends Endocrinol. Metab. 15: 73–78 [DOI] [PubMed] [Google Scholar]
- 19. Hah N, et al. 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harnish DC, Scicchitano MS, Adelman SJ, Lyttle CR, Karathanasis SK. 2000. The role of CBP in estrogen receptor cross-talk with nuclear factor-kappaB in HepG2 cells. Endocrinology 141: 3403–3411 [DOI] [PubMed] [Google Scholar]
- 21. Heldring N, et al. 2011. Multiple sequence-specific DNA-binding proteins mediate estrogen receptor signaling through a tethering pathway. Mol. Endocrinol. 25: 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holloway AF, Rao S, Chen X, Shannon MF. 2003. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J. Exp. Med. 197: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holloway JN, Murthy S, El-Ashry D. 2004. A cytoplasmic substrate of mitogen-activated protein kinase is responsible for estrogen receptor-alpha down-regulation in breast cancer cells: the role of nuclear factor-kappaB. Mol. Endocrinol. 18: 1396–1410 [DOI] [PubMed] [Google Scholar]
- 24. Hong SY, et al. 2000. Involvement of two NF-kappa B binding elements in tumor necrosis factor alpha-, CD40-, and Epstein-Barr virus latent membrane protein 1-mediated induction of the cellular inhibitor of apoptosis protein 2 gene. J. Biol. Chem. 275: 18022–18028 [DOI] [PubMed] [Google Scholar]
- 25. Hua B, et al. 2006. A splice variant of stress response gene ATF3 counteracts NF-kappaB-dependent anti-apoptosis through inhibiting recruitment of CREB-binding protein/p300 coactivator. J. Biol. Chem. 281: 1620–1629 [DOI] [PubMed] [Google Scholar]
- 26. Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jakacka M, et al. 2002. An estrogen receptor (ER)alpha deoxyribonucleic acid-binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol. Endocrinol. 16: 2188–2201 [DOI] [PubMed] [Google Scholar]
- 28. Kalaitzidis D, Gilmore TD. 2005. Transcription factor cross-talk: the estrogen receptor and NF-kappaB. Trends Endocrinol. Metab. 16: 46–52 [DOI] [PubMed] [Google Scholar]
- 29. Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. 2006. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol. Endocrinol. 20: 1479–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. King AE, et al. 2010. An additive interaction between the NFkappaB and estrogen receptor signalling pathways in human endometrial epithelial cells. Hum. Reprod. 25: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kininis M, et al. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 27: 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. 2001. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor alpha. Mol. Cell Endocrinol. 174: 151–166 [DOI] [PubMed] [Google Scholar]
- 33. Kushner PJ, et al. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74: 311–317 [DOI] [PubMed] [Google Scholar]
- 34. Lalmansingh AS, Uht RM. 2008. Estradiol regulates corticotropin-releasing hormone gene (crh) expression in a rapid and phasic manner that parallels estrogen receptor-alpha and -beta recruitment to a 3′,5′-cyclic adenosine 5′-monophosphate regulatory region of the proximal crh promoter. Endocrinology 149: 346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin CY, et al. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mahmoodzadeh S, et al. 2009. Nuclear factor-kappaB regulates estrogen receptor-alpha transcription in the human heart. J. Biol. Chem. 284: 24705–24714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mao C, et al. 2008. A new small molecule inhibitor of estrogen receptor alpha binding to estrogen response elements blocks estrogen-dependent growth of cancer cells. J. Biol. Chem. 283: 12819–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mason CE, et al. 2010. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 38: 2355–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDevitt MA, et al. 2008. New insights into the classical and non-classical actions of estrogen: evidence from estrogen receptor knock-out and knock-in mice. Mol. Cell Endocrinol. 290: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ndlovu N, et al. 2009. Hyperactivated NF-κB and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol. Cell. Biol. 29: 5488–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nettles KW, et al. 2008. CBP is a dosage-dependent regulator of nuclear factor-kappaB suppression by the estrogen receptor. Mol. Endocrinol. 22: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nissen LJ, Gelly JC, Hipskind RA. 2001. Induction-independent recruitment of CREB-binding protein to the c-fos serum response element through interactions between the bromodomain and Elk-1. J. Biol. Chem. 276: 5213–5221 [DOI] [PubMed] [Google Scholar]
- 43. Pradhan M, Bembinster LA, Baumgarten SC, Frasor J. 2010. Proinflammatory cytokines enhance estrogen-dependent expression of the multidrug transporter gene ABCG2 through estrogen receptor and NFκB cooperativity at adjacent response elements. J. Biol. Chem. 285: 31100–31106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quandt K, Frech K, Karas H, Wingender E, Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rubio MF, et al. 2006. TNF-alpha enhances estrogen-induced cell proliferation of estrogen-dependent breast tumor cells through a complex containing nuclear factor-kappa B. Oncogene 25: 1367–1377 [DOI] [PubMed] [Google Scholar]
- 46. Ryser S, Massiha A, Piuz I, Schlegel W. 2004. Stimulated initiation of mitogen-activated protein kinase phosphatase-1 (MKP-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem. J. 378: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Safe S, Kim K. 2008. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J. Mol. Endocrinol. 41: 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sorlie T, et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sorlie T, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Speir E, Yu ZX, Takeda K, Ferrans VJ, Cannon RO., III 2000. Competition for p300 regulates transcription by estrogen receptors and nuclear factor-kappaB in human coronary smooth muscle cells. Circ. Res. 87: 1006–1011 [DOI] [PubMed] [Google Scholar]
- 51. Stanculescu A, et al. 2010. Estrogen promotes breast cancer cell survival in an inhibitor of apoptosis (IAP)-dependent manner. Horm. Cancer 1: 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vega VB, et al. 2006. Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol. 7: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wissink S, van der Burg B, Katzenellenbogen BS, van der Saag PT. 2001. Synergistic activation of the serotonin-1A receptor by nuclear factor-kappa B and estrogen. Mol. Endocrinol. 15:543–552 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.