Abstract
Objective
Connexin40 (Cx40) is a gap junction protein expressed specifically in developing and mature atrial myocytes and cells of the conduction system. In this report, we identify cis-acting elements within the mouse Cx40 promoter and unravel part of the complex pathways involved in the cardiac expression of this gene.
Methods
To identify the factors involved in the cardiac expression of Cx40, we used transient transfections in mammalian cells coupled with electrophoretic mobility shift assays (EMSA) and RT-PCR.
Results
Within the promoter region, we identified the minimal elements required for transcriptional activity within 150 base pairs (bp) upstream of the transcriptional start site. Several putative regulatory sites for transcription factors were predicted within this region by computer analysis, and we demonstrated that the nuclear factors Sp1, Nkx2-5, GATA4 and Tbx5 could interact specifically with elements present in the minimal promoter region of the Cx40. Furthermore, co-transfection experiments showed the ability of Nkx2-5 and GATA4 to transactivate the minimal Cx40 promoter while Tbx5 repressed Nkx2-5/GATA4-mediated activation. Mutagenesis of the Nkx2-5 core site in the Cx40 promoter led to significantly decreased activity in rat smooth muscle cell line A7r5. Consistent with this, mouse embryos lacking Nkx2-5 showed a marked decrease in Cx40 expression.
Conclusion
In this work, we cloned the promoter region of the Cx40 and demonstrated that the core promoter was modulated by cardiac transcriptional factors Nkx2-5, Tbx5 and GATA4 acting together with ubiquitous Sp1.
Keywords: Gap junctions, Cx40, Nkx2-5, GATA4, Tbx5, Muscle gene regulation
1. Introduction
Connexin40 (Cx40, Gja5) is a member of a multigene family that encodes for at least 20 distinct proteins in mice and humans [1]. These proteins are structural subunits of gap junctions, intercellular channels that provide a pathway between cells for the exchange of ions, metabolites, second messengers and small molecules up to 1 kDa. Several roles have been attributed to this form of cellular communication including the regulation of early events during development, cell differentiation, cell growth and proliferation [2,3]. Gap junctions also play an important role in excitable tissues such as the myocardium mediating impulse propagation and, thereby, coordinating cardiac contraction [4].
In the adult heart, gap junctional channels are essential for the action potential propagation from the sinoatrial node to the working myocardium. Because fibrous tissue electrically isolates atria from ventricles, the electrical impulse preferentially uses a route known as the cardiac conduction system. The most abundantly expressed connexin in the mammalian heart is Cx43, but Cx40 is the major isoform present in the conduction system [5]. Cx40 expression is generally restricted to the conduction system and atria in adult hearts and is under strict temporal and spatial regulation during heart development [5]. Hypertension in rats leading to cardiac hypertrophy causes a three-fold increase in Cx40 expression with an increased tendency to develop arrhythmias that are associated with sudden death [6]. Additionally, Cx40 deficient mice display pronounced sinoatrial and atrioventricular conduction disturbances with increased atrial vulnerability to arrhythmogenesis [7] and display high incidence of cardiac malformations [8]. Recently, it was shown that, like many other cardiac specific genes, Cx40 expression requires highly regulated combinatorial interactions between cardiac specific and ubiquitous factors [9–11]. These findings suggest that quantitative or qualitative changes in Cx40 expression in the heart may be related to pathological conditions.
The structure of the 5′-flanking region of both mouse and rat Cx40 genes have been previously reported [12,13]. The basal promoter of the rat Cx40 gene was identified in the proximal region from −175 to +84 relative to the transcriptional initiation site. Further deletion analysis suggested critical regulatory elements were localized within two regions (−96 to −71 and +53 to +84) containing GC boxes that could interact with transcriptional factors Sp1/Sp3. Mutagenesis of these regions indicated that they were essential for basal activity of the rat Cx40 promoter [14]. By contrast, the basal promoter of the mouse Cx40 was located within 300 base pairs (bp) upstream of the transcription start site. Additionally, a nearby region between +100 to +297 contained a strong negative regulatory element [12]. In the present study, we define cis-acting elements involved in the transcriptional control of the mouse Cx40 gene and show that several cardiac and muscle specific transcription factors such as the homeobox gene Nkx2-5, GATA4 and Tbx5, are required for its regulation, providing important insights into the mechanisms driving the complex expression pattern of Cx40 during cardiogenesis.
2. Methods
The investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85-23, revised 1996).
2.1. Cloning and sequencing of the 5′-flanking region of the Cx40 gene
A 2.4 kilobase pair (kb) fragment of the 5′-flanking region of the Cx40 gene was amplified by polymerase chain reaction (PCR) using the Genome Walker Kit (Clontech) and specific primers 12AS and 8AS. The fragment was cloned, sequenced and deposited at GenBank under accession number AF375853. All primers used in this work are described in Supplementary Table 1.
2.2. Cx40 promoter-luciferase constructs
The following plasmids used in this study were previously described elsewhere [12]: −1190/+121Cx40Luc; −889/+121Cx40Luc and −558/+121Cx40Luc. The construct −203/+121Cx40Luc was obtained using Double Strand Nested deletion Kit (Amersham Pharmacia Biotech), while constructs −150/+121Cx40Luc, −50/+121Cx40Luc and −20/+121Cx40Luc were generated by PCR and cloned into the pGL3-Basic (Promega). Site directed mutagenesis of the putative Nkx2-5 site (NKE) was performed using QuickChange Site-Direct Mutagenesis Kit (Stratagene) and confirmed by sequencing.
2.3. Cell culture and transient transfections
The embryonic rat smooth muscle cell line derived from the thoracic aorta, A7r5, COS7 and CH310T1/2 fibroblasts were obtained from American Type Culture Collection (Rockville, MD). Rat neonatal cardiomyocytes were isolated as previously described [15]. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM-GIBCO) supplemented with 10% (vol/vol) fetal bovine serum (GIBCO) and maintained in a humidified 10% CO2 incubator at 37 °C. 10T1/2 cells were transfected as previously described [12]. A7r5 cells were seeded into 24-well plates to reach 70% confluence by the next day and transiently transfected using Lipofectamine 2000 (Invitrogen) with 0.5 μg of each test plasmid and 0.1 μg of the control pRL-CMV (Promega), carrying the Renilla luciferase gene. Cells were harvested after 48 h. The results were expressed as fold activity after normalization for Renilla or β-Galactosidase activity. Transfections were performed in triplicate in at least three independent experiments.
2.4. Preparation of nuclear extracts and GST fusion proteins
Nuclear extracts were prepared 48 h after transfection; 100 mm2 plates were homogenized in 300 μl of buffer I (10 mM NaCl; 20 mM Hepes pH 7.4; 1.5 mM MgCl2; 0.1% v/v TritonX-100; 20% v/v Glycerol; 1 mM DTT; 0.5 mM PMSF), chilled on ice for 10 min, and precipitated by centrifugation at 2800×g for 1 min at 4 °C. Nuclear fraction was resuspended in 300 μl of buffer C (150 mM NaCl; 50 mM Hepes pH 7.4; 1.5 mM MgCl2; 1 mM EGTA; 10 mM Na pyrophosphate; 1% v/v Triton X-100; 10% v/v glycerol; 1 mM DTT; 0.5 mM PMSF) plus 41.5 μl of 5 M NaCl and incubated for 1 h at 4 °C. The nuclear extracts were then centrifuged at 14,000×g for 10 min and stored at −70 °C. Recombinant Tbx5 or the Nkx2-5HD proteins were purified on glutathione-Sepharose columns (Amersham Pharmacia Biotech) and eluted by cleavage with 5 U/ml of thrombin (Amersham Pharmacia Biotech) generating GST-free proteins used in the EMSA. Protein concentration was determined by Bradford method (BIORAD).
2.5. Electrophoretic mobility shift assay (EMSA)
Five to ten micrograms of nuclear extracts were mixed with binding buffer (25 mM Hepes pH 7.4; 75 mM NaCl; 1 mM MgCl2; 0.2 mM EDTA; 0.1% v/v NP-40; 1 mM DTT; 10 ng BSA; 200 ng poly dI/dC) and incubated for 10 min at room temperature. Annealed oligonucleotides (5 pM) were labeled using [γ32P] ATP and T4 polynucleotide kinase (NEB); 50,000 cpm were added to each sample and incubated at room temperature for 20 min; 10 μl of the mixture was submitted to electrophoresis on a 5% PAGE gel 0.5× TBE or 1× Tris Glycine gels, dried and subjected to autoradiography. Supershift assays were performed as described above except that 0.5–2 μg of antibody against Sp1 (Sigma) or GATA4 C20 (Santa Cruz Biotechnology) were added to the reaction for 10 min before incubation with the 32P-labeled oligonucleotide.
2.6. RNA extraction and RT-PCR
RNA was extracted from hearts of wild-type or homozygous knockout Nkx2-5 mice littermates at 9 days post coitum (dpc) stage using TRIZOL method (Invitrogen), treated with RQ1 RNase-free Dnase and reverse transcribed using M-MuLV transcriptase with oligo-dT primers. Samples were amplified using specific primers. PCR conditions consisted of 40 cycles at 94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min using 5 U of Taq polymerase (Merck).
3. Results
3.1. Transcriptional activity of the Cx40 promoter
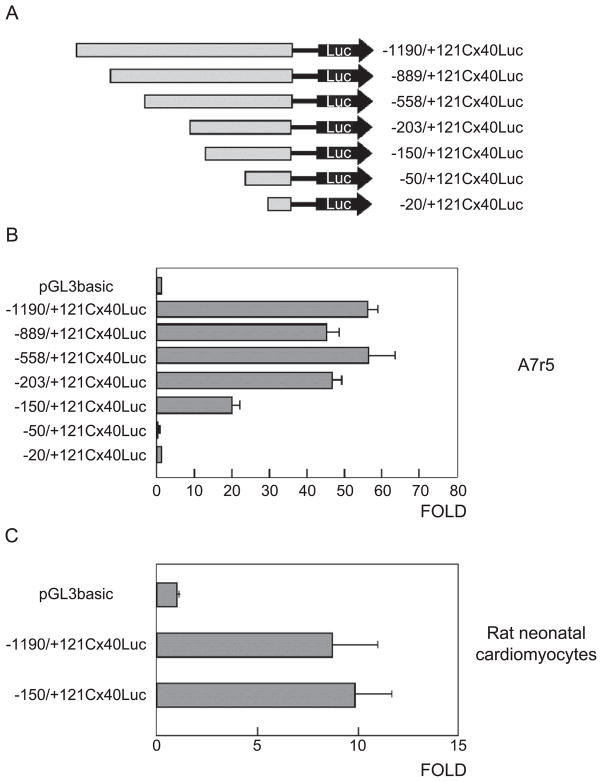
To identify cis-acting regulatory elements of the Cx40 gene, a 2.4 kb DNA fragment was amplified from a mouse genomic library and several deletions, ranging from −1190 to −20 relative to the transcription start site were linked to a luciferase reporter gene (Fig. 1A). The resulting series of constructs were transiently transfected into A7r5 cells that express high levels of Cx40 [16] and several other cardiac connexins, including Cx43 and Cx45 (Supplementary Fig. S1). An increase of 55 fold, above control plasmid, was observed for the −1190/+121Cx40Luc construct in A7r5 (Fig. 1A). Similar levels of activity were observed for 203/+121Cx40Luc (Fig. 1B), suggesting that the sequences between −203 and +121 contain important regulatory modules controlling expression of Cx40. Removal of an additional 50 bp from this construct reduced the activity of the −150/+121Cx40Luc plasmid to only 28% of the activity observed for the −1190/+121Cx40Luc construct and further deletions of 100 bp led to the complete silencing of the reporter gene. These data indicate that in A7r5 cells maximal activity for this promoter requires only the first 203 bp of the promoter and further that a basal promoter is found within the first 150 bp upstream of the transcriptional start site of this gene. Importantly, the −150/+121Cx40Luc construct displays equal activity to −1190/+121Cx40Luc when transfected into embryonic primary cardiomyocytes (Fig. 1C).
Fig. 1.
Deletion analysis of the mouse Cx40 promoter in A7r5 cells and rat neonatal myocytes. (A) Schematic diagram of the 5′ deletions of the Cx40 promoter used for transient transfections. (B) A fragment of approximately 1.2 kb upstream of the transcription start site displays 55-fold activity over pGL3basic. This activity was maintained until 203 and decreases to 20-fold upon deletion of 53 base pairs. Thus, the minimum elements required for Cx40 expression in this cell line were located in the first 150 bp upstream of the transcription start site (C). Constructs −1190/+121Cx40Luc and the minimal promoter −150/+121Cx40Luc display approximately 10-fold activity in rat neonatal myocytes. Data was normalized for transfection efficiency by co-transfection with renilla luciferase vector (pRL-CMV). The increase in luciferase activity (FOLD) was expressed relative to the reporter vector alone (pGL3basic).
3.2. Nuclear factor Sp1 binds specifically within the proximal region of the Cx40 promoter
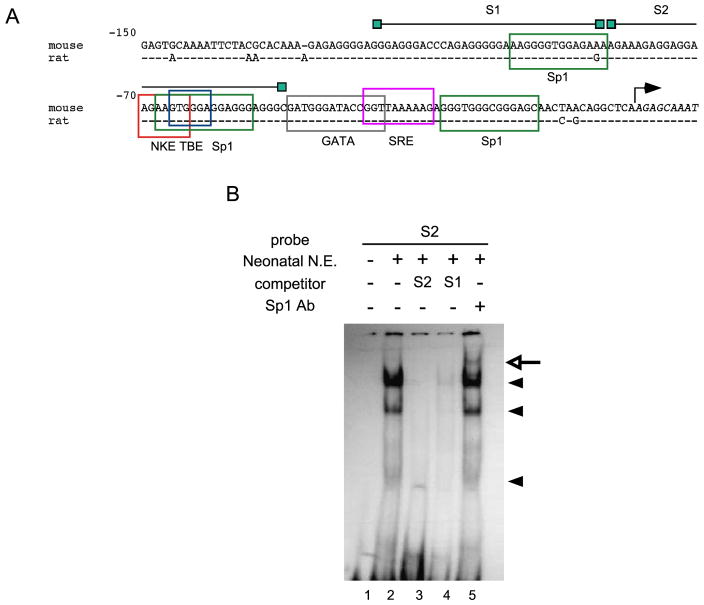
Our transfection data indicates that the elements required for the basal activity of the Cx40 promoter are located between −150 and −50 relative to the transcriptional start site. Within this region, several putative binding sites for transcription factors were predicted by computer analysis. To validate these sites functionally, EMSAs were performed using the identified sequences as probes (Fig. 2A).
Fig. 2.
Electrophoretic mobility shift analysis of the proximal Cx40 promoter: identification of an Sp1 site in the mouse Cx40 promoter. (A) Upstream sequence of the Cx40 promoter region is highly conserved between mouse and rat. Important sequences within this region are boxed. Oligonucleotides used for EMSA are represented in the upper part of the picture. (B) Three specific bands (arrowheads) were detected in neonatal mouse heart nuclear extracts that were supershifted with increased amounts of added Sp1 antibody (arrow).
Initial experiments investigated the binding site for the Sp1/Sp3 family members since a highly homologous region of the rat Cx40 promoter (Fig. 2A) was recently shown capable of specific interaction with members of the Sp1/Sp3 proteins [14]. Specific bands were identified in EMSAs of cardiomyocyte and A7r5 nuclear extracts (Fig. 2B and data not shown). Furthermore, addition of an antibody to Sp1 resulted in the appearance of a slower migrating band (Fig. 2B, arrow), confirming that these complexes contained Sp1. Thus, functional Sp1 sites are found in the promoters of both the rat and mouse Cx40.
Since Sp1 and Sp3 are ubiquitous transcriptional factors, these results raised an important question regarding Cx40 expression control: what are the unique proteins that regulate the cell-specific expression of Cx40? The role of the ubiquitous Sp1/Sp3 factors in the transcriptional activation of Cx40 appears to be related to stimulation of basal activity, indicating that additional interactions are necessary for correct transcriptional activation of the Cx40 gene in muscle cells.
3.3. Cardiac transcription factors Nkx2-5 and GATA4 confer specific Cx40 promoter transcriptional activation
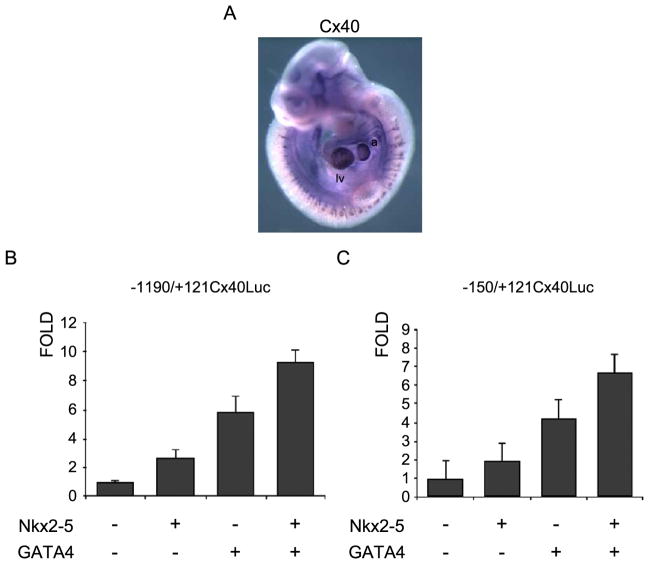
The proximal promoter of Cx40 has putative DNA-binding sites for two important cardiac specific transcriptional factors, Nkx2-5 and GATA4. Since these factors are both capable of supporting synergistic activation [15,17] and are present in the heart during the onset of Cx40 expression (Fig. 3A), we decided to investigate whether they could influence Cx40 promoter activity. The co-expression of either Nkx2-5 or GATA4 induced an approximate three- to six-fold increase in luciferase activity from −1190/+121Cx40Luc (Fig. 3B). Addition of both factors together led to a 10-fold increase in activation. The same effect was observed when the minimal promoter −150/+121Cx40Luc construct was used (Fig. 3C), indicating that this region contained the sites required for activation by both Nkx2-5 and GATA4 transcription factors.
Fig. 3.
Cardiac specific factors Nkx2-5 and GATA4 regulate Cx40 promoter in 10T1/2 cells. (A) In situ hybridization of 10.5 dpc embryos showing expression of Cx40 in the heart. Cx40 is also expressed in this developing stage in the vasculature, where it seems to be associated with smooth muscle and endothelial expression. a: atrium; lv: left ventricle. (B) Activation of the −1190/+121Cx40Luc construct or (C) the minimal promoter region −150/+121Cx40Luc by cardiac factors Nkx2-5, GATA4 or a combination of both factors.
3.4. Nkx2-5 and GATA4 are both capable of specific interaction within the minimal promoter of Cx40
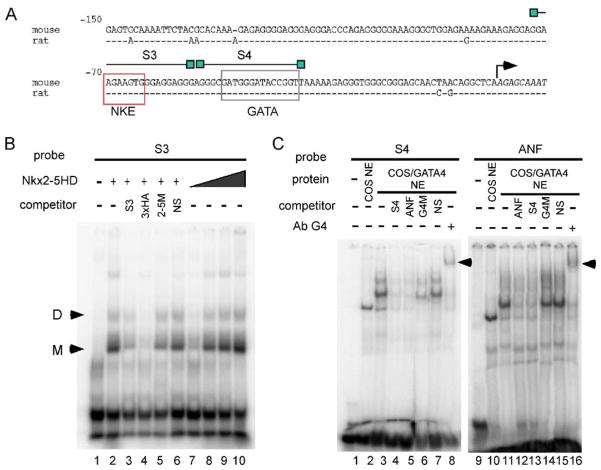
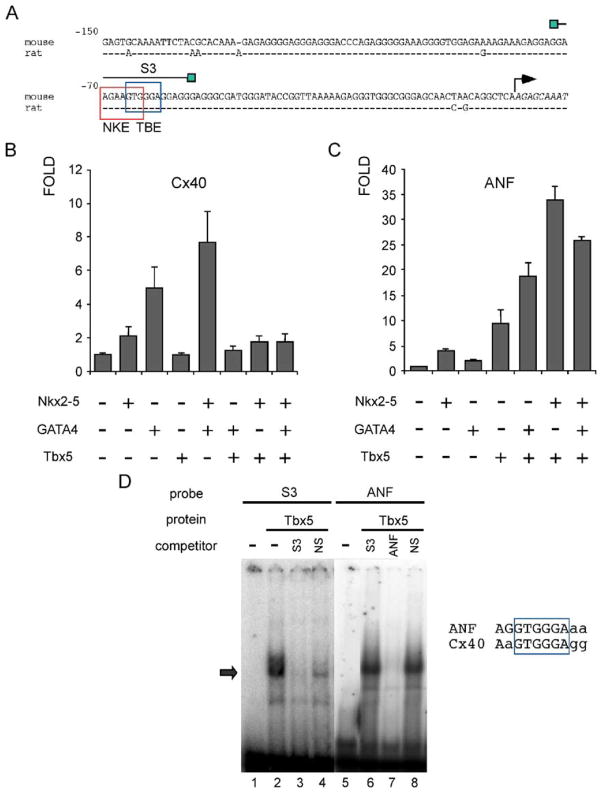
The minimal promoter contains putative sites for interaction with NK and GATA family members. To test if the cardiac factors Nkx2-5 and GATA4 could interact with these sites, EMSA was performed. The homeodomain of Nkx2-5 was capable of specific interaction with its putative site within the Cx40 proximal promoter (Fig. 4B, lanes 7–10) in a dose dependent manner. Competition experiments with an excess of unlabeled self-oligonucleotide competitor or the high affinity multimerized site for Nkx2-5, 3xHA or control oligonucleotides (NKE mutant or nonspecific oligonucleotide, Fig. 4C, lanes 2–6) demonstrated that the observed mobility shift was due to binding of the Nkx2-5 homeodomain to the corresponding recognition site in the Cx40 promoter. GATA4 was also able to interact specifically with its putative site, as indicated by the competition with self-oligonucleotide or its closely related site in the ANF promoter (Fig. 4C, lanes 3–5), and this complex was supershifted with GATA4 antibodies (Fig. 4C, lane 8).
Fig. 4.
Nkx2-5 and GATA4 are capable of specific interaction with proximal elements within the Cx40 promoter. (A) Cx40 proximal promoter sequence with NKE and GATA putative sites boxed and oligonucleotides used. (B) GST purified Nkx2-5HD interacts specifically with its putative site in S3. (C) GATA4 overexpressed in nuclear extracts of COS7 cells interacts with its proximal site in S4. The complex was supershifted by the addition of GATA4 antibody.
3.5. Tbx5 has a repressive role on the Cx40 promoter in 10T1/2 cells
T-box transcription factors are important regulators of early cell fate decision in the embryo. One member of the family, Tbx5, is expressed at high levels in the embryonic heart and is an essential component of the myogenic program. Mouse homozygous for a null Tbx5 allele die at mid gestation with severe cardiac defects, and haploinsufficiency in humans results in the autosomal dominant disorder, Holt Oram Syndrome [18].
The presence of a putative site for T-box factors that overlapped the NKE site within the proximal Cx40 promoter led us to investigate if Tbx5 could also be involved in Cx40 regulation. Tbx5 was unable to activate the Cx40 minimal promoter construct in transient transfections of 10T1/2 cells. Interestingly, however, Tbx5 strongly inhibited activation of the proximal promoter mediated by Nkx2-5/GATA4 complex (Fig. 5B). This was specific for the Cx40 promoter since Tbx5 elicited a strong activation of the ANF promoter and displayed synergism with Nkx2-5 and GATA4 or either factor alone (Fig. 5C). Despite these differences, Tbx5 was capable of specific interaction with both Cx40 and ANF proximal sites as detected by EMSA (Fig. 5D).
Fig. 5.
Tbx5 has a repression role in the Nkx2-5/GATA4 activation of the Cx40 promoter. (A) Cx40 proximal promoter sequence with overlapping NKE and TBE putative sites boxed and oligonucleotide S3 used. (B) Tbx5 expression was able to repress Nkx2-5/GATA4 activation observed for the Cx40 minimal promoter in 10T1/2 cells. (C) On the ANF promoter, the same complex of factors displayed synergistic activation. (D) EMSA showing that Tbx5 could interact with either Cx40 or ANF promoters (arrow).
3.6. Mutation of proximal NKE site impairs Cx40 expression in A7r5
To confirm the role of Nkx2-5 in the regulation of Cx40 promoter, the NKE site present in the minimal promoter −150/+121Cx40Luc was mutated (Fig. 6A) and the ability of this factor to activate transcription assessed in A7r5 cells. When this mutated region was used as competitor oligonucleotide, there was no displacement of Nkx2-5 binding from its site on the proximal promoter (Fig. 4B, lane 5). A significant decrease in promoter activity around 50% was observed when the mutant promoter was assayed in A7r5. Co-transfection of Nkx2-5 in A7r5 cells expressing the minimal promoter region led to a significant increase in promoter activity that was also abolished when the NKE mutated promoter was used (Fig. 6). Together, this data suggests that binding of Nkx2-5 is critical for normal expression of Cx40.
Fig. 6.
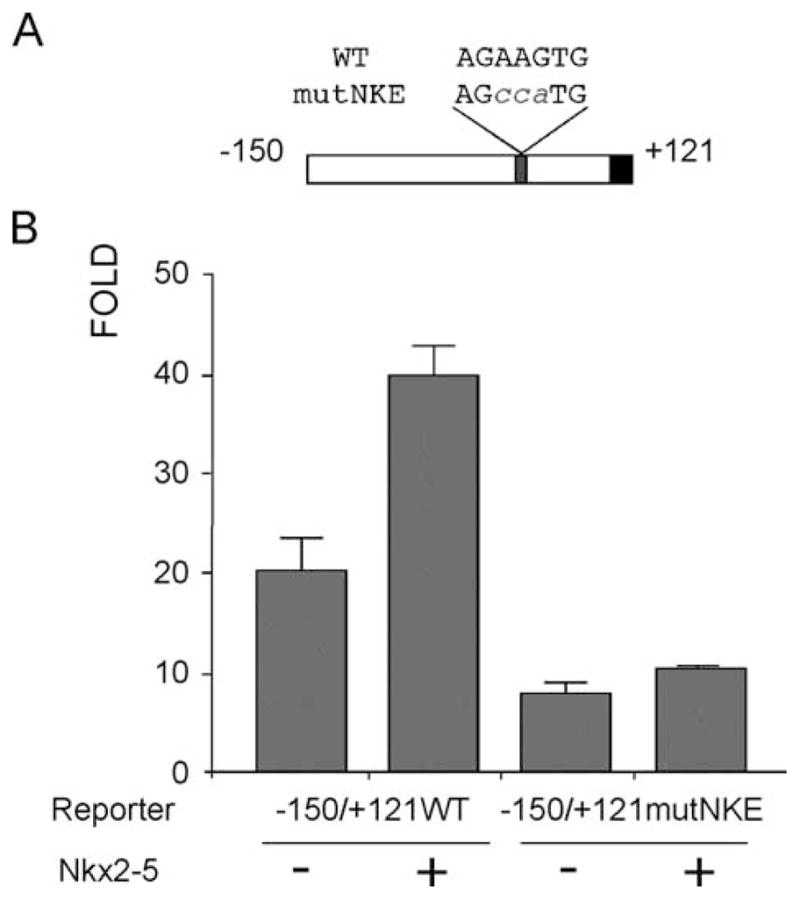
Nkx2-5 proximal site is essential for expression of the minimal promoter in A7r5 cells. (A) Schematic representation of the minimal Cx40 promoter showing NKE wild-type and mutated sequences. (B) Mutated promoter reporter construct (−150/+121mutNKE) transfected in A7r5 cells decreases basal activity by approximately 50% compared to the wild-type construct (−150/+121WT). Co-expression of Nkx2-5 activates the wild-type promoter by two-fold but has no significant effect on the mutant Cx40 promoter.
3.7. Cx40 expression is downregulated in vivo in Nkx2-5 knockout mice
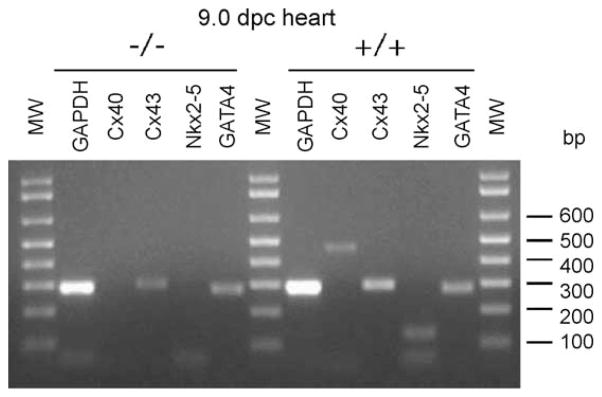
Overexpression of a dominant negative isoform of Nkx2-5 in the heart of transgenic mice leads to severe atrioventricular conduction defects that were correlated with decreased Cx40 expression [10]. To further confirm the modulation of Cx40 expression by Nkx2-5, we analyzed the levels of Cx40 mRNA in 9 dpc hearts of Nkx2-5 knockouts by RT-PCR. Indeed, Cx40 expression was markedly decreased in the knockout animals when compared with wild-type littermates (Fig. 7). Interestingly, Cx43 mRNA was also downregulated in the knockout embryos, in agreement with previous results indicating that both connexin genes might be regulated in cardiac tissues by Nkx2-5 [10].
Fig. 7.
Cx40 expression is significantly decreased in Nkx2-5 knockouts. RT-PCRs from total mRNA extracted from 9.0 dpc hearts from littermates that were homozygous for the mutant allele (−/−) or wild type (+/+) display a marked decreased expression of mRNA for Cx40 while showing no effect on GATA4 levels.
Together these data support a role for the cardiac transcriptional factors Nkx2-5 and GATA4 in the regulation of Cx40 expression.
4. Discussion
Like other tissue-specific genes, Connexin40 is under strict transcriptional regulation during development. In mice, Cx40 expression is initiated at early 9 dpc, subsequently being detected in the primitive atrial and ventricular chambers of the developing heart [5], as well as in smooth muscle and endothelial cells of the early circulatory system and the conduction system [19]. In the adult mouse heart, Cx40 mRNA is also still detected in the atria [5]. Through deletion analysis we have identified several important regions that lie within 2.4 kb of the start codon of Cx40. Furthermore, we have shown that, as for the rat Cx40 promoter, sequences proximal (within 150 base pairs) to the initiator codon contain key regulatory modules of the Cx40 promoter. This proximal region contains several putative sites for transcription factors. Recently, it has been demonstrated that at least one of these sites interacts with the factor Sp1 and that this interaction is also essential for basal Cx40 expression in the rat [14]. Using nuclear extracts derived from A7r5 cells and neonatal hearts, our work showed that Sp1 is capable of specific interaction with regions of the proximal mouse Cx40 promoter. Although functionally important, the ubiquitous expression of Sp1 suggests the requirement of other factors for the generation of the expression pattern observed. For instance, Sp1 is expressed at high levels in HeLa cells that do not express either Cx40 mRNA or protein, suggesting the existence of additional mechanisms for the tissue specific activation or repression of this promoter. Sp1 can also physically interact with several transcriptional factors including GATA1, Nkx2.1, MEF2C and SRF [20], thereby generating specificity at the transcriptional level. The presence of putative binding sites for Nkx, GATA, T-box and SRF factors in the proximal region prompted us to investigate the potential role of these factors in regulating Cx40 expression.
The transcription factors Nkx2-5 and GATA4 are essential for early cardiac differentiation and specification [21,22]. Recently, both factors were also associated with inherited congenital heart diseases, including septal defects, thus confirming its importance during normal heart development [23–25].
Nkx2-5 and GATA4 have both been shown to control the transcriptional regulation of several cardiac and smooth muscle specific genes including ANF and A1AR [15,17]. Several other binding partners have also been implicated in regulating these cardiac transcriptional complexes, including Tbx5 [9], SRF [26] and Myocardin [27] and the formation of a high order complex comprised of Nkx2-5, GATA4 and SRF seems essential for the correct expression of cardiac α-actin [20]. Our results further indicate a primary role for the interplay of these factors in regulating expression of Cx40 in the heart. In fact, Purkinje fibers of the conduction system have been shown to express high levels of both Nkx2-5 and GATA4 [28,29].
Several other families of cardiac transcriptional factors are now known to be involved in the formation of the outer-curvature of the heart that initially forms the working myocardium from which the peripheral conduction system later differentiate [30]. Members of the T-box family have been shown to cooperate with other transcription factors in the regulation of many cardiac genes [18,31,32]. Atrioventricular node defects have also been observed in humans and mice harboring mutations in Tbx5, one of the cardiac T-box genes, as well as Nkx2-5, thus confirming the role for these factors in the specification of the conduction system [29]. Other members of the T-box family also display important roles in cardiac specification. Tbx2 and Tbx3 are expressed in complementary fashion with Cx40 during cardiac development and have been shown to directly interact and repress Cx40 promoter in vitro and in vivo [33,34]. This repression was postulated to inhibit chamber differentiation allowing the formation and correct alignment of the inflow tract, atrioventricular canal (AVC) and outflow tract (OFT), regions that will generate the atrioventricular septum/heart valves and great vessels in the mature heart [33]. Tbx20, another member of the T-box family is expressed in the same region as Cx40 in early development and can interact directly with the Cx40 promoter regulating its transcription. Interestingly, different isoforms of Tbx20 are capable of transcriptional activation or repression depending on interactions with cardiac factors Nkx2-5 and GATA4 [11,35]. Recently, it was shown that Tbx5 interacts with Nkx2-5 and activates the ANF promoter [36] whereas the Tbx2/Nkx2-5 complex, in vivo, represses the same promoter in the AV canal during development [37]. Recent reports indicated that in some cases Tbx5 could bind and activate Cx40 expression together with Nkx2-5 when a 1 kb fragment of the Cx40 promoter was used [9], while we have described a strong repressive effect on the minimal promoter. Such differences might be related to the lack of important elements in the minimal promoter or a strong dependence on Nkx2-5 and Tbx5 levels for the regulation of Cx40 expression [29]. Thus, differences in the expression levels of these factors as well as complex interactions with other nuclear proteins are likely to play essential roles in coordinating the activation/repression of Cx40 in the developing heart.
We propose a model for Cx40 promoter regulation in which Sp1 interaction is necessary for basal promoter activity, while NK and GATA factors account for the observed tissue specificity. However, Cx40 expression is first detected in the developing mouse heart and dorsal aorta around 9 dpc [5], whereas Nkx2-5 and GATA4 are first detected in the precardiac mesoderm at an earlier developmental stage (around 7.0–7.5 dpc in mouse embryos) [38]. Thus other repressive factors, like Tbx2, could be active during early stages of cardiogenesis [33]. In smooth muscle cells, Cx40 expression might be under control of other related members of the NK and GATA families in the vasculature. In fact, Nkx3.2, GATA6 and SRF are detected in smooth muscle cells, cooperatively activate several smooth muscle specific promoters like the α1-integrin and SM22α and may play important roles in the regulation of the Cx40 expression on this domain [39]. Thus the same elements regulating Cx40 expression may interact with different family members of transcription factors in the smooth muscle of the vasculature and the myocardium, likely reflecting evolutionary changes that resulted in the divergence of these cell lineages. Differential spatial expression of GATA members may also be important for this specification [29,38]. In fact, other as yet uncharacterized factors must also be involved in this regulation, since Cx40 expression is later silenced in the adult ventricle, where Sp1, GATA and NK factors are still present.
Existing evidence suggests that the regulation of Cx40 is a complex and precise process reliant upon several different classes of transcription factors, both basal and tissue specific. The in vitro evidence presented here supports a role for Sp1, NK-2 homeodomains, GATA zinc finger proteins and T-box proteins in regulation of the proximal Cx40 promoter. However, several other putative regulatory regions have also been identified and remain to be studied. Continued characterization of the regulatory network and transcriptional factors involved in correct expression of Cx40 transcripts in cell lines and transgenic models should decipher the mechanisms required for correct timing and localization of this gene during development.
Supplementary Material
Acknowledgments
This work was supported by research grants from FAPERJ, PADCT and FUJB to MWC, PADCT and PRONEX to ACCC and NIH Grants HL45464 and HL59199 to ECB. ACCC is a senior investigator of CNPq. VLFL, NASA and DCM were supported by student fellowship from CNPq and CAPES. MWC is a recipient of postdoctoral fellowship from CAPES. We would like to thank Dr. Richard Harvey for intellectual inputs and financial support, Dr. Kyung H. Seul for the −1190/+121 Cx40 promoter construct and Dr. Fiona Stennard, Dr. Mark Solloway, Dr. Christine Biben and Milena B. Furtado for helpful discussions.
Abbreviations
- Cx40
connexin40
- bp
base pairs
- kb
kilobase pairs
- PCR
polymerase chain reaction
- EMSA
electrophoretic mobility shift assay
- dpc
days post coitum
- AVC
atrioventricular canal
- OFT
outflow tract
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.cardiores. 2004.09.021.
References
- 1.Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–32. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 2.White TW, Paul DL. Genetic diseases and gene knockouts reveal diverse connexin functions. Annu Rev Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- 3.Gros D, Dupays L, Alcolea S, Meysen S, Miquerol L, Theveniau-Ruissy M. Genetically modified mice: tools to decode the functions of connexins in the heart—new models for cardiovascular research. Cardiovasc Res. 2004;62:299–308. doi: 10.1016/j.cardiores.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Simon AM, Goodenough DA. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–83. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- 5.Delorme B, Dahl E, Jarry-Guichard T, Briand JP, Willecke K, Gros D, et al. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ Res. 1997;81:423–37. doi: 10.1161/01.res.81.3.423. [DOI] [PubMed] [Google Scholar]
- 6.Bastide B, Neyses L, Ganten D, Paul M, Willecke K, Traub O. Gap junction protein connexin40 is preferentially expressed in vascular endothelium and conductive bundles of rat myocardium and is increased under hypertensive conditions. Circ Res. 1993;73:1138–49. doi: 10.1161/01.res.73.6.1138. [DOI] [PubMed] [Google Scholar]
- 7.Verheule S, van Batenburg CA, Coenjaerts FE, Kirchhoff S, Willecke HJ, Jongsma HJ. Cardiac conduction abnormalities in mice lacking the gap junction protein connexin40. J Cardiovasc Electrophysiol. 1999;10:1380–9. doi: 10.1111/j.1540-8167.1999.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 8.Gu H, Smith FC, Taffet SM, Delmar M. High incidence of cardiac malformations in connexin40-deficient mice. Circ Res. 2003;93:201–6. doi: 10.1161/01.RES.0000084852.65396.70. [DOI] [PubMed] [Google Scholar]
- 9.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–21. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 10.Kasahara H, Wakimoto H, Liu M, Maguire CT, Converso KL, Shioi T, et al. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2. 5 homeoprotein. J Clin Invest. 2001;108:189–201. doi: 10.1172/JCI12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–24. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 12.Seul KH, Tadros PN, Beyer EC. Mouse connexin40: gene structure and promoter analysis. Genomics. 1997;46:120–6. doi: 10.1006/geno.1997.5025. [DOI] [PubMed] [Google Scholar]
- 13.Groenewegen WA, van Veen TA, van der Velden HM, Jongsma HJ. Genomic organization of the rat connexin40 gene: identical transcription start sites in heart and lung. Cardiovasc Res. 1998;38:463–71. doi: 10.1016/s0008-6363(97)00325-8. [DOI] [PubMed] [Google Scholar]
- 14.Bierhuizen MF, van Amersfoorth SC, Groenewegen WA, Vliex S, Jongsma HJ. Characterization of the rat connexin40 promoter: two Sp1/Sp3 binding sites contribute to transcriptional activation. Cardiovasc Res. 2000;46:511–22. doi: 10.1016/s0008-6363(00)00041-9. [DOI] [PubMed] [Google Scholar]
- 15.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–96. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyer EC, Reed KE, Westphale EM, Kanter HL, Larson DM. Molecular cloning and expression of rat connexin40, a gap junction protein expressed in vascular smooth muscle. J Membr Biol. 1992;127:69–76. doi: 10.1007/BF00232759. [DOI] [PubMed] [Google Scholar]
- 17.Rivkees SA, Chen M, Kulkarni J, Browne J, Zhao Z. Characterization of the murine A1 adenosine receptor promoter, potent regulation by GATA-4 and Nkx2.5. J Biol Chem. 1999;274:14204–9. doi: 10.1074/jbc.274.20.14204. [DOI] [PubMed] [Google Scholar]
- 18.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–9. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 19.Haefliger JA, Nicod P, Meda P. Contribution of connexins to the function of the vascular wall. Cardiovasc Res. 2004;62:345–56. doi: 10.1016/j.cardiores.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5 and serum response factor directs early cardiac gene activity. J Biol Chem. 2002;30:30. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 21.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2-5. Genes Dev. 1995;9:1654–66. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 22.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 23.Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res. 2000;87:888–95. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- 24.Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–6. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 25.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 26.Belaguli NS, Sepulveda JL, Nigam V, Charron F, Nemer M, Schwartz RJ. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol Cell Biol. 2000;20:7550–8. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–62. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 28.Thomas PS, Kasahara H, Edmonson AM, Izumo S, Yacoub MH, Barton PJ, et al. Elevated expression of Nkx-2. 5 in developing myocardial conduction cells. Anat Rec. 2001;263:307–13. doi: 10.1002/ar.1106. [DOI] [PubMed] [Google Scholar]
- 29.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–19. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- 30.Moorman AF, de Jong F, Denyn MM, Lamers WH. Development of the cardiac conduction system. Circ Res. 1998;82:629–44. doi: 10.1161/01.res.82.6.629. [DOI] [PubMed] [Google Scholar]
- 31.Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–8. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 32.Harvey RP. Patterning the vertebrate heart. Nat Rev, Genet. 2002;3:544–56. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- 33.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–70. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 34.Hoogaars WM, Tessari A, Moorman AF, de Boer PAJ, Hagoort J, Soufan AT, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–99. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Plageman TF, Jr, Yutzey KE. Differential expression and function of tbx5 and tbx20 in cardiac development. J Biol Chem. 2004;279:19026–34. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- 36.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–80. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 37.Habets PE, Moorman AF, Clout DE, Van Roon MA, Lingbeek M, Van Lohuizen M, et al. Cooperative action of Tbx2 and Nkx2. 5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–46. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JB. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–19. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- 39.Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, et al. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308–17. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.