Abstract
M1 protein contributes to Group A Streptococcus (GAS) systemic virulence by interfering with phagocytosis and through proinflammatory activities when released from the cell surface. Here we identify a novel role of M1 protein in the stimulation of neutrophil and mast cell extracellular trap formation, yet also subsequent survival of the pathogen within these DNA-based innate defense structures. Targeted mutagenesis and heterologous expression studies demonstrate M1 protein promotes resistance to the human cathelicidin antimicrobial peptide LL-37, an important effector of bacterial killing within such phagocyte extracellular traps. Studies with purified recombinant protein fragments mapped the inhibition of cathelicidin killing to the M1 hypervariable N-terminal domain. A survey of GAS clinical isolates found that strains from patients with necrotizing fasciitis or toxic shock syndrome were significantly more likely to be resistant to cathelicidin than GAS M types not associated with invasive disease; M1 isolates were uniformly resistant. We conclude increased resistance to host cathelicidin and killing within phagocyte extracellular traps contribute to the propensity of M1 GAS strains to produce invasive infections.
Key Words: Group A <italic>Streptococcus</italic>, <italic>Streptococcus pyogenes</italic>, Virulence factor, Innate immunity, M protein, Neutrophil, Mast cell, Extracellular traps, Antimicrobial peptide, Cathelicidin
Introduction
Group A Streptococcus (GAS) is a major bacterial pathogen responsible for a wide range of human disease. In the last 30 years, a striking resurgence in reports of severe, invasive GAS infections, such as necrotizing fasciitis (NF) and toxic shock syndrome (TSS), has arisen to persist in many parts of the world [1]. In the United States, Europe and Australia, recent estimates of the incidence of invasive GAS infection ranged from 2.7 to 3.5 cases per 100,000 population, with case-fatality rates of 8–14% overall, 17–24% for NF and 36–59% for TSS [2, 3, 4, 5, 6].
Protective immunity to GAS is determined in large part by antibodies directed against the M protein, a dimeric coiled-coil fibrillar protein that coats the surface of all clinical isolates. It is projected on the basis of emm sequence comparisons that there are approximately 180 known distinct GAS serotypes generated by antigenic diversity at the N-terminal hypervariable (HV) region and adjacent ‘A’ repeat domains of the M protein [7]. The antigenic diversity of M proteins across strains is dictated through HV sequences of emm genes within the GAS chromosome. The recent increase in serious GAS disease is linked epidemiologically to strains from a subset of M serotypes, with M1, M3, M28 and M12 most frequently identified [1]. Indeed, a single GAS clone of serotype M1 has disseminated to 4 continents and persisted for more than 20 years, representing the single most common isolate in invasive infections such as NF and TSS.
M protein is recognized as a critical virulence factor in GAS pathogenesis. For example, certain M proteins facilitate GAS adherence via fibronectin [8] or host glycosaminoglycans [9], and certain M protein types (1, 3, 6 and 18) have been shown to promote invasion of human pharyngeal epithelial cells or keratinocytes [10, 11]. M protein promotes GAS survival and multiplication in human blood by interacting with serum proteins that sterically interfere with complement C3b deposition and activation on the GAS surface, thereby blocking normal opsonophagocytosis [12]. Released M1 protein can complex with fibrinogen, leading to inflammatory activation of neutrophils and vascular leakage [13]; crystal structure studies identifying irregularities in the M1 coiled-coil provide insight into the molecular basis for these proinflammatory phenomena [14].
Recently, the phenomenon of neutrophil extracellular traps (NETs), DNA-based structures produced during a novel cell death process, has forced a reappraisal of the principal means by which neutrophils function in innate host defense and bacterial killing [15, 16, 17, 18]. Elimination of pathogens proceeds effectively upon their entrapment in NETs, which contain cationic histones, antimicrobial peptides and granule proteases with antibacterial properties. Extracellular trap-based bacterial killing has also been demonstrated for mast cells (MCs) and eosinophils, with ejected mitochondria rather than the nucleus providing the component DNA in the latter case [19, 20].
Here we consider for the first time the implications of extracellular traps for our understanding of GAS M protein-mediated resistance to neutrophil killing. Using targeted mutagenesis and heterologous gene expression, a unique role of the GAS M1 protein in stimulation of NET and MC extracellular trap (MCET) formation, but subsequent bacterial survival within these structures is identified. M1 protein-dependent resistance to killing by cathelicidin antimicrobial peptides is offered as a potential mechanism underlying GAS resistance to NET and MCET killing. A further association between the propensity of various GAS emm genotype strains to produce invasive infection and their relative resistance to the antimicrobial action of human cathelicidin LL-37. M1 protein interference with cathelicidin function may allow emm1 GAS strains to resist phagocytes innate immune defenses, both extra- and intracellularly, increasing their potential to produce invasive human infection.
Materials and Methods
Bacterial Strains and Mutants
GAS strain 5448 is an isolate from a patient with NF and TSS genetically representative of the globally disseminated M1T1 clone that is the leading cause of invasive GAS infections [21]. The serotype M49 strain NZ131 is a skin isolate from a patient with glomerulonephritis [22]. A panel of 56 GAS isolates of various emm genotypes and clinical associations was provided from the collections of the Centers for Disease Control and Prevention (CDC). GAS were grown in Todd-Hewitt broth (THB) or on Todd-Hewitt agar plates; for antibiotic selection, 2 μg of erythromycin (Em) or 1 μg of chloramphenicol (Cm) per ml was added to the media. Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates; antibiotic selection utilized 100 μg of ampicillin per ml, 500 μg of Em per ml, or 5 μg of Cm per ml. Lactococcus lactis NZ9000 was grown in M17 broth (Difco) supplemented with 1% glucose (GM17) or on GM17 agar plates with selection by Em at 5 μg/ml. GAS strains were rendered transformable by electroporation through growth in THB plus 0.3% glycine as described for Streptococcus agalactiae [23]. For use in neutrophil or MC experiments, bacteria were grown to the logarithmic phase in THB (OD600 = 0.4), centrifuged at 4,000 rpm for 10 min, washed with PBS and resuspended in cell culture media at the desired concentration.
MIC Testing and Antimicrobial Killing Assays
Human (LL-37) and murine (CRAMP) cathelicidin peptides were synthesized de novo by the Louisiana State University Protein Facility (Martha Juban, Director) using solid-phase Fmoc chemistry on a MilliGen 9050 Peptide Synthesizer and purified to >98% by HPLC using C4 and C18 columns on Waters Delta Prep and 600 E systems. Overnight, bacterial cultures were reinoculated into fresh THB and bacterial growth monitored to early logarithmic phase (OD600 = 0.2). The bacterial culture was then diluted in THB 1:200 (approx. 105 CFU/ml). Ninety microliters of this bacterial suspension was added to individual wells of a 96-well plate containing 10 μΐ of serially diluted AMP in distilled H2O, giving final concentrations of AMP equal to 36, 24, 18, 12, 8, 4 and 2 μM. The plate was incubated at 37°C for 18 h and MIC defined as the lowest concentration of AMP yielding no detectable bacterial growth by OD600 measurement. Killing kinetics were performed as above, except at different time points 10 μl aliquots of the culture were diluted and plated on THB agar overnight for enumeration of CFU. Growth index was calculated as recovered CFU/initial CFU. Experiments for M1 protein fragment inhibition of AMP killing were performed as for MIC testing, except the specified concentration of the M1 soluble protein fragment was added to the well at the specified concentration, and bacterial growth monitored by OD600 for 4–6 h.
Allelic Replacement of the emm1 Gene
PCR was used to amplify a 1.7-kb fragment from GAS strain 5448 that included 460 bp upstream of the emm1 gene plus the first 1,203 bp of the emm1 gene (encoding amino acid residues 1–401) using the specific primers Emm1-ExtF (5’-cggaattccgctagatgcttctgaccatcga-3’) and Emm1–401R (5’-ctcgagtgcaagttcttcagcttgtttcg-3’). The product was TA cloned into pCR2.1 TOPO (Invitrogen) and used as the template for inverse PCR. The reverse primer was designed immediately upstream of the start codon and a forward primer starting at bp 655 of the emm1 gene was used in the inverse PCR reaction. Each primer was also designed to include 25-bp 5’ extensions corresponding to the start and end of the chloramphenicol acetyltransferase gene (cat), respectively. The resultant linearized PCR product contained an in-frame deletion of the emm1 gene corresponding to amino acid residues 1–218 and was engineered to have 2 stop codons following the cat gene insertion. This product was cotransformed into E. coli Top10 cells with an approximately 650-bp PCR amplicon of the complete cat gene from pACYC184. In vivo recombination events were identified by screening for Top10 cells exhibiting resistance to both ampicillin and chloramphenicol on LB agar plates. PCR and restriction digests verified that the targeted gene segment had been replaced in frame with cat. The mutated emm1Δcat gene and flanking DNA was subcloned as EcoRI/BamHI fragment to the temperature-sensitive EmR vector pHY304 to produce knockout vector pemm1Δcat-KO. Electrocompetent M1 5448 cells were transformed with the knockout vector and single recombination events were selected at 37°C under Em selection. Selective pressure was relaxed by serial passage at 30°C without antibiotics, and double crossover events were identified as 5448 mutants exhibiting CmR and EmS. Allelic exchange of the 5’ half of the emm1 gene was confirmed in mutant 5448ΔM1 by PCR using cat primers with upstream and downstream primers as well as the absence of amplification of the wild-type gene.
Vectors for Complementation and Heterologous Expression of the emm1 and emm49 Genes
The entire emm1 and emm49 genes along with their respective ribosome-binding site was amplified by PCR and TA cloned into pCR2.1 TOPO vector (Invitrogen). The M protein genes were then subcloned as EcoRI/BamHI fragment into streptococcal/E. coli shuttle vector pDCerm [24] under control of the constitutive promoters in the plasmid backbone. The resultant vector pM1 was used to transform M49 GAS strain NZ131 and L. lactis strain under Em selection. Heterologous expression of M1 protein was confirmed by dot blot analysis, in which dilutions of overnight bacterial culture supernatants were transferred to nitrocellulose, probed with rabbit polyclonal anti-M1 antisera (1:500) followed by goat anti-rabbit antibody (1:10,000) conjugated to horseradish peroxidase, and detected using the ECL chemiluminescent reagent (Amersham). For complementation studies, pM1 or pM49 were introduced into the 5448ΔM1 mutant.
Neutrophil and MC Killing Assays
Human neutrophils were isolated from healthy donors using the PolymorphPrepTM system (Axis-Shield) and resuspended in RPMI medium and 2% autologous heat-inactivated human plasma. The human MC line HMC-1 was obtained from Joseph H. Butterfield (Rochester, Minn., USA). HMC-1 cells were cultured in IMDM supplemented with 2% FCS, 1.2 mM α-thioglycerol and 100 U/ml penicillin/100 μg/ml streptomycin (Invitrogen). MCs were washed with the same medium without antibiotics before usage in the killing experiments. Neutrophils and MCs were plated in tissue culture plates at a density of 2 × 106 and 1 × 106 cells/ml, respectively. Incubations were performed at 37°C in the presence of 5% CO2. Bacteria were added directly to cultured cells at a multiplicity of infection of 0.1 bacteria per cell, centrifuged at 800 rpm for 10 min and further incubated for 20 min. Neutrophils as well as MCs were then lysed with sterile water and serial dilutions plated on Todd-Hewitt agar for enumeration of surviving bacterial CFU. In some assays, cells were treated with cytochalasin D (Sigma-Aldrich) at a final concentration of 10 μg/ml to inhibit phagocytosis 10 min prior to infection. In extracellular trap stimulation assays using purified M1 protein, a final concentration of 3.75 μg/ml M1 was used. To induce maximal extracellular trap formation, cells were stimulated with 25 nM PMA for 4 h prior to infection. To dismantle NETs or MCETs, PMA-stimulated cells were treated with 100 U/ml DNase alone (neutrophils) or with 100 U/ml DNase and 1 μmol myeloperoxidase (MCs; Sigma-Aldrich). For quantification, extracellular traps were enumerated by counting 3 transects of 3 independent wells after staining with 2 μM Sytox orange or Syto 9 (Invitrogen).
Immunofluorescence Assays
For visualization of extracellular traps, cells were seeded on poly-L-lysine-covered cover slides, infected with Streptococcus pyogenes at a multiplicity of infection of 1:2, centrifuged at 800 rpm for 10 min and further incubated for 20 min, and the Live/Dead viability/cytotoxicity kit for mammalian cells (Invitrogen) used to visualize NETs or MCETs by fluorescence microscopy. The Live/Dead BacLightTM Bacterial Viability Kit (Invitrogen) was used to determine viability of S. pyogenes entrapped in the NETs or MCETs by fluorescence microscopy. After staining, cells were fixed with 2% paraformaldehyde, cover slides were washed and mounted onto glass slides using Prolong Gold (Invitrogen), a mounting medium that contains the DNA-staining dye DAPI (blue). Mounted samples were examined using an inverted confocal laser-scanning 2-photon microscope Olympus Fluoview FV1000 with FluoviewTM Spectral Scanning technology (Olympus). Images were obtained using 20×/0.7 or 60×/1.42 PlanApo objectives. Alternatively, images were recorded using a Zeiss Ax-iolab microscope (Zeiss 20×/0.5 Plan-Neofluor objective) with an attached Sony Digital Photo Camera DKC-5000 at calibrated magnifications.
Expression and Purification of Recombinant M1 Soluble Proteins
The emm1 PCR product encoding amino acid residues 42–453, corresponding to the mature form of the processed M1 protein, was amplified with Pfu from the genomic DNA of type M1 strain 5448. The PCR product was blunt end-ligated into the SalI site of pCR-Blunt (Invitrogen). The insert was confirmed by sequencing, then digested and ligated into pET28b (Novagen) in frame with the C-terminal His6 tag using the NcoI and XhoI sites. E. coli BL21(DE3) cells were grown in LB to an OD600 of 0.6 before inducing with 1 mM IPTG for 6 h at 37°C. The cells were harvested by centrifugation and stored at −80°C. Bacteria were sonicated in 200 mM NaCl, 100 mM phosphate buffer, pH 8, 10 mM imidazole and 0.5 mM PMSF. The clarified lysates were subjected to Ni-NTA chromatography (Qiagen) and the eluted fractions were pooled and dialyzed overnight in 50 mM NaCl, 30 mM Tris buffer, pH 7, and 1 mM EDTA in preparation of strong anion exchange chromatography (Q Sepharose Fast Flow, Poros HQ/M). Size exclusion chromatography (Superdex 200) in 50 mM NaCl and 10 mM Tris, pH 8.0, was used as a final purification step. The protein was concentrated to approximately 20 mg/ml using an Amicon filtration device, aliquoted, and flash frozen in liquid nitrogen before storing at −80°C. All subsequent constructs were subcloned from this construct and purified in the same manner. Expression and purification of M49 was performed in a similar fashion using cloned residues 42–389.
LL-37 Binding Assay for M1 Protein Fragments
LL-37 (6 μM final concentration) and M1 synthetic protein fragments (at indicated concentrations) were mixed in Luria broth medium, incubated for 15 min at 37°C, then centrifuged at 5,000 rpm for 30 min in a Jouan C412. Five microliters of the resulting supernatant was loaded on a 20–25% gradient polyacrylamide gel (Gradipore Longlife MicroGels) according to manufacturer instructions. SDS-PAGE immunoblotting and detection were performed as previously described [25].
Statistical Analysis
Data were analyzed by using Excel 2000 (Microsoft) and GraphPad Prism 4.0 (GraphPad Software). Each experiment was performed at least 3 times at independent occasions, and within each experiment all samples were processed in triplicate. Differences were analyzed using Student's t test. p ≤ 0.05 was considered significant.
Results
M1 Protein Promotes Release of Extracellular Traps from Neutrophils and MCs
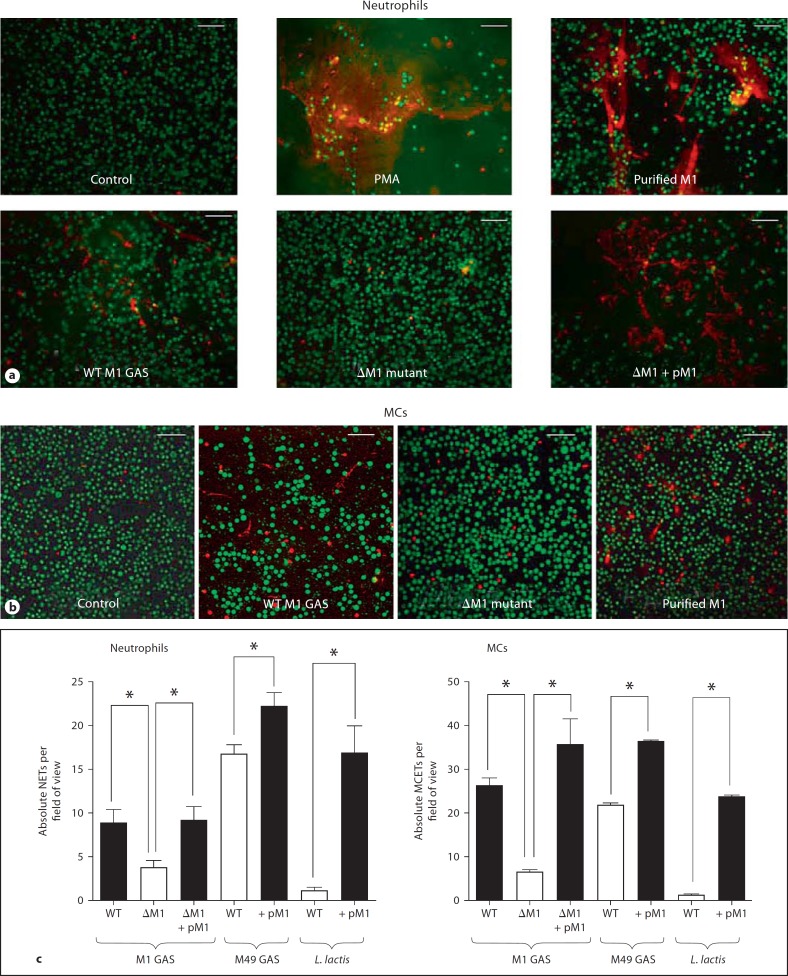
Extracellular release of DNA-based networks contributes to neutrophil and MC entrapment and killing of GAS and other leading bacterial pathogens [15, 19]. Using an extracellular DNA stain, we observed strong induction of NETs upon exposure of freshly isolated human neutrophils to a wild-type M1 GAS isolate (fig. 1a). An M1 protein-deficient mutant was then generated by allelic replacement to determine the contribution of this preeminent surface-exposed virulence factor to the phenotype. NET induction was significantly reduced in the isogenic ΔM1 mutant bacteria, restored upon complementation of the mutant with a plasmid expressing the M1 protein, and elicited with a purified preparation of the M1 protein itself (fig. 1a). Comparison of wild-type GAS to isogenic ΔM1 mutant and treatment with purified protein also demonstrated a clear contribution of M1 protein expression to induction of MCETs from a human MC line (fig. 1b). In quantitative assays, loss of M1 protein in the GAS ΔM1 mutant resulted in a significant decrease in the stimulation of NET and MCET release; conversely, heterologous expression of M1 protein in M49 GAS or L. lactis led to a significant increase in NET production by the transformed strains (fig. 1c). Baseline NET levels in the M49 GAS background are higher than the M1 GAS background because the M1 strain possesses the potent DNAse, Sda1 [26]. These paired loss-of-function and gain-of-function analyses show GAS M1 protein to be an important contributor to the NET and MCET response upon host cell engagement of the bacterial pathogen. The ability of M1 protein to form complexes with fibrinogen is key to its neutrophil activation activity [15, 16]. Consistent with this observation, addition of 2% plasma in place of 2% FCS to the MC culture increased MCET response to M1 protein (online suppl. fig. S1a, www.karger.com/doi/10.1159/000203645).
Fig. 1.
GAS M1 protein is necessary and sufficient to promote the induction of NETs and MCETs. Representative fluorescent images of neutrophils and NETs (a) and MCs and MCETs (b), stained with Live/Dead viability/cytotoxicity kit for mammalian cells after exposure to wild-type (WT) and isogenic ΔM1 mutant GAS. PMA was used as a positive control to stimulate extracellular trap formation. Note the distinctly reduced NET and MCET formation with the ΔM1 mutant strain. Scale bars = 25 μm. c Quantitative enumerations of NETs or MCETs per field of view containing approximately 150 cells. Decreased trip induction was observed upon deletion of M1 in the GAS wild-type (WT) strain, while increased induction was observed upon heterologous expression of M1 in M49 GAS or L. lactis. Experiments were performed in triplicate and repeated 3 times with similar results. One representative experiment is shown ± standard deviation. * p < 0.05 by t test.
M1 Protein Promotes GAS Survival in NETs
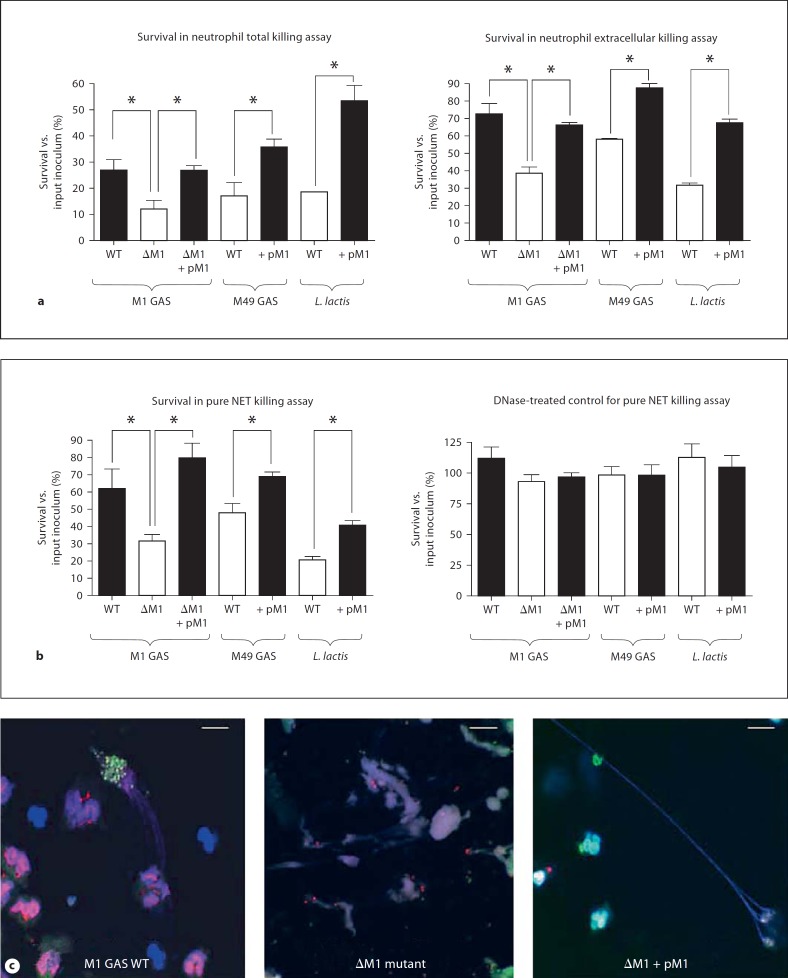
Through an increased stimulation of NETs, one might infer that GAS expressing M1 protein could be more susceptible to extracellular entrapment and killing by human neutrophils. Interestingly, just the opposite was observed. As expected, the GAS ΔM1 mutant was more susceptible to overall neutrophil killing than the wild-type parent strain, and this attenuation was reversed upon complementation of the mutant with the pM1 expression plasmid (fig. 2a, left). Yet, even when phagocytic uptake was blocked by administration of cytochalasin D, a similar contribution of M1 to GAS resistance to neutrophil extracellular killing was observed (fig. 2a, right). Control studies confirmed that NETs are produced with cytochalasin D treatment (online suppl. fig. S1b). Heterologous expression of the M1 protein in either M49 GAS or L. lactis increased the resistance of the transformed bacteria to both total and extracellular neutrophil killing (fig. 2a). To focus more specifically on NET survival, PMA was used to stimulate maximum NET production and restrict phagocytosis; here, M1 protein was again necessary and sufficient to promote bacterial resistance to NET killing in both loss-of-function and gain-of-function studies (fig. 2b, left). When DNAse treatment was used as a control to disrupt NETs, all differences in bacterial survival across the panel of mutant and transformed strains were eliminated (fig. 2b, right). Thus, we conclude that while M1 protein is itself a stimulus for NET production, it is an even more important factor in promoting GAS survival within NETs. This phenomenon is illustrated in a representative fluorescent vital stain image of viable (green) versus dead (red) GAS entrapped by NETs (fig. 2c). Here, examples of viable wild-type and complement mutant GAS expressing M1 within NETs are readily identified, whereas the vast majority of NET-entrapped ΔM1 mutant GAS are dead.
Fig. 2.
GAS M1 protein is necessary and sufficient to promote bacterial resistance to killing in NETs. a M1 protein contributes to bacterial survival in a total (intracellular + extracellular) neutrophil killing assay. b M1 protein promotes bacterial resistance to neutrophil extracellular killing when phagocytic uptake is inhibited with cytochalasin D (10 μg/ml). c Neutrophils were pre-stimulated with 25 nM PMA for 4 h before infection to induce maximal trap formation and to avoid phagocytosis. Note that M1 contributes to GAS survival and increases L. lactis survival upon 30 min co-incubation with NETs. As control, the cells were treated with 100 U/ml DNase to disrupt extracellular trap formation before infection, which completely abolished the killing of bacteria by extracellular traps. Experiments were performed in triplicate and repeated 3 times with similar results. One representative result is shown ± standard deviation. * p < 0.05 by t test. d Representative fluorescent image of viable (green) versus dead (red) GAS entrapped by NETs (DAPI stained, blue) as determined by Live/Dead BacLightTM Bacterial Viability assay. Scale bars = 10 μm. Note that M1 contributes to GAS survival within NETs. WT = Wild type.
M1 Protein Promotes GAS Resistance to MCET Killing
To determine if our findings in the neutrophil system extended to the human MC line, we repeated quantitative assays for total killing, extracellular killing and MCET-dependent killing using the full panel of isogenic GAS strains (online suppl. fig. S2). The observed pattern of results closely mirrored what was observed in the neutrophil assays, as loss of M1 protein rendered GAS more susceptible to MCET-dependent killing and enhanced resistance to MCET-dependent killing could be conferred to bacteria transformed with the pM1 expression plasmid (online suppl. fig. S2).
M1 Protein Contributes to GAS Cathelicidin Resistance
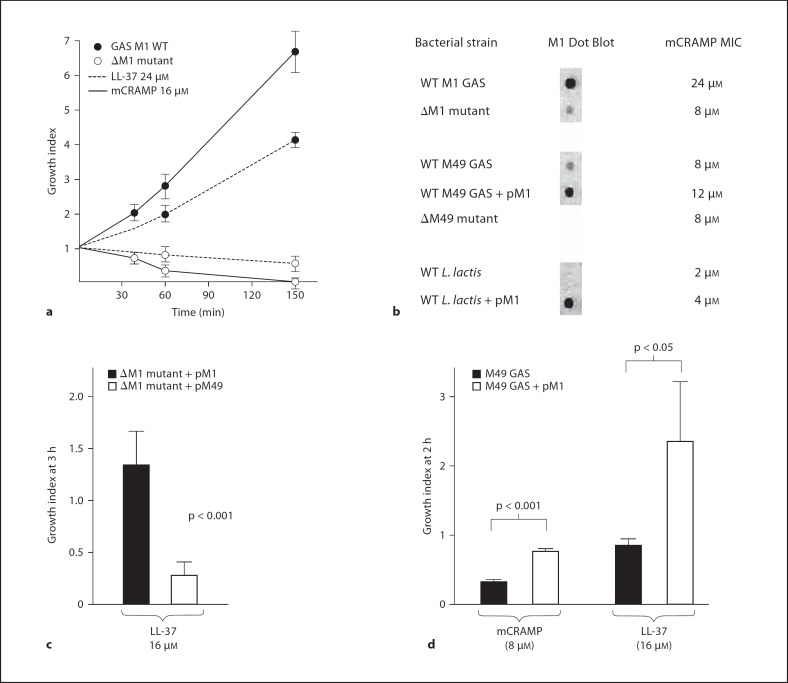
Cathelicidin antimicrobial peptides produced by epithelial cells and phagocytes, including neutrophils and MCs, are an important component of innate immune defense against GAS infection [27, 28, 29]. These amphipathic, cationic α-helical peptides are released upon phagocyte degranulation and can be identified within extracellular traps [19]. We hypothesized that resistance to cathelicidin and related AMPs could be one mechanism by which M1 protein could promote GAS survival upon entrapment within NETs or MCETs. We found that cathelicidins LL-37 (human) or mCRAMP (mouse) killed the ΔM1 mutant at concentrations where the wild-type M1 GAS strain proliferated (fig. 3a). The LL-37 MIC for the ΔM1 mutant was reduced from 24 to 8 μM (fig. 3b), a level of sensitivity similar to most noninvasive GAS strains (fig. 5). In contrast, elimination of M protein expression in a cathelicidin-sensitive M49 GAS strain did not alter the baseline LL-37 MIC of 8 μM (fig. 3b). Heterologous expression of M1 protein on a plasmid vector increased the LL-37 MIC of M49 GAS from 8 to 12 μM and of L. lactis from 2 to 4 μM (fig. 3b). The growth of the ΔM1 mutant in the presence of LL-37 was significantly enhanced when it was complemented with a plasmid expressing M1 compared to when it was complemented with a plasmid expressing the M49 protein (fig. 3c). Finally, heterologous expression of M1 protein in M49 GAS strains produced an approximately two-fold increased growth in the presence of LL-37 and mCRAMP in kinetic killing assays (fig. 3d). In sum, these studies showed that the expression of M1 protein could make a demonstrable contribution to the overall resistance of GAS to cathelicidin AMPs.
Fig. 3.
M1 protein contributes to cathelicidin resistance. a Killing kinetics for LL-37 and CRAMP against M1 wild-type (WT) or ΔM1 mutant GAS. Growth index = CFU at specified time point/CFU in initial inoculum. b Sensitivity testing to mCRAMP of isogenic M1 mutant and strains heterologously expressing M1 protein show the peptide is necessary and sufficient to promote increased cathelicidin resistance. c Complementation with an M1 protein expression plasmid promotes survival of the M1 mutant significantly greater than complementation with M49 protein. d Reduced killing of M49 GAS expressing M1 protein by LL-37 and mCRAMP. Experiments were performed in triplicate and repeated 3 times with similar results. One representative result is shown ± standard deviation.
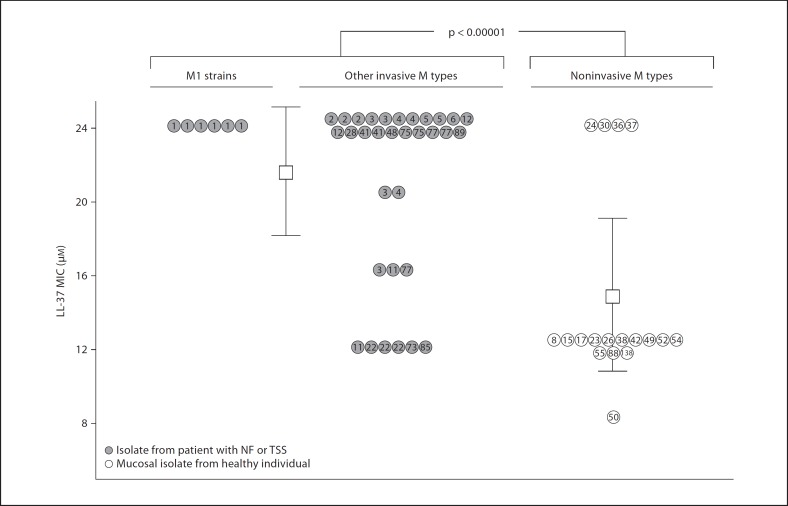
Fig. 5.
M1 and other invasive isolates of GAS show increased resistance to cathelicidin compared to colonizing strains. Sensitivity or resistance of GAS clinical isolates to inhibition by the human cathelicidin LL-37 is reported. M protein (emm) genotypes are indicated within individual circles. MIC testing was performed 3–4 times for each isolate.
Human Cathelicidin Resistance Is Associated with M1 Protein N-terminal Sequences
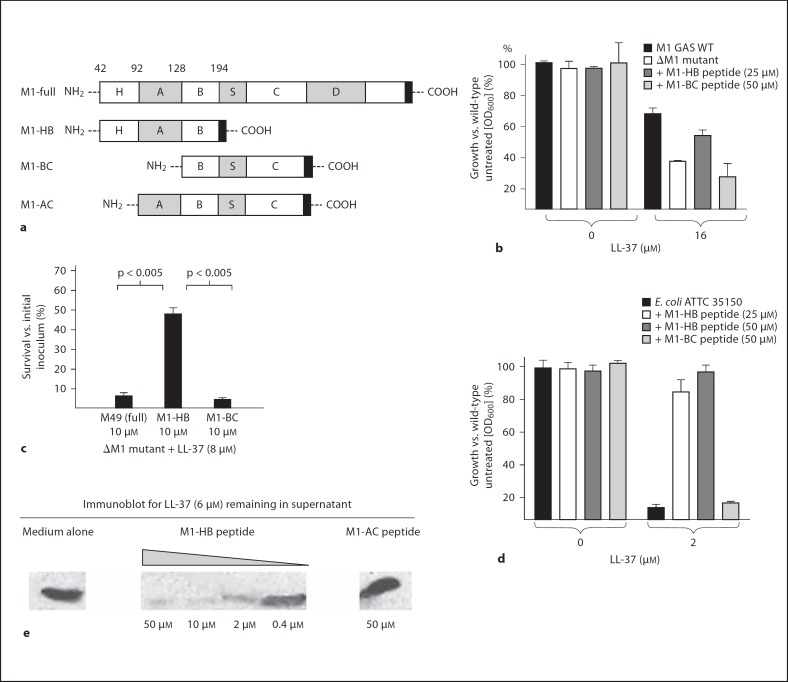
Because GAS cathelicidin resistance was associated with M1 but not M49 protein expression, we hypothesized that the resistance phenotype could be attributed to the HV N-terminus and A repeat region that encode (For figure see next page.) the type-specific immunologic epitopes of the M1 serotype. To map the cathelicidin resistance domain, we produced a recombinant protein fragment corresponding to the N-terminal 152 amino acids of mature M1 protein (HV region + A region + B repeats, M1-HB), along with control soluble protein fragments comprising the B repeats, S region and C repeat domains (M1-BC) or the A region through the C repeats (M1-AC; fig. 4a). Whereas addition of M1-HB protein fragment was able to block LL-37 killing of the sensitive ΔM1 GAS mutant, no such effect was seen with the M1-BC protein fragment (fig. 4b) nor with the complete M49 protein (fig. 4c). None of the soluble protein fragments affected the growth of GAS in the absence of cathelicidin. A similar inhibitory effect of the M1-HB protein fragment but not the M1-BC protein fragment was seen on LL-37 killing of an unrelated, highly-sensitive target bacterium, Escherichia coli (fig. 4d).
Fig. 4.
The cathelicidin inhibitory activity of M1 protein maps to its N-terminal domain. a Recombinant M1 soluble protein constructs used in these studies. b–d Exogenous M1 N-terminal protein fragment reverses increased sensitivity of ΔM1 GAS mutant to LL-37 and blocks LL-37 killing of E. coli. Experiments were performed in triplicate and repeated three times with similar results. One representative result is shown ± standard deviation. WT = Wild type. e Western blot analysis shows that the M1 N-terminal protein fragment depletes LL-37 from solution.
M1 Protein Appears to Sequester, Rather than Repel, Human Cathelicidin LL-37
One commonly described mechanism for bacterial cathelicidin resistance is modification of cell wall constituents to incorporate more positive charge, thus repelling the cationic peptide, e.g. the D-alanylation of lipoteichoic acid as described in many Gram-positive pathogens [30], including GAS [31], and which has been shown to contribute to pneumococcal survival in NETs [32]. Alternatively, bacterial proteins can bind and inactivate cathelicidin before it reaches the cell wall site of action, as has been postulated for staphylokinase of Staphylococcus aureus [33] or the serum inhibitor of complement (SIC) of GAS [34]. As a first step in mechanistic evaluation of GAS M1 interference with human cathelidicin action, we allowed different concentrations of the soluble GAS M1-HB protein fragment to incubate with a fixed amount of LL-37, then spun the mixture and assayed the supernatant by immunoblot for residual free LL-37 (fig. 4e). A dose-dependent removal of LL-37 from solution by M1-HB was observed, while no similar effect was seen with the M1-AC protein fragment. These results confirm that LL-37 inhibitory function of M1 protein maps to its N-terminal HV domain, and that a binding/sequestration mechanism rather than a repulsion mechanism is likely at play.
Correlation of Cathelicidin Resistance to Disease Phenotype among GAS Clinical Isolates
Serotype M1 strains have been the most common serotype isolated from invasive GAS infections such as NF and TSS. We sought to compare the relative susceptibility of invasive and noninvasive GAS strains to the antimicrobial action of human cathelicidin LL-37. Thirty-eight GAS sterile-site isolates from patients with NF and/or TSS were obtained from 1998–2002 surveillance studies at the CDC. This collection comprised strains from 17 emm genotypes accounting for >71% of invasive isolates and including 13 of the 16 most common emm genotypes in contemporary invasive GAS epidemiology. As a control group, we tested GAS strains of 18 different emm genotypes in the CDC collection isolated from the throat or skin of asymptomatic individuals. These ‘noninvasive’ strains are types not represented among the top 30 emm genotypes in recent invasive disease surveillance. As shown in figure 5, we found the invasive disease isolate group possessed significantly higher MIC values for LL-37 (21.3 + 4.7 μM) than the less common emm types isolated from asymptomatic patients (14.4 + 5.3 μM; p < 0.00001). Genotype emm1 strains exhibited a uniformly high MIC of 24 μM, a finding corroborated in 3 additional emm1 isolates from throat samples of patients with uncomplicated pharyngitis (data not shown). The observed correlations appear to reveal differences in cathelicidin resistance inherent to M protein lineage, source of isolate, or both.
Discussion
M protein is an immunodominant surface-anchored protein and pluripotent virulence factor of GAS, with long-appreciated roles in resistance to opsonophagocytosis and promoting epithelial cell and extracellular matrix interactions [12]. New lines of investigation have revealed that M1 protein released from the bacterial surface also functions as a potent proinflammatory toxin, contributing to neutrophil degranulation, T cell activation, vascular leakage and lung injury [13, 35, 36], via mechanisms that involve binding of M1 to fibrinogen or IgG antibodies [13, 14, 37]. Here we describe a novel AMP inhibitory effect of the M1 HV region that adds to the described immune resistance functions of the conserved M protein domains, including complement interference via binding of factor H [38], C4b-binding protein [39] or fibronectin [8], and nonimmune binding of the Fc domains of immunoglobulins A [39] or G [40]. M1 protein also interferes with antibacterial peptides derived from human β-2 glycoprotein I, a heparin-binding plasma protein [41].
Cathelicidins are important for neutrophil- and MC-mediated GAS killing and to control GAS infection in vivo [27, 29]. It is now appreciated that neutrophil and MC killing of bacteria is accomplished in significant part extracellularly, through the elaboration of DNA-based extracellular traps, and that cathelicidin peptides along with cationic histone proteins and granule proteases are critical effectors of bactericidal activity within these structures [17, 18, 19]. GAS M1 protein is a clear stimulus for NET and MCET elaboration, and thus the first specific bacterial protein identified to promote phagocyte extracellular trap production. Despite its role in inducing trap formation, we found M1 protein promoted GAS extracellular survival, at least in part due to inhibition of cathelicidin AMP killing. The invasive M1 clone of GAS thus possesses multiple complementary mechanisms to interfere with the NET-based innate defense, also including SpyCEP proteolysis of IL-8, required for maximum NET induction, and phage-encoded DNAse Sda1, which degrades NETs after their formation [26, 42, 43].
Recently, M1 protein was further shown to promote the intracellular survival of GAS within neutrophils after their phagocytic uptake [44]. A contributing mechanism appears to be an inhibitory effect on the fusion of azurophilic granules, which contain antibacterial lysozyme, myeloperoxidase and defensins, with the phagosome [45]. In contrast, GAS and M1 protein did not prevent the mobilization of specific granules [45], the vesicles in which cathelicidin peptides are stored [46]. Thus, our finding of M1 protein-mediated inhibition of cathelicidin provides a complementary mechanism to understand the intracellular survival of emm1 GAS strains within neutrophils.
We found that the intrinsic resistance of emm1 GAS strains to LL-37 can be attributed in part to an inhibitory action of the N-terminal HV domain of the M1 protein. Several Gram-positive bacterial pathogens including GAS, GBS, pneumococcus and S. aureus can resist AMP killing through incorporation of positive charge modifications into cell wall components, thereby decreasing the electrostatic attraction of the cationic AMP to the cell surface [31, 32, 47, 48]. In the case of emm1 GAS, our data suggest instead that the fimbrial-like extension of M1 protein may trap or inactivate LL-37 before the cathelicidin can reach its cell membrane target of action, thereby providing a physical shield against AMP killing. In this manner, the cathelicidin resistance phenotype of M1 protein mimics the reported LL-37 sequestering activity of another fimbrial structure, the surface-anchored pili of GBS [49].
GAS possesses additional virulence determinants that may contribute to AMP resistance. The SpeB cysteine protease of GAS can degrade LL-37, a phenomenon reversed by the SpeB-specific protease inhibitor E64 [50], and is suggested to contribute to GAS cathelicidin inactivated in the tissues of NF patients [51]. M1 strains also secrete a protein known as SIC in high quantities that can interfere with host complement function [52]. Recently, it was discovered that purified SIC binds and inactivates the human AMPs LL-37 and α- and β-defensin [34, 53]. The combined AMP-neutralizing effect of the N-terminus of the surface-anchored M1 protein and the released SIC protein may serve to make control of emm1 strains particularly problematic for host AMP-mediated innate immune defenses.
We report that emm1 and other GAS strains associated with invasive infections exhibit higher intrinsic levels of resistance to the human cathelicidin LL-37 than strains that are not associated with invasive disease. Cathelicidins are known to contribute to the epithelial barrier against infection and the killing mechanism of phagocytic cells including neutrophils and macrophages. Therefore, cathelicidin resistance among certain GAS strains could increase the likelihood that the bacteria survive host inflammatory responses to proliferate in deeper tissues or the bloodstream, whereupon additional GAS toxins and virulence factors further exert their pathogenic effects. Immunogenetic variation in human patients plays a critical role in determining the clinical outcome of invasive M1 GAS infection [54], however, potential polymorphisms in human cathelicidin genes or differences in cathelicidin gene expression among previously healthy individuals have yet to be explored.
In summary, we have combined molecular microbiologic techniques and an epidemiologic survey to ascribe a novel virulence function of the N-terminal domain of M1 protein in survival avoidance of host AMP defenses. Our studies suggest that investigations of strain variation in cathelidicin resistance/sensitivity may prove fruitful in understanding the epidemiology and pathogenesis of other bacterial infections important in human medicine.
Supplementary Material
Supplementary data
Acknowledgment
This work was supported by NIH grants AI048694 and AI077780 (V.N.), AR45676 and AI48176 (R.L.G.), and AI071167 (P.G.). X.L. was supported by a Wound Healing Foundation Fellowship Award and M.v.K.-B. was supported through a fellowship from the Deutsche Akademie der Naturforscher Leopoldina (BMBF-LPD 9901/8-187).
References
- 1.Aziz RK, Kotb M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg Infect Dis. 2008;14:1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Grady KA, Kelpie L, Andrews RM, Curtis N, Nolan TM, Selvaraj G, Passmore JW, Oppedisano F, Carnie JA, Carapetis JR. The epidemiology of invasive group A streptococcal disease in Victoria, Australia. Med J Aust. 2007;186:565–569. doi: 10.5694/j.1326-5377.2007.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 3.Darenberg J, Luca-Harari B, Jasir A, Sandgren A, Pettersson H, Schalen C, Norgren M, Romanus V, Norrby-Teglund A, Normark BH. Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis. 2007;45:450–458. doi: 10.1086/519936. [DOI] [PubMed] [Google Scholar]
- 4.O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, van Beneden C. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis. 2007;45:853–862. doi: 10.1086/521264. [DOI] [PubMed] [Google Scholar]
- 5.Vlaminckx BJ, Mascini EM, Schellekens JF. Invasive Lancefield group A streptococcal infections in the Netherlands. Ned Tijdschr Geneeskd. 2007;151:1669–1673. [PubMed] [Google Scholar]
- 6.Luca-Harari B, Ekelund K, van der Linden M, Staum-Kaltoft M, Hammerum AM, Jasir A. Clinical and epidemiological aspects of invasive Streptococcus pyogenes infections in Denmark during 2003 and 2004. J Clin Microbiol. 2008;46:79–86. doi: 10.1128/JCM.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DR, Kaplan EL, VanGheem A, Facklam RR, Beall B. Characterization of group A streptococci (Streptococcus pyogenes): correlation of M-protein and emm-gene type with T-protein agglutination pattern and serum opacity factor. J Med Microbiol. 2006;55:157–164. doi: 10.1099/jmm.0.46224-0. [DOI] [PubMed] [Google Scholar]
- 8.Cue D, Lam H, Cleary PP. Genetic dissection of the Streptococcus pyogenes M1 protein: regions involved in fibronectin binding and intracellular invasion. Microb Pathog. 2001;31:231–242. doi: 10.1006/mpat.2001.0467. [DOI] [PubMed] [Google Scholar]
- 9.Frick IM, Schmidtchen A, Sjobring U. Interactions between M proteins of Streptococcus pyogenes and glycosaminoglycans promote bacterial adhesion to host cells. Eur J Biochem. 2003;270:2303–2311. doi: 10.1046/j.1432-1033.2003.03600.x. [DOI] [PubMed] [Google Scholar]
- 10.Berkower C, Ravins M, Moses AE, Hanski E. Expression of different group A streptococcal M proteins in an isogenic background demonstrates diversity in adherence to and invasion of eukaryotic cells. Mol Microbiol. 1999;31:1463–1475. doi: 10.1046/j.1365-2958.1999.01289.x. [DOI] [PubMed] [Google Scholar]
- 11.Purushothaman SS, Wang B, Cleary PP. M1 protein triggers a phosphoinositide cascade for group A Streptococcus invasion of epithelial cells. Infect Immun. 2003;71:5823–5830. doi: 10.1128/IAI.71.10.5823-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 13.Herwald H, Cramer H, Morgelin M, Russell W, Sollenberg U, Norrby-Teglund A, Flodgaard H, Lindbom L, Bjorck L. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116:367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 14.McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol. 2007;5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 18.Wartha F, Beiter K, Normark S, Henriques-Normark B. Neutrophil extracellular traps: casting the net over pathogenesis. Curr Opin Microbiol. 2007;10:52–56. doi: 10.1016/j.mib.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 19.von Kockritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M, Medina E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111:3070–3080. doi: 10.1182/blood-2007-07-104018. [DOI] [PubMed] [Google Scholar]
- 20.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 21.Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–6369. doi: 10.1128/iai.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon D, Ferretti JJ. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;66:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 23.Framson PE, Nittayajarn A, Merry J, Youngman P, Rubens CE. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pwv01 plasmids, and mutagenesis with Tn917. Appl Environ Microbiol. 1997;63:3539–3547. doi: 10.1128/aem.63.9.3539-3547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeng A, Sakota V, Li Z, Datta V, Beall B, Nizet V. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J Bacteriol. 2003;185:1208–1217. doi: 10.1128/JB.185.4.1208-1217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaiou M, Nizet V, Gallo RL. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Dermatol. 2003;120:810–816. doi: 10.1046/j.1523-1747.2003.12132.x. [DOI] [PubMed] [Google Scholar]
- 26.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNAse expression allows the pathogen group a Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 27.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 28.Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. 2005;73:6771–6781. doi: 10.1128/IAI.73.10.6771-6781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Nardo A, Yamasaki K, Dorschner RA, Lai Y, Gallo RL. Mast cell cathelicidin antimicrobial peptide prevents invasive group A Streptococcus infection of the skin. J Immunol. 2008;180:7565–7573. doi: 10.4049/jimmunol.180.11.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus D, Peschel A. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr Top Microbiol Immunol. 2006;306:231–250. doi: 10.1007/3-540-29916-5_9. [DOI] [PubMed] [Google Scholar]
- 31.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. D-alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, Normark S, Henriques-Normark B. Capsule and D-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol. 2007;9:1162–1171. doi: 10.1111/j.1462-5822.2006.00857.x. [DOI] [PubMed] [Google Scholar]
- 33.Braff MH, Jones AL, Skerrett SJ, Rubens CE. Staphylococcus aureus exploits cathelicidin antimicrobial peptides produced during early pneumonia to promote staphylokinase-dependent fibrinolysis. J Infect Dis. 2007;195:1365–1372. doi: 10.1086/513277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 35.Pahlman LI, Olin AI, Darenberg J, Morgelin M, Kotb M, Herwald H, Norrby-Teglund A. Soluble M1 protein of Streptococcus pyogenes triggers potent T cell activation. Cell Microbiol. 2008;10:404–414. doi: 10.1111/j.1462-5822.2007.01053.x. [DOI] [PubMed] [Google Scholar]
- 36.Soehnlein O, Oehmcke S, Ma X, Rothfuchs AG, Frithiof R, van Rooijen N, Morgelin M, Herwald H, Lindbom L. Neutrophil degranulation mediates severe lung damage triggered by streptococcal M1 protein. Eur Respir J. 2008;32:405–412. doi: 10.1183/09031936.00173207. [DOI] [PubMed] [Google Scholar]
- 37.Kahn F, Morgelin M, Shannon O, Norrby-Teglund A, Herwald H, Olin AI, Bjorck L. Antibodies against a surface protein of Streptococcus pyogenes promote a pathological inflammatory response. PLoS Pathog. 2008;4:e1000149. doi: 10.1371/journal.ppat.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Casal J, Okada N, Caparon MG, Scott JR. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol Microbiol. 1995;15:907–916. doi: 10.1111/j.1365-2958.1995.tb02360.x. [DOI] [PubMed] [Google Scholar]
- 39.Carlsson F, Berggard K, Stalhammar-Carlemalm M, Lindahl G. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med. 2003;198:1057–1068. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akesson P, Schmidt KH, Cooney J, Bjorck L. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994;300:877–886. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nilsson M, Wasylik S, Morgelin M, Olin AI, Meijers JC, Derksen RH, de Groot PG, Herwald H. The antibacterial activity of peptides derived from human β-2 glycoprotein I is inhibited by protein H and M1 protein from Streptococcus pyogenes. Mol Microbiol. 2008;67:482–492. doi: 10.1111/j.1365-2958.2007.05974.x. [DOI] [PubMed] [Google Scholar]
- 42.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. DNAse Sda1 provides selection pressure for a switch to invasive group a streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 43.Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, Mishalian I, Sriskandan S, Hanski E, Nizet V. The IL-8 protease SpyCep/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe. 2008;4:170–178. doi: 10.1016/j.chom.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staali L, Morgelin M, Bjorck L, Tapper H. Streptococcus pyogenes expressing M and M-like surface proteins are phagocytosed but survive inside human neutrophils. Cell Microbiol. 2003;5:253–265. doi: 10.1046/j.1462-5822.2003.00272.x. [DOI] [PubMed] [Google Scholar]
- 45.Staali L, Bauer S, Morgelin M, Bjorck L, Tapper H. Streptococcus pyogenes bacteria modulate membrane traffic in human neutrophils and selectively inhibit azurophilic granule fusion with phagosomes. Cell Microbiol. 2006;8:690–703. doi: 10.1111/j.1462-5822.2005.00662.x. [DOI] [PubMed] [Google Scholar]
- 46.Borregaard N, Sorensen OE, Theilgaard-Monch K. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 2007;28:340–345. doi: 10.1016/j.it.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Peschel A, Collins LV. Staphylococcal resistance to antimicrobial peptides of mammalian and bacterial origin. Peptides. 2001;22:1651–1659. doi: 10.1016/s0196-9781(01)00500-9. [DOI] [PubMed] [Google Scholar]
- 48.Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol Microbiol. 2003;49:1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x. [DOI] [PubMed] [Google Scholar]
- 49.Maisey HC, Quach D, Hensler ME, Liu GY, Gallo RL, Nizet V, Doran KS. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 2008;22:1715–1724. doi: 10.1096/fj.07-093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidtchen A, Frick IM, Andersson E, Tapper H, Bjorck L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol Microbiol. 2002;46:157–168. doi: 10.1046/j.1365-2958.2002.03146.x. [DOI] [PubMed] [Google Scholar]
- 51.Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akesson P, Sjoholm AG, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 53.Fernie-King BA, Seilly DJ, Lachmann PJ. The interaction of streptococcal inhibitor of complement (SIC) and its proteolytic fragments with the human beta defensins. Immunology. 2004;111:444–452. doi: 10.1111/j.0019-2805.2004.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak MT, Khurshid A, Green K, Peeples J, Wade J, Thomson G, Schwartz B, Low DE. An immunogenetic and molecular basis for differences in outcomes of invasive group a streptococcal infections. Nat Med. 2002;8:1398–1404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data