Abstract
The development of a highly effective AIDS vaccine will likely depend on success in designing immunogens that elicit broadly neutralizing antibodies to naturally circulating strains of HIV-1. Although the antibodies induced after natural infection with HIV-1 are often directed to strain-specific or nonneutralizing determinants, it is now evident that 10%–25% of HIV-infected individuals generate neutralizing antibody responses of considerable breadth. In the past, only four broadly neutralizing monoclonal antibodies had been defined, but more than a dozen monoclonal antibodies of substantial breadth have more recently been isolated. An understanding of their recognition sites, the structural basis of their interaction with the HIV Env, and their development pathways provides new opportunities to design vaccine candidates that will elicit broadly protective antibodies against this virus.
More than a dozen broadly neutralizing monoclonal antibodies against HIV-1 have recently been isolated. Understanding the structural basis for their interaction with HIV-1 Env is an important step in designing an AIDS vaccine.
For the majority of licensed vaccines, neutralizing antibodies have provided the best correlate of vaccine efficacy. Although a variety of immune mechanisms may contribute to protection, immunity is in part caused by inactivation of the infecting virus that aborts productive replication. In the case of HIV, it has been difficult to define such antibodies because the virus has evolved a multitude of mechanisms to evade humoral immunity. Because of its error-prone DNA-dependent RNA polymerase and its ability to undergo RNA recombination, the virus has generated unprecedented diversity (Korber et al. 2000). The number of common determinants shared by naturally circulating strains is therefore diminished. In addition, HIV envelope glycoprotein (Env) displays a low spike density on the virion surface (Klein et al. 2009; Klein and Bjorkman 2010), potentially reducing the efficiency of cross-linking and the advantage of antibody avidity that enhances the neutralization of many viruses. Its high carbohydrate content further masks critical structures that may be sensitive to neutralization (Wyatt et al. 1998). Finally, other mechanisms, including conformational flexibility, strain-specific amino acid variability, and decoy forms of the HIV Env, such as the free monomer (Douek et al. 2006), stimulate nonneutralizing antibody responses to irrelevant viral structures. Thus, the definition of serotypes that has proven a successful approach for many vaccines has not been available to guide the design of broadly neutralizing antibody immunogens. These challenges have prompted efforts to understand the immunobiology of HIV-1 Env, with an emphasis on understanding the structural basis for HIV-1 Env neutralization. In addition, the definition and characterization of monoclonal antibodies that mediate such broad neutralization, as well as the structural basis for its interaction with HIV envelope, have provided opportunities for the design of HIV-1 vaccines that stimulate the production of antibodies that are directed against specific conserved regions of the virus.
Rational design of immunogens that elicit broadly reactive neutralizing antibodies is facilitated by the identification of HIV-infected individuals with broadly neutralizing sera, from which individual monoclonal antibodies can be isolated. Two strategies have led to the identification of such antibodies. First, individual B cells have been isolated, grown in microcultures, and the secreted antibodies have been tested for neutralization. Antibodies that neutralize diverse HIV-1 viruses were identified, and the immunoglobulin genes from the cells of interest were cloned and expressed. The neutralization specificity was then confirmed for the cloned expressed IgG genes. A second approach built on knowledge of structure to design resurfaced and stabilized HIV Env cores that were used as probes to select individual B cells targeted to a specific site. The immunoglobulin genes from these B cells were then rescued by PCR amplification. The neutralization breadths of expressed antibodies were defined against a panel of genetically diverse circulated viruses. Finally, targeted approaches to other specific regions of the virus, including the membrane-proximal region (MPR), CD4-induced (CD4i), and Env glycans have all provided specific targets for which immunogens can be specifically designed. Taken together, these approaches have enabled the design of probes that allow detection of antibodies to specific viral structures at the same time that they serve as prototype immunogens to elicit these responses. Nonetheless, impediments remain to the elicitation of such antibodies, including the ability to overcome the elimination of autoreactive B cells and to stimulate the relevant necessary somatic mutations that give rise to antibodies of the appropriate specificity. Finally, elucidation of the critical structures that confer relevant antigenicity while defining the determinants required for immunogenicity represents a key scientific question whose solution will facilitate the success of this rational vaccine design strategy.
ROLE OF ANTIBODIES IN PROTECTION
The design of immunogens able to elicit neutralizing antibodies (NAb) remains a major goal of HIV-1 vaccine development. Most licensed viral vaccines induce antibodies that neutralize the infecting virus, thereby protecting against infection or disease. Although the specific immune responses required to protect humans against HIV-1 infection are not known, studies of lentiviral infection in nonhuman primates (NHPs) have shown that passive infusion of antibodies can prevent infection. Specifically, antibodies that neutralize HIV-1 have been shown to prevent infection by a chimeric simian-human immunodeficiency virus (SHIV) containing the env gene of HIV-1. Early SHIV challenge studies used intravenous inoculation of the challenge virus. In this setting, high concentrations of NAbs were required to block infection, and nonneutralizing antibodies were unable to protect against infection (Mascola et al. 1999; Shibata et al. 1999). Subsequent studies, using a single oral or vaginal inoculation sufficient to infect 100% of control animals, also showed protection by NAb (Baba et al. 2000; Mascola et al. 2000; Hofmann-Lehmann et al. 2001; Parren et al. 2001). The protection against SHIV infection has been most directly associated with the neutralization potency of the infused antibodies (Nishimura et al. 2002; Mascola 2003). However, recent studies have also suggested a role of Fc-mediated antibody effector functions in conferring protection. The neutralizing monoclonal antibody (mAb) IgG1 b12 showed a diminished protective effect if the Fc region of the IgG was altered to knock out complement binding and ADCC activity (Hessell et al. 2007). Importantly, recent passive transfer studies have employed low-dose mucosal inoculation that requires multiple challenges to infect all control animals (Hessell et al. 2009a,b). This model may be more physiologically relevant to the relatively inefficient HIV-1 infection in humans. In the low-dose NHP model, approximately 10-fold less antibody was required to mediate protection against infection compared to prior studies with high-dose inoculations; serum antibody titers sufficient to mediate 90% virus neutralization at 1:5 serum dilution were associated with protection. Hence, vaccines may not need to achieve extraordinarily high levels of HIV-1 NAbs. However, the antibody response will likely need to be durable, and NAbs will have to cross-react with a genetically diverse spectrum of HIV-1 strains.
CLINICAL TRIALS OF CANDIDATE HIV VACCINES
To protect against HIV-1, antibodies must bind the viral surface envelope glycoprotein, a homotrimer composed of the gp120 surface unit and the gp41 trans-membrane domain. The earliest phase I vaccine trials included recombinant envelope products, gp120 or gp160, formulated in various adjuvants. Live recombinant vectors such as vaccinia and canary pox and nucleic acid–based vaccines have also been tested, usually including gene inserts expressing the Env glycoprotein. Recombinant protein vaccines used alone, or as a boost to vaccine vectors, generally elicited high titers of anti-Env antibodies. Initial immunogenicity studies showed that vaccine-elicited antibodies could neutralize HIV-1 in vitro, but it was soon realized that the viral neutralization was limited to prototype laboratory-adapted HIV-1 strains and did not extend to primary HIV-1 isolates (Wrin and Nunberg 1994; Mascola et al. 1996). These neutralization data generated considerable debate regarding the rationale for efficacy testing of protein-based vaccines. Despite this uncertainty, two phase III gp120 vaccine trials were conducted, each with a bivalent formulation of two strains of gp120 formulated in Alum. The VAX004 and VAX003 studies were initiated in 1998 and 1999, respectively, and the results reported in 2003. These gp120 vaccines showed no significant impact on acquisition of HIV-1 infection and had no impact on plasma viremia or peripheral CD4+ T-cell counts (Flynn et al. 2005; Gilbert et al. 2005).
The failure of these gp120 vaccines was generally viewed as evidence that a successful vaccine would need to induce more potent NAbs that can neutralize circulating strains of HIV-1. Prior to the release of the VAX003 and VAX004 results, an additional phase III trial was planned. The RV144 study included priming immunizations with an avipox vector (ALVAC) and boosting with the same bivalent gp120s used in the VAX003 study. The results of this study, released in 2009, showed that volunteers in the vaccine arm of the study acquired 31% fewer HIV-1 infections than those in the placebo arm (Rerks-Ngarm et al. 2009). This modest efficacy, although not deemed adequate for licensure, was the first indication that a vaccine could protect against HIV-1 infection. The immunologic data from the RV144 vaccine trial are still being analyzed. Immune correlates analysis will include a set of in vitro antibody assays, including traditional virus neutralization, and assays of antibody binding, ADCC, and additional measures of Fc-mediated antibody effector functions. Whether any of these measures of vaccine-elicited antibodies will correlate with protection is yet to be determined. It is also unclear why this ALVAC/gp120 trial produced a modest protective effect whereas the gp120-only VAX003 and VAX004 studies did not. Differences in the studies include the ALVAC prime in the RV144 study and substantial differences in the risk factors and routes of HIV-1 infection for the populations studied. In summary, human efficacy trials indicate that vaccine-elicited protection against HIV-1 infection is achievable, but the specific antibody responses that may contribute to protection have not been elucidated. Despite these limitations in our knowledge of immune correlates, the modest 31% protection observed in the RV144 study suggests much room for improvement. One means of achieving improved vaccine efficacy may be through the elicitation of more potent and cross-reactive NAbs.
HUMORAL IMMUNE RESPONSE DURING HIV-1 INFECTION
Most vaccines seek to mimic the immune response generated during natural infection. HIV-1, however, shows extensive genetic variability owing to error-prone reverse transcription of the viral genome and a high tolerance for mutations that mediate viral immune evasion to antibody and CD8 T-cell responses. As a consequence of chronic persistent replication in the face of immune pressure, the HIV-1 epidemic is comprised of diverse genetic variants and a poorly understood level of antigenic diversity. A critical feature of HIV-1 appears to be its general resistance to NAbs. This resistance is manifest during the early phase of HIV-1 infection, in which NAb responses to the infecting virus may not appear until several months after infection. Once such autologous NAbs arise, the virus quickly escapes, leading to an ongoing cycle of adaptive antibody responses and further viral escape (Albert et al. 1990; Montefiori et al. 1991; Richman et al. 2003; Wei et al. 2003; Flynn et al. 2005; Gilbert et al. 2005). Despite this impressive immune evasion capability, recent evidence suggests that there are some vulnerabilities in the protective armor of HIV-1 that could be exploited by rationally designed antibody-based vaccines.
The advent of high-throughput neutralization assays using recombinant Env pseudoviruses has permitted the screening of sera from relatively large cohorts of HIV-1-infected donors. These data reveal that between 10% and 25% of HIV-1-infected subjects make NAb that cross-react with a substantial portion of diverse HIV-1 strains (Dhillon et al. 2007; Li et al. 2007; Doria-Rose et al. 2009; Sather et al. 2009; Stamatatos et al. 2009). A smaller subset of these sera is able to neutralize the large majority of circulating HIV-1 isolates (Li et al. 2007; Simek et al. 2009). The observation that some individuals make broadly cross-reactive neutralizing antibodies has spurred a renewed effort to isolate neutralizing mAbs to define vulnerable epitopes on the viral Env that could serve as targets of vaccine design. Until 2009, only a handful of known neutralizing mAbs were able to neutralize primary isolates of HIV-1, and even these antibodies displayed limitations in overall potency or breadth of reactivity (reviewed in Zolla-Pazner 2004; Pantophlet and Burton 2006; Mascola and Montefiori 2010). The effort to isolate new mAbs has been bolstered by several recent technological advances. Multiparameter flow cytometry can be used to identify and sort individual HIV-1 Env-specific memory B cells (Scheid et al. 2009). The antibody heavy and light chain can then be genetically recovered from the cDNA of single B cells and the full IgG expressed. An alternate methodology involves the screening of thousands of unselected individual memory B cells, each stimulated to secrete IgG. The supernatants of these B-cell cultures are screened for HIV-1 neutralization using a high-throughput microneutralization assay (Walker et al. 2009). The application of these new B-cell technologies has resulted in the isolation of several new HIV-1 mAbs that are more potent and broadly reactive than were prior antibodies. For example, the PG9 and PG16 mAbs are directed to a previously undefined site within the V2-V3 region of the HIV-1 viral spike (Walker et al. 2009). Another example is the VRC01, VRC02, and VRC03 mAbs that bind to a functionally conserved region of gp120 that interacts with the host cell receptor CD4 (Wu et al. 2010; Zhou et al. 2010). The fairly rapid and efficient isolation of these new mAbs suggests that numerous additional neutralizing mAbs will be forthcoming. Because each category of new broadly neutralizing mAbs potentially identifies a highly conserved target of HIV-1 neutralization, the structural analysis of these antibodies bound to HIV-1 Env can provide valuable insights for vaccine design.
STRUCTURAL VIROLOGY
Structural biology provides atomic-level details about the three-dimensional organization and chemical structure of proteins. Such details facilitate an understanding of mechanism and—combined with recent advances of in silico protein design—provide a means by which to manipulate and to optimize Env-based immunogens (Nabel et al. 2011). Overall, the host-derived envelope that surrounds HIV-1 sequesters most of its proteins from antibody-mediated recognition. Only two HIV-1 proteins protrude through this protective membrane: the HIV-1 gp120 envelope glycoprotein and its gp41 trans-membrane partner. Three gp120s associate noncovalently with three gp41s to make up the viral spike (reviewed in Wyatt and Sodroski 1998).
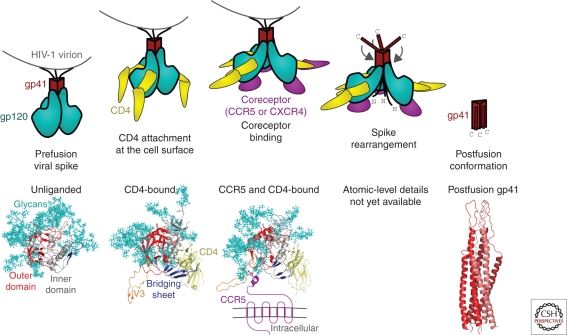
A combination of X-ray crystallography and electron microscopy has served to illuminate many of the details of the viral spike (Fig. 1). HIV-1 uses a two-receptor mechanism involving the host receptor CD4 and a coreceptor (generally CCR5 or CXCR4), along with considerable Env-based conformational change, to fuse viral and target cell membranes and to enter host cells (Wyatt and Sodroski 1998). In the first or “unliganded” state, the HIV-1 viral spike covers most of its surface with host-derived N-linked glycan (Wyatt et al. 1998). These glycans appear as “self” to the humoral immune system and therefore are mostly immunologically silent. In the unliganded state, only a few nonglycosylated sites are available for antibody-based recognition. After CD4 engages gp120 at the cell surface, the HIV-1 viral spike undergoes substantial structural rearrangements: in the CD4-bound conformation of gp120, a four-stranded bridging sheet forms and the V3 loop is sprung, and in gp41, the previously occluded N-heptad repeat becomes available for T-20 recognition (reviewed in Wyatt and Sodroski 1998). In the CD4-bound state, much more of the glycan-free viral spike surface is uncovered and potentially available for recognition by neutralizing antibody, such as V3-directed antibodies or CD4-induced (CD4i) antibodies that bind to the bridging sheet. However, the close proximity of viral and target cell membrane sterically occludes access of immunoglobulins to these epitopes (Labrijn et al. 2003). CD4 engagement induces the formation of a high-affinity site for coreceptor binding (Wu et al. 1996). Although details remain to be deciphered, coreceptor binding facilitates additional conformational changes. A transition intermediate has been hypothesized to form, in which gp41 is fully extended, with a fusion peptide thrown into the target cell membrane and the carboxy-terminal trans-membrane region embedded in the viral membrane. This transient intermediate resolves itself by forming a highly stable six-helix bundle, the postfusion state, in which fusion peptide and trans-membrane regions of gp41 are juxtaposed.
Figure 1.
Mechanistic and atomic-level details of the HIV-1 viral spike, a fusion machine that also evades humoral detection. (Top row) Entry schematic. The viral spike is composed of three gp120 envelope glycoproteins (cyan) and three gp41 trans-membrane (red). In the prefusion conformation, the outer surface of the spike is covered with N-linked glycan, which is seen as “self” by the humoral immune system therefore virtually invisible to potentially neutralizing antibody. At the cell surface, binding to CD4 (yellow) induces large structural rearrangements, which include the formation of a binding site for a second requisite coreceptor (purple). Coreceptor binding induces a transient intermediate, with the amino-terminal fusion peptide of gp41 thrown into the target cell membrane while the carboxy-terminal gp41-trans-membrane region is buried in the viral membrane. Subsequent spike rearrangements resolve into a stable postfusion conformation, with the fusion peptide and trans-membrane regions of gp41 in close proximity. (Bottom row) Atomic-level structures, with polypeptide backbones shown in ribbon representation and N-linked glycans in stick representation. The unliganded conformation of HIV-1 gp120 in its viral spike conformation is unknown, but an atomic model has been solved for an SIV core (Chen et al. 2005), in which the N-linked glycans (cyan) cluster onto one face of gp120. The CD4-bound conformation of gp120 (Kwong et al. 1998; Huang et al. 2005) contains a four-stranded bridging sheet (blue) and a protruding V3 region (orange), both of which interact with CCR5 (Rizzuto et al. 1998), although only the amino-terminal region of the bound CCR5 structure has been determined (Huang et al. 2007). The gp41 conformations of HIV-1 in prefusion and intermediate stages is unknown, but the postfusion conformation (Chan et al. 1997; Weissenhorn et al. 1998) reveals a stable six-helix bundle, with structural similarity to other type 1 viral fusion machines.
SITES OF HIV-1 VULNERABILITY
The assembled viral spike is highly protected from antibody-mediated recognition. Nonetheless, as described above, a number of broadly neutralizing antibodies have recently been discovered. The breadth of these antibodies indicates that they recognize conserved Env regions of functional importance; conversely, the susceptibility of these regions to antibody-mediated recognition indicates that they form sites of HIV-1 vulnerability to the humoral immune response.
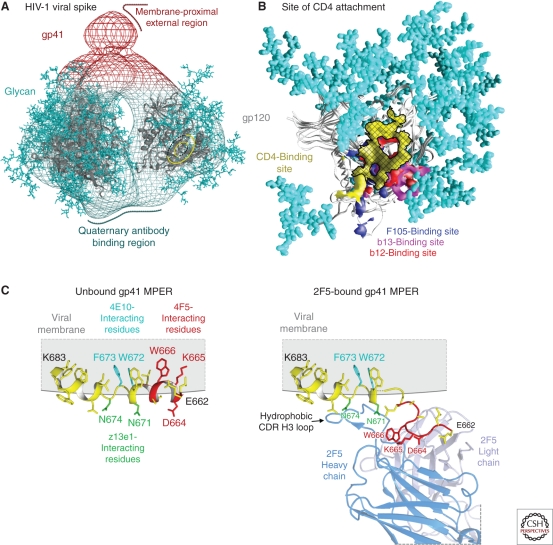
One such site of Env vulnerability is the initial site of attachment for the CD4 receptor (Fig. 2A,B) (Zhou et al. 2007; Chen et al. 2009). This site, on the outer domain of gp120, consists of roughly two-thirds of the surface HIV-1 uses to fully engage the CD4 receptor. Although the site is itself glycan-free, N-linked glycosylation and variable loops (e.g., V5) surround much of it. Moreover, most of the surrounding surface that is glycan-free is highly susceptible to conformational change; antibodies that bind to these regions induce conformations in the neighboring V1/V2 variable regions that are incompatible with the functional viral spike. Precise targeting by antibodies onto the initial site of CD4 attachment thus appears to be required for effective neutralization at this site (Chen et al. 2009).
Figure 2.
Sites of HIV-1 vulnerability to neutralizing antibody. (A) Electron tomogram of the HIV-1 viral spike (Liu et al. 2008), with docked atomic-level structure of gp120 (Pancera et al. 2010a) and with sites of vulnerability to antibody-mediated neutralization identified. (B) Initial site of CD4 attachment (Zhou et al. 2007; Chen et al. 2009). The site of vulnerability to antibody is shown as a cross-hatched yellow surface, with regions that induce conformational change and are binding sites for antibody F105 (blue) and antibody b13 (purple) or regions that extend outside of the site such as that recognized by antibody b12 (red). Neighboring N-linked glycan is shown in cyan. (C) MPER and membrane context. The MPER contains a number of highly conserved tryptophans, which are important for its entry function. (Left) Model of MPER in membrane from NMR/EPR measurements. The structure of MPER residues 662–683 is shown in the context of a DPC micelle (Sun et al. 2008), with residues required for recognition by neutralizing antibodies 2F5, z13e1, and 4E10 colored in red, green, and cyan, respectively. (Right) Model of MPER bound by broadly neutralizing antibody 2F5 as inferred from the crystal structure of the 2F5-epitope in complex with its gp41-MPER epitope (Ofek et al. 2004). The 2F5 antibody (partially shown with heavy chain in blue and light chain in gray) extracts its epitope from a helical conformation and induces an extended loop (Song et al. 2009). This has been modeled with the epitope as defined in the crystal structure (red) connected through a schematic dashed yellow line into the carboxy-terminal portion of the MPER (the structure is not known of the complete MPER when bound by the antibody nor the relative orientations of the amino- and carboxy-terminal portions of the MPER in this context). Virus neutralization by antibodies 2F5, z13e1, and 4E10, furthermore, also appears to require interactions with the surrounding membrane, likely mediated by extended CDR H3 loops, all of which contain hydrophobic motifs capable of interacting with membrane (Alam et al. 2009; Julien et al 2010; Ofek et al. 2010a,b; Scherer et al. 2010).
The first monoclonal antibody found to bind effectively to this site, the b12 antibody, was identified from a phage library (Burton et al. 1994) and found to use a heavy-chain-only means of recognition (Zhou et al. 2007), which is generally not observed in naturally elicited antibodies. The b12 antibody nevertheless displays reasonable recognition of the site (Zhou et al. 2007), although extension outside the target site, especially around the CD4-binding loop, allowed for antigenic variation, especially with non-B clade isolates (Wu et al. 2009b). As a consequence, the b12 antibody only neutralizes ∼35% of circulating isolates.
Other antibodies have been isolated more recently that also target this site. Antibody HJ16 is a natural human antibody isolated by direct assessment of neutralization coupled to single B-cell antibody sequencing (Corti et al. 2010). The atomic-level structure of this antibody in complex with gp120 has not yet been determined, but initial mapping suggests that antibody HJ16 strays into the “loop D” region, outside of the site of vulnerability (Corti et al. 2010); overall HJ16 neutralizes ∼30% of circulating isolates (Corti et al. 2010). Antibody VRC01 and related antibodies VRC02 and VRC03 appear to target the site of vulnerability more precisely. These antibodies show neutralization breadths of up to ∼90% of circulating isolates (Wu et al. 2010). The structure of VRC01 in complex with gp120 reveals an extraordinary mimicry between VRC01 heavy chain and CD4 receptor (Zhou et al. 2010).
Another site of vulnerability on the viral spike shows quaternary structural constraints, and maps to the second and third variable regions of gp120 (variable loops V2 and V3) (Fig. 2C). The structure of this portion of the Env has not yet been determined, and its functional importance is also not clear, but may relate to HIV-1 recognition of the α4β7-integrin, the gut-homing receptor for HIV-1 used by primary isolates during early stages of infection (Arthos et al. 2008). A number of antibodies have been identified that recognize this region, including the human monoclonal antibodies 2909, PG9, and PG16 and a number of rhesus antibodies (Gorny et al. 2005; Walker et al. 2009; Robinson et al. 2010). Structural analysis indicates that all of these antibodies use extended heavy chain third-complementarity determining regions, which are anionic and tyrosine sulfated (Pancera et al. 2010b; Pejchal et al. 2010; Changela et al. 2011). Despite this similarity, these antibodies vary dramatically in their neutralization breadth, with 2909 being extremely strain-specific and PG9/PG16 able to neutralize 70%–80% of current circulating isolates (Honnen et al. 2007; Walker et al. 2009). This divergence in breadth appears related to the specific immunotype of the quaternary site of vulnerability recognized, with strain-specific variants recognizing rare variants of the site, and more broadly neutralizing antibodies recognizing more common variants of the site (Wu et al. 2011).
Another critical function the HIV-1 envelope performs relates to fusion of viral and target cell membranes, which is required for virus entry. Virtually all of the functionally conserved surfaces required for fusion are occluded in the functional viral spike and available for antibody-mediated neutralization only as transient intermediates in the entry process. Even then, access is limited. For example, entry requires the N-heptad repeats of gp41 to snap back on themselves, and small molecule mimics of the carboxy-terminal heptad repeat (e.g., T-20 or Fuzeon) are effective therapeutics (Baldwin et al. 2003). Nonetheless, antibodies that recognize this potential site of vulnerability such as D5 or HK20 have weak potency and limited breadths of neutralization, properties attributed to steric occlusion at the viral membrane-target cell membrane interface (Luftig et al. 2006; Gustchina et al. 2010; Sabin et al. 2010). One area of vulnerability that appears less sterically occluded is the membrane-proximal external region (MPER) (Fig. 2C). The precise role that this region plays in viral entry is unclear, but alteration of hydrophobic residues in the MPER leads to loss of fusion capabilities (Munoz-Barroso et al. 1999; Salzwedel et al. 1999). Human antibodies 2F5, Z13e, and 4E10 have been found to recognize the MPER and to show reasonable neutralization breadths and potencies (Muster et al. 1993, 1994; Trkola et al. 1995; Stiegler et al. 2001; Zwick et al. 2001; Binley et al. 2004). Interestingly, this site of HIV-1 vulnerability includes not only the HIV-1 Env (i.e., specific amino acids in the MPER), but also the neighboring or surrounding lipid membrane. Antibodies that recognize the MPER thus require a hydrophobic membrane-binding component to neutralize virus (Ofek et al. 2004; Sun et al. 2008; Alam et al. 2009; Julien et al. 2010; Ofek et al. 2010b). Such membrane “corecognition” appears to lead to self-recognition, and the MPER-directed antibodies are generally self-reactive (Haynes et al. 2005; Alam et al. 2007). Such self-reactivity may impede antibody development (B-cell deletion or anergy), and few MPER-directed neutralizing antibodies are observed in sera from HIV-1-infected individuals (Walker et al. 2009).
A number of other sites of vulnerability can be inferred from antibodies that neutralize with different specificities. A conserved cluster of high-mannose glycans (around residues 295, 332, and 392 of gp120) is recognized by the 2G12 antibody (Sanders et al. 2002). This antibody has a highly unusual structure that involves variable-domain swapping (Calarese et al. 2003), and most sera do not competitively inhibit 2G12 binding, suggesting that antibodies against this site are very rare. A recent analysis of the elite neutralizers from the protocol G screen of sera from almost 2000 HIV-1-infected individuals, however, indicates that some of the best neutralizers recognize this face of the Env trimer (Simek et al. 2009; Walker et al. 2009, 2010a). Other antibodies directed at the V3 loop (up to 10%–30% breadth) or of CD4-induced specificity (up to 10% breadth) indicate that these regions form partial sites of vulnerability (Xiang et al. 2003; Zolla-Pazner and Cardozo 2010).
IMMUNOGEN DESIGN
CD4bs Immunogens
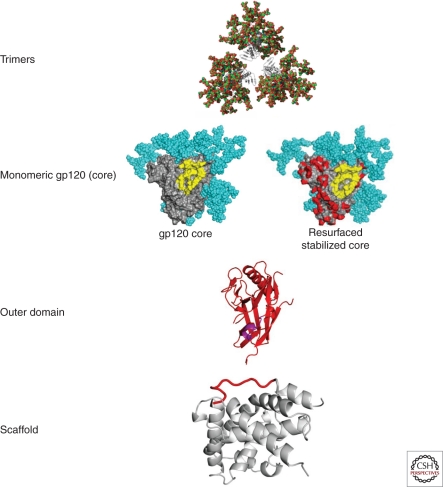
The molecular interactions of broadly neutralizing antibodies to the CD4bs have suggested at least four strategies to elicit these antibodies (Fig. 3). First, trimeric forms of HIV-1 Env have been generated by inclusion of the gp41 trimerization sites after deletion of the trans-membrane domain.The trimeric protein can be further stabilized by addition of trimerization sequences from such proteins as the fibritin protein from phage λ. Such trimers can be further stabilized with site-specific mutations previously shown to fix the core structure (Yang et al. 2002). In these prototypes, the variable V1-V3 domains are often removed to minimize immune responses to irrelevant strain-specific structures.
Figure 3.
Alternative forms of HIV Env serve as prototype immunogens for neutralizing antibody vaccines. Different forms of the HIV Env can be used to elicit neutralizing antibody responses. They range from the most complex form, the HIV trimer that most closely resembles the form found on the viral spike (upper row), monomeric forms, which include the gp120 core or resurfaced stabilized cores (second row), a region of the core that is composed primarily of the outer domain (OD) which includes the CD4-binding site (third row), or selected subdomains, such as the CD4-binding β-15 loop or MPER attached to a heterologous stable scaffold (bottom row). Modification of these prototypes by deletion of variable regions, removal, or addition of glycans, stabilization with disulfide bonds or addition of space filling mutations can serve to alter immunogenicity and elicit antibodies of the desired specificity.
An alternative approach is to develop immunogens based on a monomer structure (Fig. 3, second panel). Such proteins have been derived from stabilized core Env proteins further altered based on an understanding of structure (Zhou et al. 2007, 2010; Wu et al. 2010). Bioinformatic design has suggested mutations that replace HIV residues on the nonneutralizing inner domain of gp120 with SIV Env residues, minimizing serologic cross-reactivity with HIV-1. The surface of the conformationally stabilized Env core protein was modified and masked further with glycans. Such probes have been used to analyze antisera for the presence of broadly neutralizing antibodies and served also as prototype immunogens to elicit antibodies to this site.
A third approach focuses on generating a subdomain of the HIV-1 Env, the outer domain that contains the initial site of CD4 attachment (Fig. 3, third panel). In this protein, a large portion of the inner domain that elicits nonneutralizing antibodies is eliminated. The immune response is therefore directed to the relevant site of initial CD4 binding. In addition, by removing parts of the inner domain required for CD4 binding, potential inhibition by CD4 attachment is avoided. In theory, modified forms of the outer domain will allow targeting of the immune response to the most relevant conserved region of binding (e.g., the critical β15 loop that interacts with both the b12 and VRC01 broadly neutralizing antibodies). Although a soluble form of the outer domain that contains the β15 loop did not bind to b12 with high affinity in early studies (Yang et al. 2004), inclusion of a trans-membrane domain (Wu et al. 2009a) or further site-directed mutagenesis based on the VRC01/Env structure has stabilized this interaction and increased b12 or VRC01 binding. Additional mutations in the outer domain region have been designed to preserve high-affinity binding and are currently under evaluation both as probes of serum neutralizing antibody activity and as immunogens. A fourth approach to CD4bs immunogen development employs scaffolds based on informatics and epitope transplantion (Fig. 3, lower panel) (e.g., adding the β15 loop to an unrelated structure that presents the epitope naturally). Scaffolds have been identified that bind CD4bs antibodies and are the subject of continued investigation.
Glycan and Quaternary Immunogens
At least two types of antibodies recognize carbohydrate determinants on HIV-1 Env. Among these antibodies are those like the prototypic 2G12 mAb that recognize high-mannose structures on the outer domain (Buchacher et al. 1994; Trkola et al. 1995; Calarese et al. 2003). Although 2G12 interacts with these glycans through an unconventional arrangement of its antigen-binding sites, additional antibodies directed to this region have been described recently that display exceptionally high affinity and breadth (Walker et al. 2010a). There is concern that it will be difficult to generate antibodies to carbohydrate structures added by endogenous glycosylation machinery, and even if it is possible, there is concern that such antibodies may react with such carbohydrates on host proteins. Nonetheless, several groups have attempted to develop immunogens by chemical conjugation with nonhuman glycans or by selection in yeast (Agrawal-Gamse et al. 2011). Although some structures have been defined that react with 2G12, it has not yet been possible to elicit such neutralizing antibodies with these immunogens.
Another class of exceptionally neutralizing antibodies has been directed to glycans, and possibly peptide sequences in the V1/V2 region, with additional interactions dependent on the V3 region (Walker et al. 2009). Such immunoglobulins have been termed quaternary antibodies, although it is not certain that they are directed to complex conformational determinants from different parts of Env. Although the structure of at least one such antibody has been elucidated, the molecular details of its interaction with Env remain hypothetical. Efforts at immunogen design have focused thus far on membrane-bound trimers, which show the strongest binding to these antibodies (Walker et al. 2009); however, occasional monomeric gp120 derivatives have been identified and are also under investigation. Although this class of antibody represents ∼25% of the antibodies in the sera from subjects with broadly neutralizing responses (Walker et al. 2010b), it has not yet been possible to elicit these antibodies by vaccination in animal models or humans.
MPER, V3, and CD4i Immunogens
At least two broadly neutralizing mAbs, 2F5 and 4E10, recognize the highly conserved MPER region of Env. Both show reasonable breadth of neutralization, although their potency is generally low. Structures of these antibodies complexed to their cognate peptides have been determined and suggest that hydrophobic patches are required for stable interactions needed for neutralization. In the case of 4E10, efforts to develop immunogens based on stabilized peptides have allowed the definition of vaccine candidates with appropriate antigenic profiles but they do not elicit neutralizing antibodies. In the case of 2F5, a variety of approaches have elicited antibodies that react with 2F5 peptides but these antibodies do not neutralize diverse viral strains. An understanding of the structural interactions of 2F5 with its viral target has suggested that the structure of the 2F5 peptide in the context of gp120 as well as its interaction with the viral membrane through a hydrophobic patch are required for neutralization (Ofek et al. 2010b). Based on these findings, Ofek and colleagues have generated scaffolds that present the constrained 2F5 peptide which can elicit antibodies that interact similar to the 2F5 ab, suggesting a potential strategy to elicit such neutralizing antibodies (Ofek et al. 2010a). At the same time, it is difficult to elicit antibodies that retain the hydrophobic patch required for neutralization because these antibodies are usually polyreactive (Haynes et al. 2005) and therefore likely to undergo clonal deletion because of their autoreactivity.
The V3 and CD4i regions have also received considerable attention with respect to immunogen design and have been reviewed elsewhere. It is possible to elicit neutralizing antibodies to these V3 epitopes using modified murine retroviral gp or HIV-1 Env proteins (Chakrabarti et al. 2002; Zolla-Pazner et al. 2008). Because the V3 region is not exposed on most naturally circulating viral isolates, it is unlikely that these responses would be effective in the absence of an activity that would increase its exposure. Thus far, no such agonistic antibodies have been identified and this target therefore poses a considerable challenge. Similarly, CD4i antibodies can also be readily induced after immunization with HIV Env stabilized core proteins (Dey et al. 2009). Although such antibodies are seen frequently in HIV-infected individuals (Decker et al. 2005; Gray et al. 2007), they do not mediate neutralization, although it remains possible that they may contribute to protection through ADCC function.
Fundamental B-Cell Biology and the Antibody Response
To elicit a robust neutralizing antibody response to HIV-1, an understanding of B-cell biology is required. Immunogens must engage the appropriate naïve B-cell receptors to induce antibodies of the appropriate specificity. In addition, autoreactive B cells will be eliminated by clonal deletion, and the process of immunization likely must drive somatic mutations that are required for affinity maturation and development of high-affinity antibodies with the appropriate specificity. Critical to the success of rational vaccine design is the ability to take advantage of these factors and address the basic aspects of B-cell development that control antibody specificity and synthesis. For example, immunogens will need to engage the low-affinity germline precursors in a way that facilitates the development of high-affinity antibodies. In this regard, it is important to understand the impact of adjuvants and/or delivery matrices on the generation of antibody diversity and production. At the same time, such delivery agents must have the necessary safety and immunogenicity profiles required for widespread use. Such adjuvants may include alum, saponin-based emulsions, ASO1A and B, ASO2, MF59, nanoparticles, and multimeric viral carriers, such as Qβ. The choice of animal models for testing is also critical, because not all species show similar degrees of somatic mutation, have similar genomic precursors, nor have the ability to make long CDR3 regions as found in humans. Candidates would be optimally tested in relevant humanized mouse models and NHPs before progression into phase I human clinical trials. When possible, it is also desirable to test relevant immunogens for protection in NHP challenge studies and proof of concept and to determine effective preventive antibody levels.
CONCLUDING REMARKS
HIV-1 has evolved multiple mechanisms to evade the neutralizing antibody response. Specifically, Env evades host recognition by virtue of its sequence diversity, limited exposure to the immune system because of carbohydrate masking, and conformational flexibility. HIV-1 has been resistant to classification by traditional serotyping, suggesting that standard approaches to vaccine development are unlikely to succeed. Recent progress in the definition of broadly neutralizing antibodies, the elucidation of the structures of these antibodies complexed to HIV-1, and the utilization of structural biology to define the relationship between antigenicity and immunogenicity in vaccine candidates has catalyzed a resurgence in this area of research. An understanding of the biology of HIV-1, the human immune response to the virus, and the application of structural biology to immunogen design have provided new opportunities to advance the goal of identifying vaccines that elicit broadly neutralizing antibodies that prevent or contain infection and/or progression to AIDS.
ACKNOWLEDGMENTS
We thank Ati Tislerics for manuscript editing, Gilad Ofek for assistance with Figure 2C, and Brenda Hartman and Jonathan Stuckey for assistance with graphic arts.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
- Agrawal-Gamse C, Luallen RJ, Liu B, Fu H, Lee FH, Geng Y, Doms RW 2011. Yeast-elicited cross-reactive antibodies to HIV Env glycans efficiently neutralize virions expressing exclusively high-mannose N-linked glycans. J Virol 85: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, et al. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol 178: 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, et al. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci 106: 20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo EM 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4: 107–112 [DOI] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, et al. 2008. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 9: 301–309 [DOI] [PubMed] [Google Scholar]
- Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med 6: 200–206 [DOI] [PubMed] [Google Scholar]
- Baldwin CE, Sanders RW, Berkhout B 2003. Inhibiting HIV-1 entry with fusion inhibitors. Curr Med Chem 10: 1633–1642 [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 78: 13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein–Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10: 359–369 [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, et al. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300: 2065–2071 [DOI] [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu B-Y, Yang Z-Y, Friborg J Jr, Ling X, King SR, Montefiori DC, Nabel GJ 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol 76: 5357–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A, Wu X, Yang Y, Zhang B, Zhu J, Nardone GA, O’Dell S, Pancera M, Gorny MK, Phogat S, Robinson JE, Stamatatos L, Zolla-Pazner S, Mascola JR, Kwong PD 2011. Crystal structure of human antibody 2909 reveals conserved features of quaternary-specific antibodies that potently neutralize HIV-1. J Virol 85: 2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS 1997Core structure of gp41 from the HIV envelope glycoprotein. Cell 89: 263–273 [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433: 834–841 [DOI] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326: 1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, et al. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 5: e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201: 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, et al. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5: e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, Lee FH, Richman DD, Doms RW, Vanham G, et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1–infected donors. J Virol 81: 6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Klein RM, Manion MM, O’Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, et al. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 83: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Kwong PD, Nabel GJ 2006. The rational design of an AIDS vaccine. Cell 124: 677–681 [DOI] [PubMed] [Google Scholar]
- Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 191: 654–665 [DOI] [PubMed] [Google Scholar]
- Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, Heyward WL, Jobes DV, Popovic V, Self SG, et al. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis 191: 666–677 [DOI] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol 79: 5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol 81: 6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustchina E, Li M, Louis JM, Anderson DE, Lloyd J, Frisch C, Bewley CA, Gustchina A, Wlodawer A, Clore GM 2010. Structural basis of HIV-1 neutralization by affinity matured Fabs directed against the internal trimeric coiled-coil of gp41. PLoS Pathog 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair WE, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908 [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, et al. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449: 101–104 [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR 2009a. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 15: 951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR 2009b. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 5: e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Smith BA, Baba TW, Liska V, Ferrantelli F, Montefiori DC, McClure HM, Anderson DC, et al. 2001. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J Virol 75: 7470–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnen WJ, Krachmarov C, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J Virol 81: 1424–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310: 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, et al. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317: 1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Huarte N, Maeso R, Taneva SG, Cunningham A, Nieva JL, Pai EF 2010. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol 84: 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Bjorkman PJ 2010. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog 6: e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Gnanapragasam PN, Galimidi RP, Foglesong CP, West AP Jr, Bjorkman PJ 2009. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci 106: 7385–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288: 1789–1796 [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393: 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77: 10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med 13: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luftig MA, Mattu M, Di Giovine P, Geleziunas R, Hrin R, Barbato G, Bianchi E, Miller MD, Pessi A, Carfi A 2006. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat Struct Mol Biol 13: 740–747 [DOI] [PubMed] [Google Scholar]
- Mascola JR 2003. Defining the protective antibody response for HIV-1. Curr Mol Med 3: 209–216 [DOI] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC 2010. The role of antibodies in HIV vaccines. Annu Rev Immunol 28: 413–444 [DOI] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis 173: 340–348 [DOI] [PubMed] [Google Scholar]
- Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Dayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, et al. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol 73: 4009–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, et al. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med 6: 207–210 [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Zhou IY, Barnes B, Lake D, Hersh EM, Masuho Y, Lefkowitz LB Jr 1991. Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology 182: 635–643 [DOI] [PubMed] [Google Scholar]
- Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol 73: 6089–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67: 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol 68: 4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel GJ, Kwong PD, Mascola JR 2011. Progress in the rational design of an AIDS vaccine. Philos Trans R Soc Lond B Biol Sci (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Igarashi T, Haigwood N, Sadjadpour R, Plishka RJ, Buckler-White A, Shibata R, Martin MA 2002. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J Virol 76: 2123–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 78: 10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD 2010a. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci 107: 17880–17887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, Wyatt R, Zwick MB, Nabel GJ, Mascola JR, et al. 2010b. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity–determining region. J Virol 84: 2955–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. 2010a. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci 107: 1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, et al. 2010b. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure–function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol 84: 8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR 2006. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24: 739–769 [DOI] [PubMed] [Google Scholar]
- Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol 75: 8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci 107: 11483–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361: 2209–2220 [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci 100: 4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280: 1949–1953 [DOI] [PubMed] [Google Scholar]
- Robinson JE, Franco K, Elliott DH, Maher MJ, Reyna A, Montefiori DC, Zolla-Pazner S, Gorny MK, Kraft Z, Stamatatos L 2010. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J Virol 84: 3443–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin C, Corti D, Buzon V, Seaman MS, Lutje HD, Hinz A, Vanzetta F, Agatic G, Silacci C, Mainetti L, et al. 2010. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog 6: e1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K, West JT, Hunter E 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol 73: 2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76: 7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458: 636–640 [DOI] [PubMed] [Google Scholar]
- Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR 2010. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci 107: 1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho MW, Martin MA 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med 5: 204–210 [DOI] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83: 7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Sun ZY, Coleman KE, Zwick MB, Gach JS, Wang JH, Reinherz EL, Wagner G, Kim M 2009. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci 106: 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Morris L, Burton DR, Mascola JR 2009. Neutralizing antibodies generated during natural HIV-1 infection: Good news for an HIV-1 vaccine? Nat Med 15: 866–870 [DOI] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 17: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28: 52–63 [DOI] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF III, Burton DR, Ho DD 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol 69: 6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Chan-Hui P, Ramos A, Simek M, Falkowska E, Doores K, Hammond P, Wrin T, Mosley B, Olsen O, et al. 2010a. High through-put functional screening of activated B cells from 4 African elite neutralizers yields a panel of novel broadly neutralizing antibodies [abstract]. AIDS Res Hum Retroviruses 26: A-149–A-150 [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR 2010b. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6: e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM 2003. Antibody neutralization and escape by HIV-1. Nature 422: 307–312 [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 2: 605–616 [DOI] [PubMed] [Google Scholar]
- Wrin T, Nunberg JH 1994. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS 8: 1622–1623 [DOI] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384: 179–183 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou T, Yang ZY, Svehla K, O’Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, et al. 2009a. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol 83: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, O'Dell S, Wyatt RT, Kwong PD, Mascola JR 2009b. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody b12 that effectively targets the site of CD4 attachment. J Virol 83: 10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329: 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Changela A, O'Dell S, Schmidt SD, Pancera M, Yang Y, Zhang B, Gorny MK, Phogat S, Robinson JE, Stamatatos L, Zolla-Pazner S, Kwong PD, Mascola JR 2011. Immunotypes of a quaternary site of HIV-1 vulnerability and their recognition by antibodies. J Virol 85: 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J 1998. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 280: 1884–1888 [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393: 705–711 [DOI] [PubMed] [Google Scholar]
- Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315: 124–134 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol 76: 4634–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol 78: 12975–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445: 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329: 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol 4: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cardozo T 2010. Structure–function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol 10: 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S 2008. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology 372: 233–246 [DOI] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, et al. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75: 10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]