Abstract
Expansion of trinucleotide repeats (TNRs) is responsible for a number of human neurodegenerative disorders. The molecular mechanisms that underlie TNR instability in humans are not clear. Based on results from model systems, several mechanisms for instability have been proposed, all of which focus on the ability of TNRs to form alternative structures during normal DNA transactions, including replication, DNA repair and transcription. These abnormal structures are thought to trigger changes in TNR length. We have previously shown that transcription-induced TNR instability in cultured human cells depends on several genes known to be involved in transcription-coupled nucleotide excision repair (NER). We hypothesized that NER normally functions to destabilize expanded TNRs. To test this hypothesis, we bred an Xpa null allele, which eliminates NER, into the TNR mouse model for spinocerebellar ataxia type 1 (SCA1), which carries an expanded CAG repeat tract at the endogenous mouse Sca1 locus. We find that Xpa deficiency does not substantially affect TNR instability in either the male or female germline; however, it dramatically reduces CAG repeat instability in neuronal tissues—striatum, hippocampus and cerebral cortex—but does not alter CAG instability in kidney or liver. The tissue-specific effect of Xpa deficiency represents a novel finding; it suggests that tissue-to-tissue variation in CAG repeat instability arises, in part, by different underlying mechanisms. These results validate our original findings in cultured human cells and suggest that transcription may induce NER-dependent TNR instability in neuronal tissues in humans.
INTRODUCTION
Expansion of CAG•CTG repeats in specific human genes cause several neurodegenerative and neuromuscular diseases, including Huntington disease (HD), myotonic dystrophy type 1 (DM1) and several spinocerebellar ataxias (SCAs) (1–3). Long CAG repeat tracts in disease genes tend to be unstable in the germline, giving rise to progeny that carry either longer repeat tracts (expansions) or shorter ones (contractions). The typical bias toward expansion generally leads to a more debilitating disease phenotype in the affected offspring, with earlier onset and more severe symptoms (4). CAG repeat instability, however, is not confined to the germline. As individuals age, the ongoing expansion-biased instability in somatic tissues, especially in the pathologically important tissues of the brain, may hasten the onset of neuron dysfunction and death, exacerbating the disease phenotype (5,6). The extent, or rate, of instability often differs in the male and female germlines, and typically varies from tissue to tissue. The mechanisms of repeat instability in germline and somatic tissues have been the subject of intense investigation, but the underlying processes in humans remain uncertain.
Numerous studies in model systems, including bacteria, yeast, flies and human cells, have identified potential contributors to repeat instability in humans that encompass all the basic DNA transactions: replication, DNA repair, recombination and transcription (4,7,8). These processes expose single strands of repeats, which can form secondary structures such as hairpins and slipped-strand duplexes (9,10) that are thought to be the key intermediates that lead to repeat instability. In addition, epigenetic modifications, chromatin structure and local sequence effects also contribute to repeat instability (11,12). The diversity of mechanisms identified in model systems makes it difficult to be certain which ones account for the instability seen in human germline and somatic tissues. Relevance to humans is usually tested by experiments in mice, where effects in germline and somatic tissues can be assayed. This approach has shown, for example, that the major maintenance DNA methyltransferase (Dnmt1) specifically affects CAG repeat stability in the germline (13), that the glycosylase Ogg1 selectively affects CAG repeat instability in somatic tissues (14,15) and that the mismatch repair (MMR) proteins Msh2 and Msh3 affect CAG instability in both germline and somatic tissues (16–18).
Using a selectable system for CAG repeat contraction in cultured human cells, we have shown that transcription promotes repeat contraction in a way that does not depend on DNA replication (19–21). A role for transcription in repeat instability has been confirmed in human cells for GAA repeats (22,23) and for very long CAG repeats, where transcription was shown to promote expansions as well as contractions (24). Transcription-induced CAG repeat instability is a complex process. It requires the MMR recognition complex formed by Msh2 and Msh3, which together can bind and stabilize CAG and CTG hairpins (25,26), but not the entire MMR pathway (27); it depends on genes involved in the transcription-coupled nucleotide excision repair (TC-NER) pathway, which can remove DNA structures that block RNA polymerase II (RNAPII) (28); it is enhanced by DNA–RNA hybrids (R-loops), which readily form in CAG repeat tracts (29–31); and it needs transcription factor TFIIS, which promotes backtracking of stalled RNAPII complexes (32,33), and the BRCA1/BARD1 E3 ligase and the proteasome complex, which may be responsible for the removal of stalled RNAPII complexes by ubiquitination and degradation (34–36). Interfering with any of these processes in our human cell assay decreases the frequency of transcription-induced CAG repeat contractions. Recently, we have shown that topoisomerase 1 (TOP1), tyrosyl DNA phosphodiesterase 1 (TDP1) and single-strand break repair (SSBR) collaborate to restrict transcription-induced CAG repeat instability; interference with the TOP1-TDP1-SSBR pathway increases contraction frequencies (37). Thus, the transcription-induced pathway for CAG repeat instability encompasses an extensive network of DNA repair and related processes.
Here, we sought to test whether transcription-induced CAG repeat instability might play a role in germline and somatic instability in a mouse model. If the pathway for transcription-induced CAG instability operates as we have shown in human cells, knocking out NER should alter repeat stability in germline or somatic tissues, or both. To test the involvement of NER, we chose to analyze Xpa-deficient spinocerebellar ataxia type 1 (SCA1) mice, which carry 145 CAG repeats at the Sca1 locus. We find that Xpa deficiency dramatically reduces the instability of CAG repeats in some somatic tissues, especially in the brain, but causes minimal effects on repeat instability in the germline. These results validate our original findings using an assay for CAG repeat contraction in human cells, and they suggest that transcription may induce NER-dependent CAG instability in specific somatic tissues in humans.
RESULTS
Effects of Xpa deficiency on germline repeat instability
To test for a role of NER in CAG repeat instability during germline transmission, we bred Xpa−/− mice with SCA1 mice, which carry an expanded CAG repeat in one Sca1 allele. From these crosses, we made use of Xpa+/− SCA1 and Xpa−/− SCA1 littermates. Since Xpa+/− mice possess a normal capacity for NER, even though they express only 50% of normal levels of Xpa (38,39), we expected that Xpa+/− SCA1 mice would behave like wild-type SCA1 mice. To assess instability in both germlines, we bred males and females of each genotype to wild-type mice. Male and female Xpa+/− SCA1 and Xpa−/− SCA1 mice were bred repeatedly to assess the effects of Xpa deficiency on the age-dependent germline instability that occurs at the Sca1 locus (13,40).
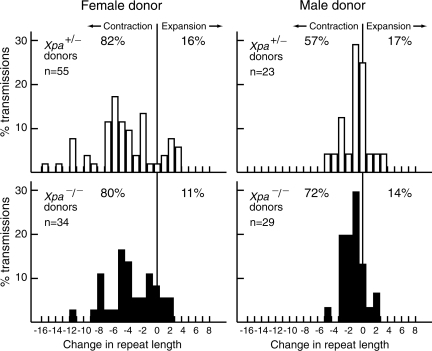
We identified progeny that carried the expanded allele, and compared the length of the repeat tract in progeny tail DNA taken at weaning to that of the SCA1 donor parent. Progeny from all ages of parent (from 10 weeks to >30 weeks of age) are grouped together in Figure 1. The CAG instability we observed in the male and female germlines of Xpa+/− SCA1 mice was indistinguishable from that reported previously for Xpa+/+ SCA1 mice (13,40). The distributions of tract length changes in the progeny of Xpa−/− SCA1 mice are similar to those observed in the progeny of Xpa+/−SCA1 mice (Fig. 1). Analysis using the nonparametric Mann–Whitney test indicates that the distributions are not significantly different for transmission through the male germline (P= 0.22) or the female germline (P= 0.27). From these studies, we conclude that Xpa deficiency does not significantly affect male or female germline instability of the expanded CAG repeat at the Sca1 locus.
Figure 1.
Intergenerational changes in CAG tract length in progeny of Xpa+/− SCA1 and Xpa−/− SCA1 mice. Percent transmission is the number of alleles with a given repeat length divided by the total number of alleles (n) multiplied by 100%. Change in repeat length is defined as the number of repeats in a progeny mouse minus the number of repeats in its donor parent.
Effects of Xpa deficiency on repeat instability in somatic tissues
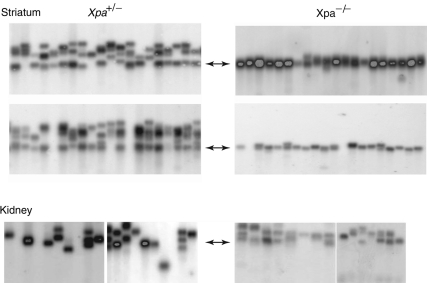
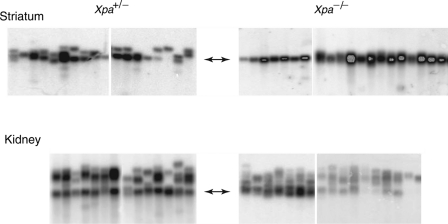
To test whether Xpa deficiency affects repeat instability in somatic tissues, we initially carried out small-pool PCR analysis of three tissues—cerebellum, kidney and striatum, which, respectively, display low, moderate and high instability of the CAG repeat tract in SCA1 mice (40). We analyzed tissue DNA from 45–50-week-old Xpa+/− SCA1 and Xpa−/− SCA1 mice that had served as parents in the germline instability assay described above. For all somatic tissues analyzed in this study, Xpa+/− SCA1 mice yielded CAG repeat instabilities that were indistinguishable from those previously reported for tissues from Xpa+/+ SCA1 mice (13,40). We initially analyzed old mice because of the age-dependence of somatic instability at the Sca1 locus (13,40). Small-pool PCR analysis of cerebellar DNA from both Xpa+/− SCA1 and Xpa−/− SCA1 mice revealed that CAG instability was too low to be informative (data not shown) (13,40). In contrast, repeat instability was clearly evident in the kidney and striatal samples from two different Xpa+/− SCA1 mice (Fig. 2). Remarkably, in samples from two Xpa−/− SCA1 mice, repeat instability in the striatum was virtually eliminated, whereas the instability in kidney samples was indistinguishable from that in Xpa+/− SCA1 mice (Fig. 2). We eliminated the trivial possibility that cells with expansions were dead and gone in these old Xpa−/− SCA1 mice by examining kidney and striatal samples from two 30-week-old mice, which showed the same dramatic difference as their older counterparts (Fig. 3). Thus, these results indicate that Xpa deficiency alters CAG repeat instability in some, but not all, somatic tissues.
Figure 2.
Somatic instability of CAG repeats at the Sca1 locus in the striatum and kidney from 45–50-week-old Xpa+/− SCA1 and Xpa−/− SCA1 mice. Thirty small-pool PCR reactions were prepared for each striatum and kidney sample from two different Xpa+/− SCA1 and Xpa−/− SCA1 mice, and the products were separated by gel electrophoresis and visualized by hybridization with a radioactive probe. Representative sections of the resulting phosphorimages are shown. Two-headed arrows indicate the length of the CAG repeat tract in tail DNA from the analyzed mice, which was included as a control, but is not shown here.
Figure 3.
Somatic instability of CAG repeats at the Sca1 locus in the striatum and kidney from 30-week-old Xpa+/− SCA1 and Xpa−/− SCA1 mice. Small-pool PCR reactions were prepared for each tissue sample from two different Xpa+/− SCA1 and Xpa−/− SCA1 mice and analyzed and displayed as described in Figure 2.
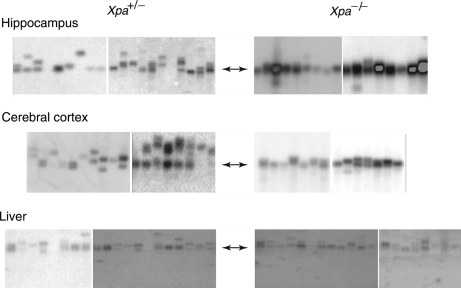
To extend these observations, we examined three additional tissues—liver, hippocampus and cerebral cortex—from two additional 45–50-week-old Xpa+/− SCA1 and Xpa−/− SCA1 mice. These tissues display moderate levels of CAG repeat instability (40). As shown in Figure 4, a similar tissue-specific difference was observed. CAG instability was dramatically reduced in the hippocampus and cerebral cortex from Xpa−/− SCA1 mice, but was not noticeably affected in the liver. Collectively, these results indicate that CAG repeat instability at the Sca1 locus is strikingly affected by Xpa deficiency in three brain tissues—striatum, hippocampus and cerebral cortex—but not in kidney or liver. To our knowledge, this is the first example of a genetic deficiency that differentially affects CAG repeat instability in different somatic tissues.
Figure 4.
Somatic instability of CAG repeats at the Sca1 locus in the hippocampus, cerebral cortex and liver from 45–50-week-old Xpa+/− SCA1 and Xpa−/− SCA1 mice. Small-pool PCR reactions were prepared for each tissue sample from two different Xpa+/− SCA1 and Xpa−/− SCA1 mice and analyzed and displayed as described in Figure 2.
Expression of Sca1 and Xpa in different somatic tissues
We originally chose to test the effect of Xpa deficiency on repeat instability in mice because we had linked transcription-dependent CAG repeat contraction to the activities of several genes known to be involved in TC-NER in human cells. In principle, the differential effects of Xpa deficiency on repeat instability in brain tissues versus kidney and liver could arise as a consequence of natural tissue-specific differences in sense or antisense transcription through the CAG repeat tract at the Sca1 locus or in the expression levels of the Xpa gene. To test these possibilities, we analyzed RNA from the Sca1 and Xpa genes in normal mouse tissues. As can be seen in Table 1, there is no obvious correlation between the expression levels of these genes and the tissues in which repeat instability is affected by Xpa deficiency. It is possible that the long CAG repeat could alter the natural proportions of the sense and antisense transcripts at the Sca1 locus. It is known, for example, that similar-length CAG repeats at the mouse HD locus decrease sense transcription ∼30% (41,42). However, much more extensive studies in mice with normal and expanded CAG repeats at the HD locus have demonstrated a similar lack of correlation between the level of stable transcripts from the Huntingtin gene, or from a variety of DNA repair genes, and the levels of tissue-specific CAG instability (43).
Table 1.
Expression of Sca1 and Xpa in various mouse tissues
| Tissue | Sca1 sensea ( × 102) | Sca1 antisensea ( × 102) | Xpa sensea ( × 102) |
|---|---|---|---|
| Whole brain | 56 | 0.35 | 2.5 |
| Cerebellum | 130 | 0.44 | 5.5 |
| Striatum | 19 | 0.42 | 1.0 |
| Hippocampus | 24 | 0.32 | 1.8 |
| Cerebral cortex | 23 | 0.51 | 1.0 |
| Kidney | 20 | 0.37 | 1.1 |
| Liver | 21 | 0.26 | 1.2 |
aLevels of Sca1 and Xpa RNA were measured relative to β-actin RNA. Values are multiplied by 100 for ease of comparison. The primers used for antisense detection gave a defined product in the presence of reverse transcriptase, but not in its absence. In addition, the detected level of antisense was ∼5-fold above the detection limit of the real-time RT–PCR assay.
DISCUSSION
Here, we have tested the hypothesis that NER modulates CAG repeat instability in a mouse model of SCA1. Our rationale for this study derives from two observations. First, we showed that several genes involved in the TC-NER pathway were required for the transcription-dependent contraction of CAG repeats in a selection assay in human cells (19,20). Second, Drosophila lines mutant for Mus201, the homolog of the NER Xpg gene, showed significantly reduced transcription-dependent CAG repeat instability in the fly germline (44). We chose to test the effects of Xpa deficiency because it is an essential component of NER and it does not have recognized activities outside that pathway (45). We show here that the expanded CAG repeat tract in Xpa−/− SCA1 mice displays dramatically reduced instability in several regions of the brain, but not in the kidney or liver. In contrast, expanded CAG repeats exhibit normal instability in both the male and female germlines of Xpa-deficient mice. Although individual genetic deficiencies have been found to differentially affect germline and somatic CAG repeat instability—for example, Dnmt1 deficiency affects only germline instability (13) and Ogg1 deficiency is specific for somatic tissues (14,15)—Xpa is the first example of a gene that modulates CAG instability in a selected set of somatic tissues.
The variation in CAG repeat instability from tissue to tissue remains one of the most puzzling features of CAG repeat instability in mouse models and human patients (46). We do not know the basis for this variability, whether the same or distinct mechanisms operate in different tissues, or whether the mechanisms of instability are the same at different repeat loci. A survey of tissue-specific instabilities reported for mouse models of DM1, HD, SCAs 1 and 7 and dentatorubral-pallidoluysian atrophy concluded that the patterns of tissue-specific repeat instability across diseases are similar (46). The kidney and liver typically show moderate instability, whereas repeats in muscle, heart and blood are fairly stable. Likewise, the striatum typically exhibits very high repeat instability, whereas the cerebral cortex and hippocampus display intermediate instability, and the cerebellum, very low instability. Within this overall similarity, specific differences exist; for example, in HD, CAG repeats in liver are more unstable than they are in the kidney, whereas in SCA1 the opposite is true (40,43). Nevertheless, the general similarity in the patterns of instability suggests that key elements of CAG repeat instability may be tissue-specific, rather than disease-locus-specific.
The striking tissue-specific differences in patterns of CAG repeat instability fostered a number of studies to identify the underlying cause. Studies on repeat instability in bacteria and yeast revealed a prominent role for replication (4,7,47), but in mouse models there is no obvious correlation between cell proliferation and CAG repeat instability: it is high in nonproliferating striatal neurons (48–51), does not correlate with tissue-specific cell proliferation rates (52–54) and occurs in meiotically arrested oocytes (55). Studies in bacteria and mammalian cells have shown that transcription through repeat tracts strongly destabilizes them (19–21,24,56–58). Nevertheless, tissue-specific measurements of mRNA from the affected genes in DM1 (52), HD (43) and SCA1 (Table 1) show that stable mRNA levels do not correlate with tissue-specific CAG repeat instability. Previous studies of CAG repeat instability in mouse somatic tissues have shown a critical dependence on the DNA repair genes Msh2, Msh3, Pms2 and Ogg1 (15,16,18,59–63), and it has been proposed that instability might depend on expression levels of particular DNA repair genes (40). Comparisons of tissue-specific gene expression and repeat instability in HD mouse models, however, revealed no correlation between expression of particular DNA repair genes and repeat instability (43). Indeed, extensive pathway analysis of gene expression data suggested that multiple tissue factors combine to give the observed levels of somatic instability in different tissues (43). Our analysis of the effects of Xpa deficiency in a mouse model of SCA1 provides genetic support for the concept that multiple mechanisms contribute to the tissue-specific CAG repeat instability in mice, and by extension in patients. Consistent with our results, a recent study in a DM1 mouse model indicates that mechanistic diversity likely extends to the germline, as well; Lig1 deficiency reduced CAG expansions in the female germline, but did not alter instability in the male germline (64).
Since the only known function of Xpa is as a component of the NER pathway (45), we conclude that NER is critical for CAG repeat instability in specific brain tissues in the SCA1 mouse model. Given our previous demonstration that transcription-dependent CAG repeat contraction in human cells depends on several genes involved in TC-NER (19,20), these results suggest that CAG repeat instability in these tissues may be induced by transcription at the Sca1 locus in a TC-NER dependent pathway. It was previously shown that deficiency of Xpc, a gene specific for global genome repair (a subpathway of NER that does not depend on transcription to trigger repair), did not affect germline transmission or instability in the striatum in an HD mouse model that carried 102 CAG repeats at the mouse homolog of the HD gene (63). These results are consistent with our observations in human cells, which showed that XPC knockdown did not affect transcription-dependent CAG repeat contraction (20).
A recent paper investigated the role of Cockayne syndrome B (Csb) protein, which is a component of NER that is specific for TC-NER (14). Using the transgenic R6/1 HD mouse model (50), which carries 130–140 CAG repeats in a randomly integrated 1.9 kb fragment of the human HD gene, the authors report that Csb−/− R6/1 mice display significantly increased repeat expansion in the germline, but no significant change in somatic instability in the whole brain: results contrary to the ones reported here. The basis for the difference is unclear. Knockdowns of XPA and CSB in a human cell system have given identical results in assays of CAG repeat contraction (19,37). The different results in mice could relate to differences in the systems—Sca1 knock-in versus R6/1 transgene, or SCA1 disease versus HD—or in the specific experimental approaches. For example, the authors’ conclusions about the germline effects of Csb deficiency are based on results with 10 progeny from Csb−/− R6/1 mice versus 8 progeny from Csb+/+ R6/1 mice, which constitutes a small study that tests only the male germline. In contrast, the results for Xpa deficiency compare 63 progeny from Xpa−/− SCA1 mice with 78 progeny from Xpa+/− SCA1 mice, with 52 coming through the male germline and 90 through the female germline. On the other hand, the differences could arise because Csb, unlike Xpa, has additional functions beyond its involvement in NER. As the authors point out, Csb possesses chromatin remodeling activity (65), affects transcription (66), binds to active transcription sites (67) and associates with RNA polymerase complexes (68–70). Resolution of the basis for the differences between results with Xpa and Csb awaits future studies.
Based on results with siRNA knockdowns of DNA repair components in human cells, we initially proposed that transcription-induced CAG repeat instability arose via stalling of RNAPII at repeat slip-outs, which then triggered TC-NER (19,20). Although our human cell system specifically assays for CAG contractions, we proposed a pathway that could also generate the expansions typically observed in somatic tissues (8,19). Transcription-dependent expansions were recently shown to occur at very long CAG repeat tracts in human fibroblasts (24). Elegant in vitro studies using transcription of slip-out substrates in HeLa nuclear extracts have now confirmed the central tenant of our model—that RNAPII can stall at repeat slip-outs (71). Moreover, the authors demonstrate that slip-outs on either the transcribed strand or the nontranscribed strand are effective in stalling RNAPII (71). They propose that either kind of stalling event could trigger TC-NER, and suggest that slip-outs on the nontranscribed strand may lead to repair-induced expansions, whereas those on the transcribed strand may lead to no change or to repair-induced contractions (71). Biochemical confirmation that TC-NER is induced by slip-outs, and that it can remove trinucleotide repeat (TNR) hairpins and restore transcription, would greatly enhance our mechanistic understanding of TNR instability. Potentially, the different consequences of TC-NER-stimulated slip-out repair on the transcribed and nontranscribed strands could underlie the NER-dependent, expansion-biased CAG repeat instability that we have documented here in the brain tissue of a mouse model of SCA1.
In summary, we have shown that Xpa deficiency substantially reduces the age-dependent CAG repeat instability at the mouse Sca1 locus in several tissues of the brain, but does not affect instability in the kidney or liver. Why neuronal tissues should be more sensitive to Xpa deficiency is not clear, but it may indicate that the transcription-dependent pathway for CAG repeat instability, which is dependent on genes involved in TC-NER (19,20), is the key pathway for CAG instability in neurons. In any case, these studies constitute the first example of a genetic defect that affects CAG repeat instability in a selected set of somatic tissues, and they support the notion that CAG instability in somatic tissues arises by multiple mechanisms (43). If ongoing expansion-biased CAG instability in the somatic tissues responsible for disease pathology hastens the onset of neuron dysfunction (5,6,72), then genes such as Xpa might be productive targets for therapeutic approaches to prevent CAG expansions in target tissues. XPA might be an especially relevant target in diseases such as HD that arise due to dysfunction in the striatum, a region of the brain that is highly susceptible to age-dependent Xpa-dependent repeat expansion (5,6).
MATERIALS AND METHODS
Mice
All animal procedures were carried according to protocols approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Xpa-deficient mice were generated by gene targeting in embryonic stem (ES) cells derived from the F1 hybrid of a CBA by C57BL/6 cross, and then outcrossed to CD-1 mice (38). The Xpa null allele in these mice carries a neomycin cassette in exon 4 (38). The knock-in allele of Sca1 with a long CAG repeat tract was generated from AB2.2 ES cells (derived from the 129Sv/Ev mouse strain) and backcrossed to C57BL/6 females for more than 10 generations before crossing to Xpa-deficient mice (73). Males heterozygous for the knock-in allele of Sca1 with 145 CAG repeats were crossed to Xpa−/− mice to generate Xpa+/− Sca1+/145Q. We refer to Sca1+/145Q mice as SCA1 mice. Xpa+/− SCA1 male mice were crossed to Xpa−/− mice and the progeny were genotyped to identify Xpa+/− SCA1 and Xpa−/− SCA1 mice. Male and female Xpa+/− SCA1 and Xpa−/− SCA1 mice were then crossed to C57BL/6 mice to assess intergenerational repeat stability. Progeny SCA1 mice were obtained from parental mice at a variety of ages from 10 weeks to >30 weeks and were fairly evenly distributed with respect to the age of the parents. For female Xpa+/− SCA1 donors, 13 progeny came from donors <15 weeks, 20 from donors between 15 and 25 weeks and 22 from donors >25 weeks. For female Xpa−/− SCA1 donors, 9 progeny came from donors <15 weeks, 14 from donors between 15 and 25 weeks and 11 from donors >25 weeks. For male Xpa+/− SCA1 donors, 5 progeny came from donors <15 weeks, 12 from donors between 15 and 25 weeks and 6 from donors >25 weeks. For male Xpa−/− SCA1 donors, 7 progeny came from donors <15 weeks, 12 from donors between 15 and 25 weeks and 10 from donors >25 weeks. The repeat tract lengths in the parents and offspring were determined by sequencing tail DNA at weaning. The repeat tract lengths in Xpa+/− SCA1 male donors (n= 8) ranged from 140 to 146; in Xpa+/− SCA1 female donors (n= 12), from 140 to 148; in Xpa−/− SCA1 male donors (n= 14), from 139 to 149; and in Xpa−/− SCA1 female donors (n= 10), from 142 to 145.
Six parental Xpa+/− SCA1 and six parental Xpa−/− SCA1 mice were ultimately sacrificed to measure repeat tract instability in various somatic tissues. Two of each genotype (one male and one female) were sacrificed at 30 weeks to examine CAG repeat instability in the striatum and kidney. Two of each genotype (one male and one female) were sacrificed at 45–50 weeks to examine instability in the striatum, cerebellum, kidney and liver. Two of each genotype (one male and one female) were sacrificed at 45–50 weeks to examine instability in the striatum, hippocampus, cerebral cortex, kidney and liver. In all cases, the results were comparable with those shown in Figures 2–4.
PCR primers and sequencing
The Xpa deficient mice were genotyped by PCR of tail DNA. Two primers were used: Xp3 (5′-tta atc tct ttc cag aga tgc tga) and Xp4 (5′-gcc ctt act aga cac ctg ta). Genotyping primers for the Sca1 locus were oVIN-24F (5′-aac atg ggc agt ctg agc cag) and oVIN-24R (5′-agc cct gct gag gtg ctg ctg). To determine repeat size at weaning, we amplified the repeat tract and sequenced it, as previously described (13). The CAG repeat was amplified with oVIN-106F (5′-cgt gta ccc tcc tcc tca gt) and oVIN-24R, and the PCR products were sequenced directly using oVIN-95F (5′-ggc cac cac tcc atc aca gc). Sequencing was performed by the Baylor College of Medicine Sequencing Core.
Small-pool PCR
Small-pool PCR was carried out as described previously (13). Briefly, genomic DNA was digested with BanII to fragment it and to cleave the wild-type allele, which carries an intact BanII site at the point where the expanded CAG repeat was inserted in the knock-in allele (73). Initial reactions were set up using 100, 50, 25 and 12.5 pg of genomic DNA from each tissue sample to determine the amount of DNA that would give two to five amplifiable genomes per reaction. For each tissue sample, 30 small-pool PCR reactions were carried out and the products were analyzed by gel electrophoresis. Representative sections of those gels are shown in Figure 2–4. Samples were amplified in 10 µl reactions with 5% DMSO, using ChromaTaq (Denville Scientific) and primers oVIN-25F (5′-gtc acc agt gca gta gcc tca g) and oVIN-25R (5′-atg tac tgg ttc tgc tgg gtg), which primed 111 bp upstream and 51 bp downstream of the repeat, respectively. The PCR program was 94°C for 2 min for initial denaturation, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min. The PCR products were separated by electrophoresis on a 2% agarose TAE gel at 80 V for 16 h, transferred to a nylon membrane and hybridized with a 5′ 32P-labeled (CAG)10 probe. The resulting blots were analyzed by phosphorimaging. Tail DNA from the same mouse as the tissue sample was included on either side of each set of reactions to serve as a marker against which to judge expansion and contraction.
Real-time RT–PCR
A panel of tissue total RNA from 30-week-old C57BL/6 mice was purchased from Zyagen (San Diego, CA, USA). To measure the level of Sca1 sense transcript, we first synthesized cDNA from the sense transcript, using reverse transcriptase and primer mSCA1-4SSP (CGATGCTTGGACAGCCTGCCTCTGGGTTGAAGTTCTCG), which links the Sca1 sense-strand-specific sequence (CCTCTGGGTTGAAGTTCTCG) to the SSP universal primer (CGATGCTTGGACAGCCTG), whose sequence is not present in the genome. Reverse transcriptase was then inactivated by incubation at 95°C for 10 min. The cDNA was amplified using primer mSCA1-3 (TATGCTGGTGGTCTGCC) and the SSP universal primer, which together amplify a 170 bp segment of Sca1 exon 7, just downstream of the position of the CAG repeats. The amplified fragment was quantified by real-time PCR. For the measurement of antisense transcripts, we synthesized cDNA from the antisense transcript, using reverse transcriptase and the primer mSCA1-3SSP (CGATGCTTGGACAGCCTGTATGCTGGTGGTCTGCC), in which the universal SSP primer is linked to the Sca1 antisense-strand-specific sequence (TATGCTGGTGGTCTGCC). The antisense cDNA was then amplified using a mixture of the mSCA1-4 (CCTCTGGGTTGAAGTTCTCG) primer and the universal SSP primer, which amplifies the same 170 bp segment of the Sca1 mRNA, and quantified by real-time PCR. For the antisense-strand-specific assays, we showed that amplification was not observed if reverse transcriptase was omitted from the reactions, indicating a lack of significant DNA contamination. An ethidium bromide-stained gel of the products of extensive amplification of the antisense transcript from various tissues is shown in Supplementary Material, Figure S1. To measure Xpa transcript levels, we used primers mXPA-1 (ACTGATGCTGCGCCAG) and mXPA-2 (CATGAACAATGTTTCCAATCTCGTG), which yield a 165 bp fragment. In all cases, results were normalized to the concentration of β-actin RNA, which was determined the same way using primers mActin3 (ATTGTTACCAACTGGGACGA) and mActin4 (ATCTGGGTCATCTTTTCACG), which yield a 142 bp fragment. Conditions for real-time PCR were 95°C for 15 min, followed by 45 cycles of 94°C for 15 s, 50°C for 30 s and 72°C for 30 s. Double-stranded DNA was detected with SYBR green chemistry, using a BioRad C1000 Thermal Cycler.
Statistics
Distributions of tract-length changes during germline transmission were compared using the non-parametric Mann–Whitney test, which does not assume a normal distribution.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health grants (grant number 1F31HG004918) to L.H. (grant number GM38219) to J.H.W., and by a Natural Sciences and Engineering Research Council of Canada postgraduate scholarship D to V.D.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr John Fryer for help with mouse tissue dissections, Dr B.V. Prasad for providing remote bench space for small-pool PCR, Dr Huda Zoghbi for SCA1 mice, Dr John DiGiovanni for Xpa-deficient mice, and members of the J.H.W. laboratory for helpful discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Pearson C.E., Edamura K.N., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 5.Shelbourne P.F., Keller-McGandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J., Vonsattel J.P., Wexler N.S., Arnheim N., Augood S.J. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 6.Swami M., Hendricks A.E., Gillis T., Massood T., Mysore J., Myers R.H., Wheeler V.C. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y., Hubert L., Jr, Wilson J.H. Transcription destabilizes triplet repeats. Mol. Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gacy A.M., Goellner G., Juranic N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 10.Pearson C.E., Wang Y.H., Griffith J.D., Sinden R.R. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n*(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary J.D., Pearson C.E. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 12.Dion V., Wilson J.H. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dion V., Lin Y., Hubert L., Jr, Waterland R.A., Wilson J.H. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovtun I.V., Johnson K.O., McMurray C.T. Cockayne Syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging (Albany, NY) 2011;3:509–514. doi: 10.18632/aging.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovtun I.V., Liu Y., Bjoras M., Klungland A., Wilson S.H., McMurray C.T. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 17.Savouret C., Garcia-Cordier C., Megret J., te Riele H., Junien C., Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Broek W.J., Nelen M.R., Wansink D.G., Coerwinkel M.M., te Riele H., Groenen P.J., Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y., Wilson J.H. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y., Dion V., Wilson J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y., Leng M., Wan M., Wilson J.H. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol. Cell. Biol. 2010;30:4435–4451. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ditch S., Sammarco M.C., Banerjee A., Grabczyk E. Progressive GAA*TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soragni E., Herman D., Dent S.Y., Gottesfeld J.M., Wells R.D., Napierala M. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamori M., Pearson C.E., Thornton C.A. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum. Mol. Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson C.E., Ewel A., Acharya S., Fishel R.A., Sinden R.R. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 26.Owen B.A., Yang Z., Lai M., Gajec M., Badger J.D., II, Hayes J.J., Edelmann W., Kucherlapati R., Wilson T.M., McMurray C.T. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y., Wilson J.H. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair (Amst.) 2009;8:878–885. doi: 10.1016/j.dnarep.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tornaletti S., Patrick S.M., Turchi J.J., Hanawalt P.C. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J. Biol. Chem. 2003;278:35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M. R loops stimulate genetic instability of CTG*CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy K., Tam M., Bowater R.P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y.H., Pearson C.E. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIvor E.I., Polak U., Napierala M. New insights into repeat instability: Role of RNA-DNA hybrids. RNA Biol. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svejstrup J.Q. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 33.Donahue B.A., Yin S., Taylor J.S., Reines D., Hanawalt P.C. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starita L.M., Horwitz A.A., Keogh M.C., Ishioka C., Parvin J.D., Chiba N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J. Biol. Chem. 2005;280:24498–24505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 35.Kleiman F.E., Wu-Baer F., Fonseca D., Kaneko S., Baer R., Manley J.L. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H.S., Li H., Cevher M., Parmelee A., Fonseca D., Kleiman F.E., Lee S.B. DNA damage-induced BARD1 phosphorylation is critical for the inhibition of messenger RNA processing by BRCA1/BARD1 complex. Cancer Res. 2006;66:4561–4565. doi: 10.1158/0008-5472.CAN-05-3629. [DOI] [PubMed] [Google Scholar]
- 37.Hubert L., Jr, Lin Y., Dion V., Wilson J.H. Topoisomerase 1 and single-strand break repair modulate transcription-induced CAG repeat contraction in human cells. Mol. Cell. Biol. 2011;31:3105–3112. doi: 10.1128/MCB.05158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakane H., Takeuchi S., Yuba S., Saijo M., Nakatsu Y., Murai H., Nakatsuru Y., Ishikawa T., Hirota S., Kitamura Y., et al. High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature. 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 39.Koberle B., Roginskaya V., Wood R.D. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair (Amst.) 2006;5:641–648. doi: 10.1016/j.dnarep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Watase K., Venken K.J., Sun Y., Orr H.T., Zoghbi H.Y. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum. Mol. Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 41.Dixon K.T., Cearley J.A., Hunter J.M., Detloff P.J. Mouse Huntington's disease homolog mRNA levels: variation and allele effects. Gene Expr. 2004;11:221–231. doi: 10.3727/000000003783992234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lloret A., Dragileva E., Teed A., Espinola J., Fossale E., Gillis T., Lopez E., Myers R.H., MacDonald M.E., Wheeler V.C. Genetic background modifies nuclear mutant huntingtin accumulation and HD CAG repeat instability in Huntington's disease knock-in mice. Hum. Mol. Genet. 2006;15:2015–2024. doi: 10.1093/hmg/ddl125. [DOI] [PubMed] [Google Scholar]
- 43.Lee J.M., Zhang J., Su A.I., Walker J.R., Wiltshire T., Kang K., Dragileva E., Gillis T., Lopez E.T., Boily M.J., et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung J., Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 45.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 46.Lin Y., Dion V., Wilson J.H. In: Genetic Instabilities and Neurological Diseases. Wells R.D., Ashizawa T., editors. Academic Press, Waltham, Massachusetts; 2006. pp. 691–704. [Google Scholar]
- 47.Wells R.D., Dere R., Hebert M.L., Napierala M., Son L.S. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy L., Shelbourne P.F. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington's disease? Hum. Mol. Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 49.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 50.Mangiarini L., Sathasivam K., Mahal A., Mott R., Seller M., Bates G.P. Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat. Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler V.C., Auerbach W., White J.K., Srinidhi J., Auerbach A., Ryan A., Duyao M.P., Vrbanac V., Weaver M., Gusella J.F., et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum. Mol. Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 52.Lia A.S., Seznec H., Hofmann-Radvanyi H., Radvanyi F., Duros C., Saquet C., Blanche M., Junien C., Gourdon G. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet. 1998;7:1285–1291. doi: 10.1093/hmg/7.8.1285. [DOI] [PubMed] [Google Scholar]
- 53.Fortune M.T., Vassilopoulos C., Coolbaugh M.I., Siciliano M.J., Monckton D.G. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 54.Gomes-Pereira M., Fortune M.T., Monckton D.G. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001;10:845–854. doi: 10.1093/hmg/10.8.845. [DOI] [PubMed] [Google Scholar]
- 55.Kaytor M.D., Burright E.N., Duvick L.A., Zoghbi H.Y., Orr H.T. Increased trinucleotide repeat instability with advanced maternal age. Hum. Mol. Genet. 1997;6:2135–2139. doi: 10.1093/hmg/6.12.2135. [DOI] [PubMed] [Google Scholar]
- 56.Mochmann L.H., Wells R.D. Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats. Nucleic Acids Res. 2004;32:4469–4479. doi: 10.1093/nar/gkh787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schumacher S., Pinet I., Bichara M. Modulation of transcription reveals a new mechanism of triplet repeat instability in Escherichia coli. J. Mol. Biol. 2001;307:39–49. doi: 10.1006/jmbi.2000.4489. [DOI] [PubMed] [Google Scholar]
- 58.Bowater R.P., Jaworski A., Larson J.E., Parniewski P., Wells R.D. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savouret C., Brisson E., Essers J., Kanaar R., Pastink A., te Riele H., Junien C., Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomes-Pereira M., Fortune M.T., Ingram L., McAbney J.P., Monckton D.G. Pms2 is a genetic enhancer of trinucleotide CAG*CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 61.Foiry L., Dong L., Savouret C., Hubert L., Riele H.T., Junien C., Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 62.Wheeler V.C., Lebel L.-A., Vrbanac V., Teed A., te Riele H., MacDonald M.E. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 63.Dragileva E., Hendricks A., Teed A., Gillis T., Lopez E.T., Friedberg E.C., Kucherlapati R., Edelmann W., Lunetta K.L., MacDonald M.E., et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tome S., Panigrahi G.B., Lopez Castel A., Foiry L., Melton D.W., Gourdon G., Pearson C.E. Maternal germline-specific effect of DNA ligase I on CTG*CAG instability. Hum. Mol. Genet. 2011;20:2131–2143. doi: 10.1093/hmg/ddr099. [DOI] [PubMed] [Google Scholar]
- 65.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H., Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balajee A.S., May A., Dianov G.L., Friedberg E.C., Bohr V.A. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le May N., Mota-Fernandes D., Velez-Cruz R., Iltis I., Biard D., Egly J.M. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 68.van Gool A.J., Citterio E., Rademakers S., van Os R., Vermeulen W., Constantinou A., Egly J.M., Bootsma D., Hoeijmakers J.H. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tantin D., Kansal A., Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selby C.P., Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salinas-Rios V., Belotserkovskii B.P., Hanawalt P.C. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr429. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomes-Pereira M., Monckton D.G. Chemical modifiers of unstable expanded simple sequence repeats: what goes up, could come down. Mutat. Res. 2006;598:15–34. doi: 10.1016/j.mrfmmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 73.Watase K., Weeber E.J., Xu B., Antalffy B., Yuva-Paylor L., Hashimoto K., Kano M., Atkinson R., Sun Y., Armstrong D.L., et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.