Abstract
Cardiac failure is associated with increased levels of oxidized DNA, especially mitochondrial (mtDNA). It is not known if oxidized mtDNA contributes to cardiac dysfunction. To test if protection of mtDNA can reduce cardiac injury, we produced transgenic mice with cardiomyocyte-specific overexpression of the DNA repair enzyme 8-oxoguanine DNA glycosylase 1 (OGG1) isoform 2a. In one line of mice, the transgene increased OGG1 activity by 115% in mitochondria and by 28% in nuclei. OGG1 transgenic mice demonstrated significantly lower cardiac mitochondrial levels of the DNA guanine oxidation product 7,8-dihydro-8-oxoguanine (8-oxo-dG) under basal conditions, after doxorubicin administration, or after transaortic constriction (TAC), but the transgene produced no detectable reduction in nuclear 8-oxo-dG content. OGG1 mice were tested for protection from the cardiac effects of TAC 13 wk after surgery. Compared with FVB-TAC mice, hearts from OGG1-TAC mice had lower levels of β-myosin heavy chain mRNA but they did not display significant differences in the ratio of heart weight to tibia length or protection of cardiac function measured by echocardiography. The principle benefit of OGG1 overexpression was a significant decrease in TAC-induced cardiac fibrosis. This protection was indicated by reduced Sirius red staining on OGG1 cardiac sections and by significantly decreased induction of collagen 1 and 3 mRNA expression in OGG1 hearts after TAC surgery. These results provide a new model to assess the damaging cardiac effects of 8-oxo-dG formation and suggest that increased repair of 8-oxo-dG in mtDNA decreases cardiac pathology.
Keywords: mitochondria, transgene
oxidization of DNA by reactive oxygen species (ROS) can produce nucleotide mutations potentially involved in carcinogenesis and aging (40). Guanine is the nucleic acid base with the lowest oxidation potential, rendering it the most easily oxidizable by hydroxyl radicals and singlet oxygen (31). Therefore, oxidized guanine [7,8-dihydro-8-oxoguanine (8-oxo-dG)] is the most abundant DNA lesion upon oxidative exposure. During DNA replication, 8-oxo-dG will frequently mispair with adenine, leading to formation of G:C to T:A transversions (4, 10, 25). DNA base excision is a major pathway to repair oxidized nucleotides, and the enzyme 8-oxoguanine DNA glycosylase 1 (OGG1) has a major role in removal of 8-oxo-dG (7). For example, OGG1 deficiency in Saccharomyces cerevisiae gives rise to a spontaneous mutator phenotype (11); a 10-fold increase of G:C to T:A transversion frequency in DNA from liver cells was found in homozygous OGG1 knockout (OGG−/−) mice (19); and human OGG1 gene mutations and polymorphisms were found to be associated with head, neck, lung, and kidney cancers (29, 30).
Mitochondrial DNA (mtDNA) damage is more abundant and longer lasting than nuclear DNA (nDNA) damage following exposure to oxidative stress (40, 43). A case in point is the aging rat where levels of 8-oxo-dG in nDNA do not change from 6 to 23 mo of age while 8-oxo-dG increases 2.5-fold in mtDNA with age (15) and 8-oxo-dG levels in mtDNA are an order of magnitude higher than in nDNA (13). One of the reasons mtDNA is more sensitive to ROS mediated damage than nDNA is that mtDNA has no histone protection that can serve as a barrier against ROS. Secondly, mitochondria are the major sources for superoxide production from the electron transport chain (ETC), thereby placing mtDNA in close proximity to ROS production. Lastly, mitochondria have low capacity to repair mutagenesis. The level of 8-oxo-dG in mtDNA correlates with mitochondria dysfunction in many disorders associated with oxidative stress such as aging and neurodegeneration (23), and levels of 8-oxo-dG are inversely correlated to mammalian life span (3). Therefore, 8-oxo-dG in mtDNA is an important lesion that is at least a marker and possibly a cause for development of various diseases associated with oxidative stress.
Oxidative stress is increased in heart failure patients (16) and is frequently associated with quantitative and qualitative defects in mtDNA (17). In heart failure, mitochondria produce more superoxide than normal (38). A vicious cycle can develop for mitochondrial damage in which leakage of superoxide from the ETC promotes mtDNA damage resulting in more ETC damage that then produces more ROS and mtDNA damage. We hypothesized that this destructive cycle in the heart could be disrupted by enhancing mtDNA repair via overexpression of the repair enzyme OGG1. To test this hypothesis, we produced transgenic mice with cardiomyocyte overexpression of OGG1 and subjected them to transaortic constriction (TAC).
MATERIALS AND METHODS
Construction of the OGG1 transgenic mouse model with cardiac-specific overexpression of OGG1.
First-strand cDNA was synthesized from human fibroblast total RNA by superscript II and oligo dT primers (Invitrogen). The cDNA was amplified by PCR using primers designed for the human mitochondrial isotype 2a of OGG1 (5, 26). The sense primer extends from nucleotide 78 of the OGG1–2a mRNA in the 5′-untranslated region and included an additional Sal I restriction site (gacttagtcgaccgggagaagataagtcgcaa). The antisense primer extended to position 1,782 in the OGG1–2a mRNA 3′-untranslated region and contained an additional Hind III restriction site (actactaagcttccagagggccacataatgtt). This PCR reaction produced a fragment containing all of the coding sequence of the human OGG1–2a isoform. The OGG1–2a isoform is predominantly localized in mitochondria (26) due to the presence of a putative mitochondrial targeting sequence and absence of a nuclear localization signal from exon 7 of the OGG1 gene. The purified OGG1 cDNA fragment was subcloned behind a 5.5-kb DNA fragment from the α-myosin heavy chain (MHC) promoter (12) that produces transcription exclusively in cardiac myocytes and in front of a 500 bp of poly(A) DNA sequence derived from the rat insulin II gene. The sequence of the entire subcloned OGG1 cDNA fragment was confirmed. The transgene was released from the plasmid by Not I digestion and microinjected into single cell fertilized FVB mouse embryos by standard embryo microinjection procedures. Male mice between the ages of 90 and 120 days were used for subsequent experiments. This age was chosen so that animals were large enough for surgery and to reduce variation between mice. All animal procedures conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Pub. No. 85-23, Revised 1996) and were approved by the United State Department of Agriculture-Certified Institutional Animal Care Committee.
TAC surgery.
TAC surgery was performed by a modification of a previously published technique (2). Mice were anesthetized with avertin (0.4 g/kg), maintained on a 37°C pad, and ventilated with 100% oxygen. An incision at the left second intercostal space was made to open the chest. A chest retractor was applied to facilitate the view. The thymus was pulled away, and transverse aorta was dissected from surrounding tissues. A 6–0 silk suture was passed around the transverse aorta and tightened against a 26-G needle. The needle was immediately removed to provide a lumen with a stenotic aorta. Lungs were inflated, and the chest cavity, muscles, and skin were closed layer by layer with 6–0 silk sutures. The whole procedure lasted 20–30 min. After surgery, mice were warmed with a heating lamp and received an intraperitoneal injection of 0.5 ml of saline at 37°C. For 48 h, mice were given subcutaneous injections of buprenorphine, 0.1 mg/kg every 12 h. Sham mice were done same as TAC animals except that banding of the transverse aorta was omitted.
Echocardiographic assessment of cardiac function.
Transthoracic echocardiography of the left ventricle (LV) was performed using a 15-MHz linear array transducer (15L8) interfaced with a Sequoia C512 system (Siemens, Malvern, PA) as previously described (42). Mice were anesthetized with 2% isoflurane, maintained under anesthesia with 1.25% isoflurane, and examined. Ventricular parameters were measured in M-mode with a sweep speed of 200 mm/s. The echocardiograms were captured from short-axis views of the LV at the midpapillary level. LV percent fractional shortening (LV%FS) was calculated according to the following equation: LV%FS = [(LVEDD − LVESD)/LVEDD] × 100, where LVEDD and LVESD are LV end diastolic and systolic diameter, respectively. All data were calculated from 10 independent cardiac cycles per experiment.
Mitochondrial and nuclear protein and DNA.
Mitochondria and nuclei were isolated by modification of a previously described protocol (34). A fresh heart was cut into small pieces and homogenized by a motor-driven Teflon pestle (Wheaton Scientific, Millville, NJ) in 5 ml of isolation buffer containing 10 mM HEPES, 200 mM mannitol, 70 mM sucrose, and 1 mM EGTA, pH 7.4. Nuclei were obtained by centrifugation at 700 g for 10 min at 4°C. The supernatant was moved to a new tube for centrifuging at 8,000 g for 15 min at 4°C. The pellet was collected and washed again with isolation buffer. The final mitochondrial pellet was used to prepare mitochondrial DNA and mitochondrial protein. The purity of nuclei and mitochondria was assessed by Western blots with antibodies to lamin A (Abcam, Cambridge, MA) and cyclooxygenase IV (Abcam). For purification of DNA, freshly isolated mitochondria or nuclei were lysed and extracted by QIAprep miniprep kit (Qiagen. Hilden, Germany). The concentration of DNA was determined with a spectrophotometer, and the quality of mtDNA was confirmed by Sac II digestion and agarose gel electrophoresis. Protein was isolated from freshly isolated mitochondria or nuclei by lysis in buffer containing 2% SDS, 10% glycerol, and 62.5 mmol/l Tris·HCl (pH 7.0), sonication, and centrifuging at 11,000 g at 4°C for 15 min. The supernatant containing protein was collected, and the concentration was determined by the Lowry method (Pierce kit, Rockford, IL). Western blot (41) protein was probed with rabbit anti-hOGG1 antibody at 1:1,000 dilution (Abcam) at 4°C overnight. The secondary antibody was incubated with the membrane for another hour at room temperature. Finally, the antigen-antibody complexes were visualized with use of an enhanced chemiluminescence kit (ECL; GE Healthcare, Niskayuna, NY).
Assay of OGG1 activity.
The fresh isolated mitochondrial or nuclear pellets were lysed in buffer containing 20 mM HEPES-KOH (pH 7.4), 1 mM EDTA, 1 mM DTT, 300 mM KCl, 5% glycerol, and 0.5% Triton X-100 on ice for 10 min. The lysate was centrifuged at 11,000 g for 30 min at 4°C (36). The supernatant was collected, and the concentration of protein was determined by the Lowry method. OGG1 activity was determined by excising 8-oxoG from a 32P-labeled oligonucleotide substrate as described previously (36) with modification. Briefly, a synthetic oligonucleotide containing an 8-oxogunine adduct (5′-gaactagtgOatcccccgggctg-3′ where O is 8-oxoguanine; Trevigen, Gaithersburg, MD) was labeled with γ−32P-ATP by T4 kinase (Invitrogen, Carlsbad, CA) and the labeled oligonucleotide was annealed with its complementary oligonucleotide (5′-gcagcccgggggatccactagttc-3′; Invitrogen). Unincorporated label was removed by passage over a NAP-5 column (GE Healthcare, Niskayuna, NY). Fifty micrograms of protein were incubated with 0.1 pmol of the 32P-labeled duplex oligonucleotide in excision buffer [20 mM HEPES-KOH (pH 7.6), 5 mM EDTA, 1 mM DTT, 100 mM KCl, and5% glycerol] for 3 h at 37°C in a volume of 20 μl. After incubation, the reaction was terminated by addition of proteinase K (250 μg/ml) and SDS (0.5%) and heating at 50°C for 15 min. Olignucleotides were precipitated with 70% ethanol, 45 mM ammonium acetate, and 0.01 μg/μl glycogen and redissolved in loading buffer containing 70% formamide, 30 mM NaOH, and 0.05% bromophenol blue. The substrate and cleaved oligonucleotide product were separated on a gradient (4–15%) polyacrylamide gel containing 7 M urea (Invitrogen). Radioactivity in the separated DNA bands was quantitated with a PhosphoImager and Imagequant software (Molecular Dynamics, Sunnyvale, CA).
Quantitative RT-PCR.
Cardiac RNA was extracted with Trizol reagent. The total RNA was transcribed to cDNA with Superscript II enzyme and random oligonucleotide primers (Invitrogen). The primers, probes, and reaction buffer for RT-PCR were purchased from AB (Applied Biosystems, Carlsbad, CA) including hOGG1 (Hs00213454_m1), α-MHC (Mm01313844_mH), β-MHC (Mm00600555_m1), atrial natriuretic peptide (ANP) (Mm01255748_g1), brain natriuretic peptide (BNP; Mm01255770_g1), procollagen 1α1(Mm01302043_g1), procollagen 3α1(Mm01254476_m1), 18S RNA (Hs99999901_s1), and 2× Master buffer. RT-PCR was carried out on AB 7300 thermocycler with 35 cycles, each cycle consisted of 95°C for 15 s, 55°C for 15 s, and 75°C for 30 s. 18S RNA was used as endogenous control. Relative abundance of transcripts was determined by the ΔΔCT method.
Histological experiments.
Cryostat sections (5 μm) were fixed in 10% formalin for 15 min and washed three times with PBS. The cryostat slides were incubated with a saturated solution of picric acid containing 0.1% Sirius red for staining collagen and 0.1% fast green for staining noncollagen proteins. Staining was performed in the dark for 2 h. The slides then were rinsed with distilled water, dehydrated with alcohol, and mounted with permount. The sections were visualized and photographed by a blinded observer. Interstitial fibrosis in the sections was scored by a blinded observer against reference images using a scale of 1 to 4 based on the severity of fibrosis with scores of 1 for low, 2 for mild, 3 for moderate, and 4 for severe.
Measurement of 8-oxo-dG content.
Content of 8-oxo-dG was measured using a commercial enzyme-linked immunosorbent assay kit (Trevigen, Gaitherburg MD) as described by Gao et al. (9) with modification. In brief, 0.5 μg of sample DNA or 8-oxo-dG standards from 0 to 60 ng/ml were mixed with anti-8-oxo-dG antibody overnight at 4°C, and the sample DNA and 8-oxo-dG standards were transferred to a 96-well microplate previously coated with albumin:8-oxo-dG adduct and kept in the dark at room temperature for 2 h. The secondary antibody coupled with peroxidase was added to each well and incubation followed for 1 h in the dark, and the plate was washed. After six rounds of washing, the substrate tetramethylbenzidene was added and the reaction continued for 15 min in the dark at room temperature before a stop solution was used to terminate the reaction. The wavelength of 450 nm was set to measure the absorbance of the final solution in each well. Results were calculated according to the standard curve.
RESULTS
Characterization of OGG1 transgenic mice.
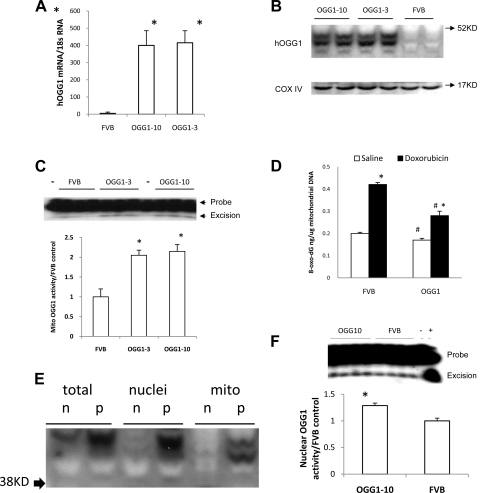
Transgenic mice carrying a cardiac targeted transgene for overexpression of human OGG1 were produced as described in the materials and methods. A total of four positive founders were obtained from 22 pups. The founder lines were named OGG1–3, OGG1–6, OGG1–7, and OGG1–10. Lines OGG1–3 and OGG1–10 were fertile and passed on the transgene to ∼50% of offspring. These lines were tested for expression of human OGG1 mRNA and protein and OGG1 activity. The expression of human OGG1 mRNA in heart was significantly elevated (Fig. 1A; P < 0.01 vs. FVB control). Western blots showed that human OGG1 protein was markedly increased in mitochondria of both transgenic lines (Fig. 1B). A possible explanation for the two immunoreactive bands in mitochondria is that the larger band (∼45 kDa) derives from the full human OGG1 protein and the smaller band (∼42 kDa) comes from the OGG1 protein after removal of the mitochondrial targeting sequence. OGG1 activity measured in extracts of cardiac mitochondria was over twofold elevated over FVB in both transgenic lines (Fig. 1C; P < 0.01). Lines OGG1–3 and OGG1–10 had similar human OGG1 expression by all parameters, and line OGG1–10 was used for subsequent experiments. To confirm OGG1 activity in vivo, OGG1–10 mice were injected with 11 mg/kg doxorubicin or saline and 8-oxo-dG content of mtDNA was measured 48 h postinjection. As shown in Fig. 1D, doxorubicin increased 8-oxo-dG content in transgenic and control mtDNA. The OGG1 transgene produced significant reductions of 25% and 31 in 8-oxo-dG content in saline- and doxorubici- treated mice, respectively. These results established cardiac overexpression of active OGG1 in mitochondria of transgenic mice.
Fig. 1.
Elevation of 8-oxoguanine DNA glycosylase 1 (OGG1) mRNA, protein, and enzyme activity in OGG1 transgenic mice. A: human OGG1 (hOGG1) mRNA level, detected by RT-PCR in cardiac RNA of OGG1–10 and OGG1–3 mice was significantly increased vs. FVB control (*P < 0.001 vs. FVB control by one-way ANOVA analysis; n = 5 for each group). B: Western blot of expression of hOGG1 protein in isolated mitochondria. No hOGG1 band was detected in FVB control mice in contrast to strong bands evident for the 2 OGG1 lines. Cyclooxygenase IV (COX IV) was used as loading control. C: OGG1 enzyme activity was measured by cleavage of a 32P-end-labeled, 7,8-dihydro-8-oxoguanine (8-oxo-dG) containing oligonucleotide substrate using 50 μg of mitochondrial protein (position of the 8-oxo-dG containing oligonucleotide probe and the excision product are indicated by arrows, and minus indicates lanes in which no protein was added to the probe). Activity of OGG1 in the 2 lines was 2-fold higher than that of FVB control (*P < 0.01, vs. FVB control by one-way ANOVA analysis; n = 6 for each group). D: content of 8-oxo-dG in mtDNA from mice 48 h after treatment with 11 mg/kg doxorubicin. Doxorubicin increased 8-oxo-dG content (*P < 0.05) in FVB and OGG1 mice. OGG1 transgene reduced 8-oxo-dG content in saline- and doxorubicin-treated mice compared with FVB mice (#P < 0.05, by two-way ANOVA; n = 5 per group). E: transgenic OGG1 was present in transgenic nuclei and mitochondria (mito). Results are typical of separate blots obtained from 3 different transgenic mice. In this blot, fractions from control mice are indicated by n and transgenic samples are indicated by p. F: OGG1 enzyme activity was significantly increased in OGG10 transgenic nuclei by 28% (*P < 0.05, by t-test). Lane marked with a plus symbol shows product obtained with pure OGG1 enzyme.
The subcellular localization of human OGG1 was assessed on Western blots loaded with total, nuclear, and mitochondrial protein extracts from hearts of control and OGG1–10 transgenic mice (Fig. 1E). Mitochondria contained the largest amount of the small 42-kDa band. Unexpectedly, nuclei from OGG1 hearts contained a strong OGG1 band of ∼46 kDa that was due to the transgene. The reason this was unexpected is that the amino acid sequence of the OGG1–2a isoform (26) used to construct the OGG1 transgene does not contain a nuclear localization signal. The additional OGG1 protein present in transgenic nuclei resulted in a 28% increase in OGG1 enzyme activity (Fig. 1F) compared with FVB nuclei.
Evaluation of heart size and function.
TAC mimics human aortic stenosis with development of pressure-overload-induced left ventricular hypertrophy. TAC or sham surgery was performed on OGG1 and FVB mice and mice were killed for evaluation after 13 wk. As shown in Table 1, TAC surgery significantly increased heart to tibia length and heart to body wt ratio in FVB and OGG1 mice. When compared by heart to body weight ratio, there was less hypertrophy in OGG1 mice (P < 0.05). However, this was partially due to differences in body weight. When heart weight was compared with tibia length, the difference in hypertrophy of OGG1 and FVB mice did not quite reach significance (P = 0.07).
Table 1.
Measurement of heart weight, body weight, tibia length, and the ratios of heart weight to body weight and to tibia length in FVB and OGG1 mice 13 wk after TAC or sham surgery
| Parameters | FVB-Sham | FVB-TAC | OGG1-Sham | OGG1-TAC |
|---|---|---|---|---|
| Heart weight, mg | 124.8 ± 2.9 | 186.9 ± 9.1* | 128.9 ± 3.0 | 168.8 ± 8.0* |
| Body weight, g | 30.8 ± 0.9 | 31.8 ± 0.7 | 31.8 ± 0.7 | 33.7 ± 1.0 |
| Tibia length, mm | 18.14 ± 0.16 | 18.29 ± 0.07 | 18.24 ± 0.07 | 18.32 ± 0.07 |
| HW/BW | 4.09 ± 0.15 | 5.91 ± 0.31* | 4.01 ± 0.14 | 5.09 ± 0.32*† |
| HW/TL | 6.88 ± 0.14 | 10.22 ± 0.50* | 7.07 ± 0.16 | 9.22 ± 0.45* |
Values are means ± SE. OGG1, 8-oxoguanine DNA glycosylase 1; TAC, transaortic constriction; HW/BW, heart weight-to-body weight ratio; HW/TL, heart weight-to-tibia length ratio.
P < 0.05, FVB-sham vs. FVB-TAC or OGG1-sham vs. OGG1-TAC.
P < 0.05, OGG1-TAC vs. FVB-TAC by two-way ANOVA analysis (n = 11 for sham and n = 14 for TAC groups).
Echocardiography was used to assess the effect of TAC and the OGG1 transgene on cardiac function 13 wk after TAC. The data reported in Table 2 show that TAC mice had significantly impaired cardiac function compared with sham mice. This was evident by several parameters including increased left ventricular end diastolic and systolic diameter as well as reduced fractional shortening. However, there were no significant differences between FVB and OGG1 mice, indicating that OGG1 overexpression did not alter cardiac function nor protect it from the effect of TAC.
Table 2.
Cardiac function measured by echocardiography in FVB and OGG1 mice 13 wk after TAC or sham surgery
| Parameter | FVB-Sham | FVB-TAC | OGG1-Sham | OGG1-TAC |
|---|---|---|---|---|
| LVEDD, mm | 3.81 ± 0.08 | 4.15 ± 0.05* | 3.81 ± 0.10 | 4.03 ± 0.08* |
| LVESD, mm | 2.36 ± 0.10 | 2.76 ± 0.08* | 2.27 ± 0.09 | 2.59 ± 0.08* |
| IVS(d), mm | 0.91 ± 0.02 | 1.02 ± 0.03* | 0.89 ± 0.03 | 1.0 ± 0.02* |
| IVS(s), mm | 1.18 ± 0.03 | 1.29 ± 0.03* | 1.19 ± 0.03 | 1.24 ± 0.02 |
| PWTh(d), mm | 0.80 ± 0.04 | 0.89 ± 0.03* | 0.76 ± 0.03 | 0.84 ± 0.03 |
| PWTh(s), mm | 1.16 ± 0.04 | 1.25 ± 0.02 | 1.17 ± 0.05 | 1.25 ± 0.03 |
| IVS%Th | 30.87 ± 2.78 | 29.26 ± 3.50 | 33.74 ± 2.67 | 24.72 ± 2.73 |
| PW%Th | 40.67 ± 5.27 | 40.32 ± 3.08 | 48.51 ± 3.31 | 49.03 ± 4.08 |
| IVS/PW | 1.16 ± 0.05 | 1.13 ± 0.03 | 2.2 ± 1.01 | 1.19 ± 0.04 |
| %FS | 38.6 ± 1.7 | 33.8 ± 1.1a | 40.59 ± 1.40 | 34.47 ± 1.06* |
| HR, beats/min | 487 ± 10 | 516 ± 11 | 513 ± 12 | 535 ± 12 |
Values are means ± SE. LVEDD, left ventricular end diastolic diameter; LVESD, left ventricular end systolic diameter; IVS(d), interventricular septum thickness at diastole; IVS(s), interventricular septum thickness at systole; PWTh(d), postwall thickness at diastole; PWTh(s), postwall thickness at systole; IVS%Th, interventricular septum % thickening; PW%Th, posterior wall % thickening; IVS/PW, interventricular septum to posterior wall thickness ratio; %FS, percent fractional shortening; HR, heart rate.
P < 0.05, FVB-sham vs. FVB-TAC or OGG1-sham vs. OGG1-TAC by two-way ANOVA analysis (n = 12 for FVB-sham, n = 10 for OGG1-sham, n = 14 for FVB-TAC, and n = 13 for OGG1-TAC group).
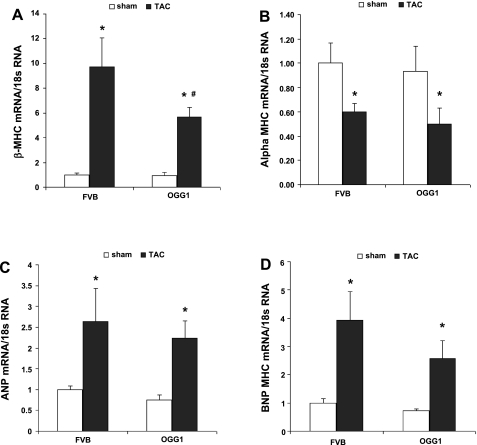
Hypertrophic mRNA markers β-MHC, α-MHC, ANP, and BNP were measured to determine whether OGG1 could reduce the fetal pattern of RNA expression induced by pressure overload and hypertrophy (Fig. 2). In sham-treated mice, there were no differences between OGG1 and FVB RNA expression. In both OGG1 and FVB mice, TAC produced the expected increases in expression for β-MHC, ANP, and BNP as well as the expected reduction in the expression of α-MHC. Compared with FVB-TAC mice, OGG1-TAC mice had 41% lower expression of β-MHC (P < 0.05) and OGG1-TAC mice tended toward lower BNP expression (35% lower than FVB-TAC; P = 0.12).
Fig. 2.
Hypertrophic biomarkers β-myosin heavy chain (MHC), α-MHC, atrial natriuretic peptide (ANP), and brain natriuretic peptide (BNP) mRNA were analyzed by RT-PCR in OGG1 and FVB hearts 13 wk after transaortic constriction (TAC) or sham surgery. A: β-MHC mRNA was significantly increased after TAC in both FVB and OGG1 hearts; however, the β-MHC mRNA level in OGG1-TAC hearts was significantly lower than that in FVB-TAC hearts. B: α-MHC mRNA level was significantly deceased in both FVB and OGG1 after TAC and no significant difference was found between FVB and OGG1 mice with or without TAC. ANP mRNA (C) and BNP (D) mRNA were increased significantly after TAC compared with sham mice. No significant difference was found between OGG1 and FVB in sham or TAC groups. Statistical analysis was done by two-way ANOVA (*P < 0.05, sham vs. TAC; #P < 0.05, OGG1-TAC vs. FVB-TAC; n = 5 for FVB-sham, FVB-TAC, and OGG1-sham; n = 8 for OGG1-TAC).
TAC induction of cardiac fibrosis.
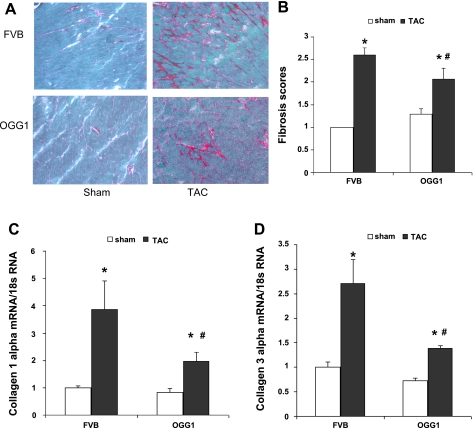
Semiquantative analysis showed that 13 wk after TAC surgery, fibrosis was significantly increased in both OGG1 and FVB mice (Fig. 3, A and B; P < 0.05). This analysis also showed that interstitial fibrosis staining was lower in OGG1-TAC mice compared with FVB-TAC mice (P < 0.05). The staining analysis was supported by quantitative RT-PCR assays of cardiac collagen mRNA expression (Fig. 3, C and D). Compared with FVB-TAC mice, expression of collagen 1α1 mRNA and collagen 3α1 mRNA was 49 and 48% lower in OGG1-TAC mice, respectively (P < 0.05 for both mRNAs).
Fig. 3.
Fibrosis in FVB and OGG1 hearts 13 wk after TAC. A: representative Sirius red staining for fibrosis in FVB and OGG1 hearts. B: semiquantitative scores for fibrosis staining performed as described in materials and methods by a blinded observer. Staining of fibrosis in OGG1-TAC hearts was significantly lower than that in FVB-TAC hearts (#P < 0.05, OGG1-TAC vs. FVB- TAC; *P < 0.05, sham vs. TAC by two-way ANOVA; n = 5 for each group). Expression of collagen 1α1 mRNA (C) and collagen 3α1 mRNA (D) measured by RT-PCR was significantly lower in OGG1-TAC hearts than in FVB-TAC hearts (#P < 0.05, OGG1-TAC vs. FVB-TAC; *P < 0.05, sham vs. TAC by two-way ANOVA; n = 5 for FVB-sham, FVB-TAC, and OGG1-sham; n = 8 for OGG1-TAC).
8-oxo-dG content in DNA after TAC.
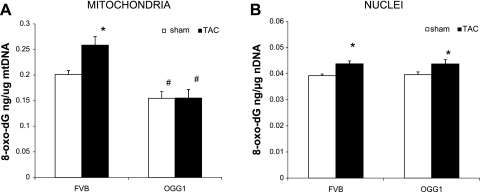
Thirteen weeks after surgery, we tested whether TAC increased 8-oxo-dG content in nDNA and mtDNA and whether OGG1 overexpression could reduce 8-oxo-dG content. In mitochondria (Fig. 4A), TAC produced a 30% increase in 8-oxo-dG (P < 0.05) of FVB mice. This TAC-induced increase in mtDNA 8-oxo-dG content was completely prevented by overexpression of OGG1. In addition, OGG1-sham mice had 25% lower 8-oxo-dG content in mtDNA than FVB-sham mice (P < 0.05). In nuclear DNA 8-oxo-dG levels were much lower than in mtDNA (Fig. 4B). TAC produced a significant 10% increase in 8-oxo-dG content in both types of mice but there were no significant differences for nuclear DNA 8-oxo-dG content between OGG1 and FVB mice. Overall, these results show that OGG1 provides protection against some components of cardiac dysfunction in the TAC model including reductions in fibrosis, β-MHC expression, and 8-oxo-dG content in mtDNA.
Fig. 4.
Content of 8-oxo-dG content in mitochondrial and nuclear DNA 13 wk after TAC. A: in FVB mitochondria TAC increased 8-oxo-dG content (*P < 0.05, for FVB-sham vs. FVB-TAC). OGG1 lowered sham levels of 8-oxo-dG and prevented the TAC-induced increase in 8-oxo-dG content (#P < 0.05, for OGG1-sham vs. FVB-sham and for OGG1-TAC vs. FVB-TAC; n = 6 for each group). B: in nuclei TAC increased 8-oxo-dG content in FVB and OGG1 mice (*P < 0.05, for sham vs. TAC; n = 5 per group, all statistics by two-way ANOVA). OGG1 transgene did not alter 8-oxo-dG content in nuclear DNA of sham or TAC mice.
DISCUSSION
Damage to cardiac DNA, especially mtDNA by reactive oxygen has been proposed to be a contributing factor in the development of heart failure (3, 37). Human studies (17) and animal models (8, 21, 27, 33) have demonstrated that there is a strong association between cardiac injury and oxidative damage to mtDNA. This relationship has been shown following various treatments including exposure to doxorubicin (21, 33), angiotensin (32), or isoproterenol (18), the process of aging (6, 15), and knockout of mitochondrial SOD2 (24). However, no cardiac-specific models have been created to determine if reducing mtDNA damage can protect the heart, thereby testing whether the association between oxidative mtDNA damage and heart failure is actually a causal relationship. In this study, we describe successful production of OGG1 transgenic mice that have cardiac-specific overexpression of active OGG1 enzyme, which catalyzes the first essential step in the repair pathway of oxidized DNA. OGG1 transgenic mice were shown to have increased hOGG1 mRNA, elevated hOGG1 protein, 28% increased nuclear OGG1 activity, and a 115% increase of OGG1 enzyme activity in mitochondria. In vivo efficacy of the transgene was demonstrated by reduced levels of mitochondrial 8-oxo-dG content in transgenic mice under basal conditions, after doxorubicin treatment, and after TAC surgery. Overexpression of OGG1 produced no measurable detrimental effects on the heart. Thus this transgenic line provides a suitable model for performing direct tests for a causal role of 8-oxo-dG mutations in cardiac pathology.
The efficacy of OGG1 overexpression was tested by challenge with TAC surgery. Transgenic mice demonstrated essentially complete protection from TAC-induced elevation of 8-oxo-dG, but despite this protection, we saw little benefit of the OGG1 transgene either to reduce cardiac hypertrophy or to improve cardiac function measured by echocardiography 13 wk after surgery. There are multiple potential explanations for this lack of effect. One explanation may be that mechanisms mediating stress-induced cardiac hypertrophy such as growth factor pathways (14) or kinase-mediated actin rearrangements (20, 22) are not closely linked to the condition of mtDNA. If these pathways are not linked to mtDNA damage, then the reduction in mtDNA mutations produced by the OGG1 transgene cannot protect from TAC-induced dysfunction. However, the fact that OGG1 transgenic mice had reduced β-MHC mRNA expression after TAC suggests that mtDNA mutations have at least a modest impact on the cardiac hypertrophy response or that decreasing fibrosis limited the elevation in β-MHC expression. An alternative reason for the lack of a larger OGG1 effect may be that we did not wait long enough after surgery. The 13-wk time point after TAC may not be long enough for sufficient mtDNA mutation to accumulate and disrupt mitochondrial function in many cardiomyocytes. Over a longer period mutated mtDNA may spread within individual myocytes from heteroplasmy towards homoplasmy through relaxed replication (6). Accumulation of mtDNA mutations appears to have a high threshold before essential mitochondrial functions are compromised: in OGG1−/− mice elevation of 8-oxo-dG content in mtDNA did not produce a measurable impairment of mitochondrial function including respiratory rate and ATP synthetic activity in heart or liver (35) despite the fact that some threshold of 8-oxo-dG accumulation can be expected to impair mitochondrial respiration. Also, our unpublished studies revealed that the OGG1 transgene had no effect on the amount of hydrogen peroxide generated by isolated mitochondria using substrates for Complex I or II and in the presence or absence of the respiratory chain inhibitors rotenone, antimycin, and myxothiazol. This finding is consistent with the previous report of Stuart et al. (35) that protein oxidation is not increased in cardiac mitochondria of OGG1−/− despite elevated 8-oxo-dG content. Sufficient oxidative damage to mtDNA is associated with increased apoptosis (28). Therefore, additional study of these transgenic mice may reveal that OGG1 can reduce apoptosis after TAC treatment and that this could be responsible for the lower level of cardiac fibrosis. Potentially more time after TAC surgery or long-term aging studies may reveal greater benefit of the OGG1 transgene.
Apart from decreased mitochondrial 8-oxo-dG, the principle benefit of OGG1 overexpression was a significant decrease in TAC-induced cardiac fibrosis. This protection was first indicated by reduced Sirius red staining on OGG1 cardiac sections. The OGG1 effect on fibrosis was confirmed by measuring decreased induction of collagen 1 and 3 mRNA expression in OGG1 hearts after TAC surgery. Cardiac fibrosis is primarily mediated by collagen secreting myofibroblasts (39). Myofibroblasts cannot express the OGG1 transgene due to the transgene's regulation by the cardiomyocyte-specific α-MHC promoter (1). Therefore, cardiomyocyte overexpression of OGG1 apparently decreased a profibrotic signal that originated in cardiomyocytes and then acted on myofibroblasts to stimulate production of collagen matrix. It is also surprising that the OGG1 transgene had only a small effect on cardiac hypertrophy but a larger effect on fibrosis. This may imply that the profibrotic-signaling process is more sensitive to mtDNA oxidation than the hypertrophic response. Alternatively, the fact that many myofibroblasts can be stimulated by a diffusible molecule may result in amplification of the profibrotic signal produced by the most damaged cardiomyocytes. The large antifibrotic effects of the OGG1 transgene may be due to production of fewer severely injured cardiomyocytes or less profibrotic signaling from these cardiomyocytes.
A limitation of this study is that OGG1 levels were increased in nuclei as well as in mitochondria. This makes it impossible to rule out a role for nuclear OGG1 in the beneficial phenotype produced by the transgene. While this possibility must be considered when interpreting our results, it seems more plausible that mitochondrial OGG1 was the primary actor. OGG1 activity was increased 115% in mitochondria but only by 28% in nuclei and most importantly mitochondrial 8-oxo-dG was significantly reduced by the transgene with or without TAC treatment but there was no change in nuclear 8-oxo-dG under either condition.
In summary, cardiac overexpression of OGG1 reduced mtDNA content of 8-oxo-dG in normal and stressed hearts. After TAC, the protection of mtDNA was associated with a slightly reduced hypertrophic response and clearly reduced cardiac fibrosis. These findings support the concept that mtDNA oxidation contributes to LV remodeling during pressure overload hypertrophy.
GRANTS
This work was supported by National Institutes of Health Grants DK-073586 (to P. N. Epstein), HL-094419 and HL-083320 (to S. P. Jones), and NCRR COBRE P20-RR-024489.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Aikawa R, Huggins GS, Snyder RO. Cardiomyocyte-specific gene expression following recombinant adeno-associated viral vector transduction. J Biol Chem 277: 18979–18985, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci USA 103: 10086–10091, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J 14: 312–318, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Boiteux S, Radicella JP. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie 81: 59–67, 1999 [DOI] [PubMed] [Google Scholar]
- 5. Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys 377: 1–8, 2000 [DOI] [PubMed] [Google Scholar]
- 6. Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet 360: 1323–1325, 2002 [DOI] [PubMed] [Google Scholar]
- 7. de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA Ddepends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res 61: 5378–5381, 2001 [PubMed] [Google Scholar]
- 8. Ferreira AL, Salvadori DM, Nascimento MC, Rocha NS, Correa CR, Pereira EJ, Matsubara LS, Matsubara BB, Ladeira MS. Tomato-oleoresin supplement prevents doxorubicin-induced cardiac myocyte oxidative DNA damage in rats. Mutat Res 631: 26–35, 2007 [DOI] [PubMed] [Google Scholar]
- 9. Gao X, Campian JL, Qian M, Sun XF, Eaton JW. Mitochondrial DNA damage in iron overload. J Biol Chem 284: 4767–4775, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet 9: 246–249, 1993 [DOI] [PubMed] [Google Scholar]
- 11. Guibourt N, Boiteux S. Expression of the Fpg protein of Escherichia coli in Saccharomyces cerevisiae: effects on spontaneous mutagenesis and sensitivity to oxidative DNA damage. Biochimie 82: 59–64, 2000 [DOI] [PubMed] [Google Scholar]
- 12. Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem 266: 9180–9185, 1991 [PubMed] [Google Scholar]
- 13. Hamilton ML, Guo Z, Fuller CD, Van RH, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res 29: 2117–2126, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hill JA, Olson EN. Cardiac plasticity. N Engl J Med 358: 1370–1380, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res 29: 573–579, 1998 [DOI] [PubMed] [Google Scholar]
- 16. Ide T, Tsutsui H, Kinugawa S, Suematsu N, Hayashidani S, Ichikawa K, Utsumi H, Machida Y, Egashira K, Takeshita A. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ Res 86: 152–157, 2000 [DOI] [PubMed] [Google Scholar]
- 17. Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res 106: 1541–1548, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim T, Thu VT, Han IY, Youm JB, Kim E, Kang SW, Kim YW, Lee JH, Joo H. Does strong hypertrophic condition induce fast mitochondrial DNA mutation of rabbit heart? Mitochondrion 8: 279–283, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA 96: 13300–13305, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuppuswamy D, Kerr C, Narishige T, Kasi VS, Menick DR, Cooper G., 4th Association of tyrosine-phosphorylated c-Src with the cytoskeleton of hypertrophying myocardium. J Biol Chem 272: 4500–4508, 1997 [DOI] [PubMed] [Google Scholar]
- 21. Lebrecht D, Setzer B, Ketelsen UP, Haberstroh J, Walker UA. Time-dependent and tissue-specific accumulation of mtDNA and respiratory chain defects in chronic doxorubicin cardiomyopathy. Circulation 108: 2423–2429, 2003 [DOI] [PubMed] [Google Scholar]
- 22. Lu H, Fedak PW, Dai X, Du C, Zhou YQ, Henkelman M, Mongroo PS, Lau A, Yamabi H, Hinek A, Husain M, Hannigan G, Coles JG. Integrin-linked kinase expression is elevated in human cardiac hypertrophy and induces hypertrophy in transgenic mice. Circulation 114: 2271–2279, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Mandavilli BS, Santos JH, Van HB. Mitochondrial DNA repair and aging. Mutat Res 509: 127–151, 2002 [DOI] [PubMed] [Google Scholar]
- 24. Melov S, Coskun P, Patel M, Tuinstra R, Cottrell B, Jun AS, Zastawny TH, Dizdaroglu M, Goodman SI, Huang TT, Miziorko H, Epstein CJ, Wallace DC. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci USA 96: 846–851, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA 89: 7022–7025, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell 10: 1637–1652, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palmeira CM, Serrano J, Kuehl DW, Wallace KB. Preferential oxidation of cardiac mitochondrial DNA following acute intoxication with doxorubicin. Biochim Biophys Acta 1321: 101–106, 1997 [DOI] [PubMed] [Google Scholar]
- 28. Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Oxidative stress, mitochondrial bioenergetics, and cardiolipin in aging. Free Radic Biol Med 48: 1286–1295, 2010 [DOI] [PubMed] [Google Scholar]
- 29. Paz-Elizur T, Ben-Yosef R, Elinger D, Vexler A, Krupsky M, Berrebi A, Shani A, Schechtman E, Freedman L, Livneh Z. Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Cancer Res 66: 11683–11689, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Paz-Elizur T, Krupsky M, Blumenstein S, Elinger D, Schechtman E, Livneh Z. DNA repair activity for oxidative damage and risk of lung cancer. J Natl Cancer Inst 95: 1312–1319, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med 49: 587–596, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol 294: C413–C422, 2008 [DOI] [PubMed] [Google Scholar]
- 33. Serrano J, Palmeira CM, Kuehl DW, Wallace KB. Cardioselective and cumulative oxidation of mitochondrial DNA following subchronic doxorubicin administration. Biochim Biophys Acta 1411: 201–205, 1999 [DOI] [PubMed] [Google Scholar]
- 34. Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287: E896–E905, 2004 [DOI] [PubMed] [Google Scholar]
- 35. Stuart JA, Bourque BM, de Souza-Pinto NC, Bohr VA. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic Biol Med 38: 737–745, 2005 [DOI] [PubMed] [Google Scholar]
- 36. Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci USA 100: 10670–10675, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsutsui H. Oxidative stress in heart failure: the role of mitochondria. Intern Med 40: 1177–1182, 2001 [DOI] [PubMed] [Google Scholar]
- 38. Tsutsui H, Kinugawa S, Matsushima S. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc Res 81: 449–456, 2009 [DOI] [PubMed] [Google Scholar]
- 39. van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat Rev Cardiol 7: 30–37, 2010 [DOI] [PubMed] [Google Scholar]
- 40. Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 5: 145–152, 2006 [DOI] [PubMed] [Google Scholar]
- 41. Wang J, Song Y, Elsherif L, Song Z, Zhou G, Prabhu SD, Saari JT, Cai L. Cardiac metallothionein induction plays the major role in the prevention of diabetic cardiomyopathy by zinc supplementation. Circulation 113: 544–554, 2006 [DOI] [PubMed] [Google Scholar]
- 42. Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM, Long BW, Prabhu SD, Xuan YT, Jones SP. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci USA 107: 17797–17802, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yakes FM, Van HB. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA 94: 514–519, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]