Abstract
Objectives
Apolipoprotein E (APOE) is the most statistically significant genetic risk factor for late-onset Alzheimer’s disease (LOAD). The linkage disequilibrium pattern around the APOE gene has made it difficult to determine whether all of the association signal is derived from APOE or if there is an independent signal from a nearby gene. In this study we attempted to replicate a recently reported association of APOE 3-TOMM40 haplotypes with risk and age at onset.
Design
We used standard techniques to genotype several polymorphisms in the APOE-TOMM40 region in a large case-control series, in a series with cerebrospinal fluid biomarker data and in brain tissue.
Results
We failed to replicate the previously reported association of the polyT polymorphism (rs10524523) with risk and age at onset. We found a significant association between rs10524523 and risk for LOAD among APOE 33 homozygotes but in the opposite direction to the previously reported association (the very-long allele was underrepresented in cases compared to controls in our study (allele frequency: 0.41 vs. 0.48 respectively; p=0.004)). We found no association between rs10524523 and CSF tau or Aβ42 levels or TOMM40 or APOE gene expression.
Conclusions
Although we were not able to replicate the earlier association between the APOE 3-TOMM40 haplotypes and age at onset, we did observe that the polyT polymorphism is associated with risk for LOAD among APOE 33 homozygotes in a large case-control series, but in the opposite direction to the previous report. Additional studies in very large samples will be needed to confirm this association.
Introduction
The most statistically significant signals in genome-wide association studies (GWAS) for late-onset Alzheimer’s disease (LOAD) are detected with single nucleotide polymorphisms (SNPs) in the region encoding Apolipoprotein E (APOE) and TOMM40 (translocase of outer mitochondrial membrane 40 homolog).1–10 For several reasons it is difficult to determine whether the signals in this area are due solely to the APOE genotype. First the SNPs that code for the APOE ε2, ε3 and ε4 isoforms are not included in the most popular genome-wide SNPs chips. Second, extensive linkage disequilibrium (LD) in this region of the genome makes it difficult to definitively determine the genetic variant(s) that drive the association. Third, several SNPs up to 50 Kb from APOE exhibit very significant associations with LOAD.
Prior studies have explored this issue using case-control datasets, endophenotypes and genome-wide pathway analyses.11–13 Roses et al. used DNA sequencing and an evolutionary network approach to demonstrate that a polyT polymorphism in intron 6 of TOMM40 (rs10524523) is associated with age at onset (AAO) in APOE 33 and 34 individuals.12 Roses et al. found that the very-long allele of rs10524523 is associated with increased risk and lower AAO for LOAD.
In this study we attempted to replicate the findings from Roses et al.12 We analyzed a large case control sample to test whether APOE 3-TOMM40 haplotypes or TOMM40 alleles exhibit an APOE independent effect on risk for disease, AAO of LOAD, CSF biomarker levels and expression of TOMM40/APOE in the brain.
Material and Methods
Subjects
Risk for disease and age at onset analyses were performed in a total of 1594 LOAD cases (474 APOE 33 homozygous) and 1190 cognitively normal controls (701 APOE 33 homozygous) that were matched for age, gender and ethnicity. These samples were obtained from the Knight Alzheimer’s Disease Research Center at Washington University (WU-ADRC) (759 cases, 345 controls) and the National Institute of Aging Late Onset Alzheimer Disease Family Study (NIA-LOAD Family Study) (835 cases and 845 controls). Each of the cases received a diagnosis of dementia of the Alzheimer’s type (DAT), using criteria equivalent to the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer’s Disease and Related Disorders Association for probable AD.14, 15 Individuals with a clinical dementia rating (CDR) of 0.5 who did not meet clinical criteria for probable Alzheimer’s disease were not included in the analyses. Controls received the same assessment as the cases but were cognitively normal. All individuals were of European descent and written consent was obtained from all participants.
Expression studies were carried out using cDNA obtained from the parietal lobes of 82 AD cases and 39 cognitively normal individuals (CDR=0) obtained through the WU-ADRC Neuropathology Core.
Association with CSF tau, tau phosphorylated at threonine 181 (ptau181), Aβ42 andAβ40 levels was tested in an independent series of 474 samples from the WU-ADRC and 259 samples from the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Table 1). CSF was collected and biomarker measurements obtained as described previously.16–18
Table 1.
Demographics of the samples
| Sample | n | Age (yrs) Mean±SD(range) |
Male (%) | APOE ε4+ (%) | CDR |
|---|---|---|---|---|---|
| WU-ADRC CC | |||||
| cases | 759 | 73±9 (44–102) | 45 | 57 | >0 |
| controls | 345 | 77±8 (60–103) | 40 | 41 | =0 |
| NIA-LOAD CC | |||||
| cases | 835 | 71±47 (48–98) | 33 | 76 | >0 |
| controls | 845 | 75±9 (42–101) | 39 | 30 | =0 |
| WU-ADRC brain series | |||||
| cases | 82 | 86±7 (72–102) | 45 | 41 | >0 |
| controls | 39 | 85±9 (64–107) | 41 | 23 | =0 |
| WU-ADRC-CSF | 474 | 68 ± 11 (45–94) | 39 | 40 | 0=72%:>0=28% |
| ADNI-CSF | 259 | 75 ± 6 (56–91) | 56 | 47 | 0=40%:>0=60% |
For cases the age at the last assignment and for controls the age at onset is shown
Abbreviations: CC, Case-Control series; CDR, Clinical Dementia Rating
A summary of the demographics of all subjects is shown in Table 1.
Genotyping
Rs7412 and rs429358 which define the APOE ε2/ε3/ε4 isoforms, rs1160985, and rs4420638 (TOMM40) were genotyped using Kaspar and Taqman genotyping technologies. The APOE genotype for the NIA-LOAD and ADNI series were provided by NIA-LOAD or ADNI. The polyT repeat in intron 6 of TOMM40 (rs10524523) was genotyped using fluorescence-based fragment size analysis (Fig. S1).19 A detailed explanation of the fluorescence-based fragment size genotyping, quality control steps, allele frequency and linkage disequilibrium between the studied polymorphisms can be found in supplementary materials.
Genotype calls
The polyT repeat (rs10524523) genotypes were placed into categories modeled after those reported by Roses et al.12 “short” (246–267 bases pairs (bp)), “long” (268–279 bp) and “very long” (280–289 bp) (Fig. S2). The bp numbers do not correspond to the ones provided by Roses et al. because our numbers refer to the total length of the PCR product, not the number of polyT repeats. See supplementary materials for quality control and call comparison between our study and previous reports.
Gene Expression
Quantification of gene expression was done by real-time PCR as explained previously. 20 We also used the GEO dataset GSE1522221 for replication. See supplementary materials for a detailed explanation.
Phylogenetic Analyses
Because the polyT repeat is reported as the key variant to define TOMM40 clades A and B,12 we used this marker and APOE isoform information to perform analyses based on phylogenetic groups as described by Roses et al.12 Haplotype phase was estimated using PHASE.22 The phylogeny, which represents the evolutionary relatedness of the haploytpes, was estimated using neighbor-joining with 10,000 bootstrapping replicates in the CLC DNA workbench (CLC BIO Aarhus, Denmark; Fig. S3). We tested for differences in mean AAO between APOE 3-TOMM40 clade A and B haplotypes using a t-test. Association of APOE 3-TOMM40 clade A and B haplotypes with case control status was performed using a Fisher’s Exact Test.
Analyses
Additional association tests between the polyT repeat and disease status, age at onset, TOMM40 and APOE brain expression and CSF tau and Aβ42 levels were performed using UNPHASED v3.1.4 and SAS v9.2. Several analyses were restricted to APOE 33 homozygotes, thus removing uncertainty in haplotype phasing as a possible confounding factor. See supplementary materials for a detailed description of the statistical analyses.
Multiple test correction
We tested a total of four SNPs for association with two phenotypes. A conservative threshold for multiple test correction would be to set the significance at p<0.0062, which would be the Bonferroni correction for 4×2 tests. The SNPs that were associated with risk for disease or age at onset were tested for association with CSF biomarker levels and gene expression to investigate potential pathogenic mechanisms. In this case, no multiple test correction was applied because only one or two SNPs with specific hypotheses were tested for association.
ADNI Material and Methods
“Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). The ADNI was launched in 2003 by the NIA, NIBIB, the FDA, private pharmaceutical companies and non-profit organizations. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. The Principal Investigator of this initiative is Michael W. Weiner, MD. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, to participate in the research.” For up-to-date information, see www.adni-info.org.
Results
The main aim of this study was to attempt to replicate the association of the APOE 3-TOMM40 polyT repeat (rs10524523) haplotype groups with AAO and risk reported by Roses et al.12 Frequencies of APOE 3 alleles and APOE genotypes in TOMM40 clades A and B in our phased haplotype data were consistent with those reported12 (Fig. S2). We also included rs4420638 and rs1160985 (TOMM40), that have been reported to be associated with risk for disease or age at onset independent of APOE genotype.23
Association with age at onset
When using gender, but not APOE genotype, as a covariate we found a significant association between the polyT repeat (rs10524523) and AAO in the WU-ADRC+NIA-LOAD case-control series (p=1.03×10−19, gender as covariate). To discern whether this association was driven by the poly-T repeat or by APOE genotype we performed two additional analyses: including APOE genotype as a covariate in the model and analyzing the polyT association in the APOE 33 stratum. When APOE genotype was included as a covariate, the p-value dropped to 0.11 (Table 2), indicating that the association with AAO was driven by the APOE polymorphisms. In our analyses the very long allele carriers had a higher, but not significantly different, AAO than the short allele carriers.
Table 2.
Minor Allele Frequency and p-values for association with risk and age at onset in the entire series and in the APOE 33 substratum
| WU-ADRC CC1 | Entire series | APOE 33 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Minor Allele | MAF | p-value | Minor Allele | MAF | p-value | |||||
| cases | controls | Status | AAO | cases | controls | Status | AAO | |||
| APOE 4+ (rs429358) | C | 0.33 | 0.14 | 1.69×10−26 | 9.78×10−9 | NA | NA | NA | NA | NA |
| rs10524523-polyT | L | 0.32 | 0.14 | 0.14 | 0.04 | VL | 0.37 | 0.44 | 0.10 | 0.04 |
| rs1160985 | T | 0.38 | 0.46 | 0.32 | 0.02 | C | 0.41 | 0.45 | 0.14 | 0.05 |
| rs4420638 | G | 0.36 | 0.20 | 0.29 | 0.35 | G | 0.07 | 0.08 | 0.84 | 0.003 |
| NIA LOAD2 | ||||||||||
| APOE 4+ (rs429358) | C | 0.46 | 0.16 | 3.74×10−68 | 7.08×10−12 | NA | NA | NA | NA | NA |
| rs10524523-polyT | L | 0.48 | 0.17 | 0.55 | 0.96 | VL | 0.44 | 0.49 | 0.11 | 0.69 |
| rs4420638 | G | 0.51 | 0.19 | 0.57 | 0.87 | G | 0.08 | 0.05 | 0.02 | 0.94 |
| WU-ADRC+ NIA3 | ||||||||||
| APOE 4+ (rs429358) | C | 0.41 | 0.16 | 6.07×10−92 | 6.28×10−21 | NA | NA | NA | NA | NA |
| rs10524523-polyT | L | 0.41 | 0.16 | 0.08 | 0.11 | VL | 0.41 | 0.48 | 0.004 | 0.19 |
| rs4420638 | G | 0.43 | 0.20 | 0.82 | 0.30 | G | 0.08 | 0.06 | 0.08 | 0.01 |
MAF= minor allele frequency. AAO= Age at onset. NA=Not applicable. VL: very long
For association with disease status age, gender, and APOE genotype were included as covariates in the entire series and gender and age in the APOE 33 substratum. APOE genotype was not included as a covariate when rs429358 was tested for association.
For association with age at onset, gender and APOE genotype were included as covariates in the entire series and gender in the APOE 33 substratum.
For association with disease status age, gender, APOE genotype, and first to the third principal components factors were included as covariates in the entire series and gender, age and PC1-3 in the APOE 33 substratum. APOE genotype was not included as a covariate when rs429358 was tested for association.
For association with age at onset, gender, APOE genotype and PC1-3 were included as covariates in the entire series and gender and PC1-3 in the APOE 33 substratum.
For association with disease status gender, age and site were included as covariates in the APOE 33 substratum.
For association with age at onset gender and site were included as covariates in the APOE 33 substratum.
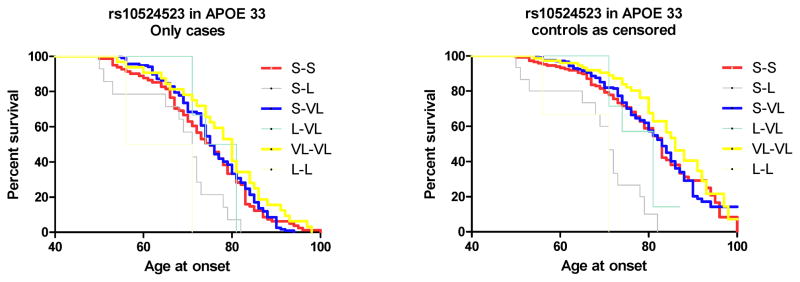
When the analyses were restricted to individuals with APOE 33 genotype, the polyT repeat showed no association with AAO in the WU-ADRC+NIA-LOAD case-control series (p= 0.19, Table 2). The same result was found when the controls were included as censored data in the Kaplan-Meier analyses: in the APOE 33 stratum the very long allele carriers have a higher, but not significantly different AAO than the short allele carriers (Fig. 1). We also found the same pattern among APOE 34 carriers: carriers of the long and very-long alleles had a slightly higher, but not significant, age at onset than carriers of the short alleles (Fig. S4). Haplotype analyses showed similar results. We found a trend toward association between APOE 3-TOMM40 haplotypes and AAO (p=0.057). Individuals with APOE 3-Clade A haplotypes had a mean AAO of 73.31y versus a mean AAO of 72.93y for APOE 3-Clade B. Thus, in our much larger study (total cases=1594, total controls=1190; total APOE 33 cases=474, total APOE 33 controls=701) than the original study (N =34), we found a trend toward association, but in the opposite direction than previously reported. In the APOE 33 stratum rs4420638 showed the most significant association with AAO (p=0.01), but this did not pass multiple test correction.
Figure 1. rs10524523 is not associated with age at onset of Late Onset Alzheimer’s Disease in APOE 33 carriers.
A) Age at onset was analyzed for association with rs10524523 in 282 APOE 33 LOAD cases, from the WU-ADRC series, by the Kaplan-Meier method and tested for significant differences by Log-rank test. The short-short, short-very long and very long-very long genotypes are highlighted because they were the most frequent in this stratum. No significant differences in the survival curves were found (p>0.05). B) Age at onset was analyzed for association with rs10524523 in 282 LOAD cases and 213 controls from the WU-ADRC series with an APOE 33 genotype of by the Kaplan-Meier method and tested for significant differences by Log-rank test. The short-short, short-very long and very long-very long genotypes are highlighted because they were the most frequent in this stratum. No significant differences in the survival curves were found (p>0.05).
Association with risk for disease
We also analyzed whether the TOMM40 polyT repeat (rs10524523) was associated with risk for LOAD. We found an allelic association (p=4.14×10−88) when gender and age, but not APOE genotype were included as covariates. The polyT repeat showed a trend to association with risk for LOAD in the WU-ADRC+NIA-LOAD case-control series when APOE genotype was included in the model (Table 2, p=0.08). When we restricted this analysis to individuals with APOE 33 genotype and used age and gender as covariates, there was a significant association with risk (p=0.004, Table 2). The frequency of the very-long allele of the polyT repeat (rs10524523) was significantly lower in cases compared to controls in the WU-ADRC+NIA-LOAD series (0.41 vs 0.48; p=0.004; OR= 0.78; 95%CI= 0.65–0.95; Table 2). In this case the association passed the multiple test correction threshold (α=0.006). No other studied SNP showed a significant association with risk.
Evaluation of possible mechanisms of disease risk: Association with CSF biomarker levels and gene expression
To determine a possible mechanism underlying the observed disease risk associated with the polyT repeat (rs10524523) we examined several endophenotypes including CSF Aβ and tau levels and APOE and TOMM40 gene expression in the brain. A very strong association was observed between rs10524523 and CSF Aβ42 levels, when CDR, age and gender, but not APOE genotype, were included as covariates (p= 4.50×10−8 for the WU-ADRC CSF series, and p= 2.42×10−12 in the WU-ADRC+ADNI CSF series). However this association was driven by APOE genotype because inclusion of APOE genotype as a covariate in the model eliminated the association between rs10524523 and CSF Aβ42 levels (p=0.49, for the WU-ADRC CSF series, and p= 0.40 in the WU-ADRC+ADNI CSF series; Table S2) suggesting that the rs10524523 association reflects LD with APOE genotype. We also failed to detect association between rs10524523 and CSF Aβ42, tau, or ptau181 in the entire series and in the APOE 33 stratum. For the WU-ADRC-CSF samples we also had CSF Aβ40 but we found no association between the polyT repeat and this phenotype in the entire series or in the APOE 33 stratum (Table S2).
Lastly, we tested whether these polymorphisms are associated with variability in APOE or TOMM40 mRNA expression in human parietal cortex. There was a marginal correlation between the cDNA levels of APOE and TOMM40 with a p-value of 0.0006 and a Pearson correlation coefficient of −0.33. Since our brain samples are derived from both cognitively normal CDR=0 and demented individuals (CDR>0.5) we first tested whether there was an association between mRNA levels and CDR. We found no association between APOE cDNA levels and CDR (p=0.63; age, gender and postmortem interval as covariates). We found a very significant association between TOMM40 cDNA levels and CDR (p=3.55×10−3; age, gender, APOE genotype and postmortem interval as covariates), in the WU-ADRC neuropathology series (82 AD cases and 39 cognitively normal individuals). However, we failed to replicate this finding in the GEO dataset GSE1522221. In this dataset the TOMM40 cDNA levels in cases (n=176) and controls (n=188) are not significantly different (p=0.174).
We found no association between any studied SNP and either TOMM40 or APOE cDNA levels (Table S3). We also did not detect association between APOE or TOMM40 cDNA expression and APOE genotype (p=0.45 and p=0.63, respectively). The association between TOMM40 cDNA levels and CDR led us to stratify the samples by CDR for further analyses but we failed to detect association between any SNP in either cases or controls (Table S3). We also analyzed the APOE 33 stratum alone but found no associations (data not shown).
Discussion
It is not clear whether all the association with risk for LOAD found in the APOE-TOMM40 gene region in the GWAS1–10 is driven by APOE genotype. Identification of new polymorphism/genes that modify risk for LOAD could provide a better understanding of the pathways involved in LOAD, as well as identify new drug targets for AD treatment. In this study we attempted to replicate the recent report by Roses et al.12 that reported an association of APOE 3-TOMM40 polyT polymorphism (rs10524523) haplotypes are associated with age at onset and risk of AD. We also performed extensive analyses in APOE 33 individuals and analyzed several endophenotypes for LOAD to investigate different potential effects of the TOMM40 polymorphisms. We failed to find a significant association between the polyT polymorphism (rs10524523) and age at onset despite the fact that our large series of 2784 individuals (1175 APOE33s) provides high statistical power. Indeed, we found that among APOE 33 and 34 individuals the longer alleles of the poly-T polymorphism are associated with a later onset and a protective effect, which is in the opposite direction to the association in the original report.12 We also studied two SNPs in TOMM40 that have been suggested to modify risk for AD or age at onset23, but found no significant association when APOE genotype was included as a covariate or when the APOE 33 stratum was analyzed alone.
We found that the frequency of the very-long alleles of the polyT is significantly lower in cases compared to controls in two independent series (7% and 5% lower for the WU-ADRC and NIA-LOAD case-control series, respectively). Only when we combined the two independent case-control series were we able to find a significant association after multiple test correction in the APOE 33 stratum. In the joint analyses, allele frequency for the very-long rs10524523 allele was 0.41 and 0.48 in cases and controls, respectively (p=0.004; OR= 0.78; 95% CI= 0.65–0.95), which is opposite that reported by Roses et al.12 While the magnitude of the observed effect is stronger than that observed for the GWAS significant SNP in CLU, rs11136000, (0.36 vs 0.40; OR=0.88, 95% CI=0.86–0.91) a 4 fold bigger sample size would be required to detect a genome-wide significant association for rs10524523 among the APOE33 homozygous (i.e., 6376 unselected cases and 4760 unselected controls or 1896 APOE 33 cases and 2804 APOE 33 controls).
Our results suggest that the TOMM40 polyT repeat may be associated with risk for disease. TOMM40 is in close proximity to APOE, but it is unknown whether the polyT repeat affects risk for AD through an APOE-dependent mechanism or a totally independent mechanism. We used CSF biomarker phenotypes to test whether the polyT repeat increases risk for LOAD through an Aβ42 (like APOE,20, 25–27 ) or a tau-dependent mechanism, but our results suggest that the polyT repeat may influence risk for AD through another mechanism. We also found no association of the polyT repeat with APOE or TOMM40 mRNA expression in parietal cortex. We were unable to find evidence for any obvious potential mechanism that could explain the association with risk, but the power of these analyses are limited and studies in larger series should be performed to identify the potential disease mechanism.
In conclusion our data do not support the findings reported by Roses et al.12, as we observed no association between APOE 3-TOMM40 clade A and B haplotypes or the polyT repeat (rs10524523) and age at onset. We did observe an association between the polyT repeat and risk for disease, but in the opposite that reported previously.12. It is unclear whether or not these results represent a type 1 error but highlight the importance of using large series, particularly when evaluating APOE subgroups, which requires sample stratification reducing power. Confirmation that rs10524523 is independently associated with AD risk will require a much larger sample. This could potentially be accomplished by imputation of rs1160985, a SNP which is in high LD (r2=0.93) with the polyT variant among the APOE 33 carriers, in the large GWAS datasets 9, 10, 28.
TOMM40 codes for a mitochondrial protein, suggesting that mitochondrial integrity and/or energy metabolism could play an important role in LOAD. Mitochondrial morphology is altered in AD brains and several studies have reported deficiencies in energy-related enzymes. The hypothesis that mitochondria may play an important role in LOAD is also supported by the fact that we did not find any association between the TOMM40 polymorphism and CSF Aβ42 and tau levels. However more genetic and molecular studies are necessary to determine whether or not the reported genetic association with rs10524523 in TOMM40 is real.
Supplementary Material
Acknowledgments
This work was supported by grants: NIH (AG16208, P01AG03991, P50AG05681, P01AG026276, AG23185, AG05136), the Barnes-Jewish Hospital Foundation and Ford Foundation. The authors thank the Clinical Core of the Knight ADRC for clinical and cognitive assessments of the participants, the Genetics Core of the Knight ADRC for APOE genotypes and the Biomarker Core of the Adult Children Study for the CSF collection and assays. CC has a fellowship from “Fundacion Alfonso Martin Escudero”.
Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We thank contributors, including the Alzheimer’s Disease Centers who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
“Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.”
References
- 1.Grupe A, Abraham R, Li Y, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007 Apr 15;16(8):865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 2.Coon KD, Myers AJ, Craig DW, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007 Apr;68(4):613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 3.Reiman EM, Webster JA, Myers AJ, et al. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007 Jun 7;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram L, Lange C, Mullin K, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008 Nov;83(5):623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Wetten S, Li L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008 Jan;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 6.Carrasquillo MM, Zou F, Pankratz VS, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009 Feb;41(2):192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feulner TM, Laws SM, Friedrich P, et al. Examination of the current top candidate genes for AD in a genome–wide association study. Mol Psychiatry. 2009 Jan 6; doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- 8.Beecham GW, Martin ER, Li YJ, et al. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009 Jan;84(1):35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009 Oct;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009 Oct;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 11.Yu CE, Seltman H, Peskind ER, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007 Jun;89(6):655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roses AD, Lutz MW, Amrine-Madsen H, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2009 Dec 22; doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong MG, Alexeyenko A, Lambert JC, Amouyel P, Prince JA. Genome-wide pathway analysis implicates intracellular transmembrane protein transport in Alzheimer disease. J Hum Genet. 2010 Oct;55(10):707–709. doi: 10.1038/jhg.2010.92. [DOI] [PubMed] [Google Scholar]
- 14.Berg L, McKeel DW, Jr, Miller JP, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer’s disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998 Mar;55(3):326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 15.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984 Jul;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 16.Fagan AM, Mintun MA, Mach RH, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006 Mar;59(3):512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 17.Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007 Mar;64(3):343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 18.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009 Mar 18; doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imle P. Fluorescence-based fragment size analysis. Methods Mol Biol. 2005;311:139–146. doi: 10.1385/1-59259-957-5:139. [DOI] [PubMed] [Google Scholar]
- 20.Cruchaga C, Kauwe JS, Mayo K, et al. SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer’s Disease. PLoS Genet. 2010;6:9. doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webster JA, Gibbs JR, Clarke J, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009 Apr;84(4):445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003 Nov;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huentelman MJ, Corneveaux J, Myers A, et al. ICAD-2010. ICAD-2010. Hawaii: Alzheimer’s Association; 2010. Genome-Wide Association Study for Alzheimer’s Disease Risk in a Large Cohort of Clinically Characterized and Neuropathologically Verified Subjects; p. 16. Vol Hot Topics Addendum. [Google Scholar]
- 24.Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007 Dec;39(12):1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 25.Kauwe JS, Cruchaga C, Mayo K, et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc Natl Acad Sci U S A. 2008 Jun 10;105(23):8050–8054. doi: 10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauwe JS, Jacquart S, Chakraverty S, et al. Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer’s disease presenilin 1 mutation. Ann Neurol. 2007 May;61(5):446–453. doi: 10.1002/ana.21099. [DOI] [PubMed] [Google Scholar]
- 27.Kauwe JS, Cruchaga C, Bertelsen S, et al. Validating predicted biological effects of Alzheimer’s disease associated SNPs using CSF biomarker levels. J Alzheimers Dis. 2010;21(3):833–842. doi: 10.3233/JAD-2010-091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun G, Naj AC, Beecham GW, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010 Dec;67(12):1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.