Abstract
Fanconi anemia (FA) is a rare genetic disorder characterized by bone marrow failure and an increased risk for leukemia and cancer. Fifteen proteins thought to function in the repair of DNA interstrand crosslinks (ICLs) comprise what is known as the FA-BRCA pathway. Activation of this pathway leads to the monoubiquitylation and chromatin localization of FANCD2 and FANCI. It has previously been shown that FANCJ interacts with the mismatch repair (MMR) complex MutLα. Here we show that FANCD2 interacts with the MMR proteins MSH2 and MLH1. FANCD2 monoubiquitylation, foci formation and chromatin loading are greatly diminished in MSH2-deficient cells. Human or mouse cells lacking MSH2 or MLH1 display increased sensitivity and radial formation in response to treatment with DNA crosslinking agents. Studies in human cell lines and Drosophila mutants suggest an epistatic relationship between FANCD2, MSH2 and MLH1 with regard to ICL repair. Surprisingly, the interaction between MSH2 and MLH1 is compromised in multiple FA cell lines, and FA cell lines exhibit deficient MMR. These results suggest a significant role for MMR proteins in the activation of the FA pathway and repair of ICLs. In addition, we provide the first evidence for a defect in MMR in FA cell lines.
INTRODUCTION
Fanconi anemia (FA) is a rare autosomal or X-linked recessive genetic disorder characterized by congenital abnormalities, bone marrow failure and an increased susceptibility to cancer and leukemia. Fifteen FA genes have now been identified that when mutated result in hypersensitivity to DNA crosslinking agents such as mitomycin C (MMC) or cisplatin (CDDP). For this reason, the proteins encoded by these genes are thought to function in the removal and repair of DNA interstrand crosslinks (ICLs) (1–3). Because of the complex nature of ICLs, several repair pathways are thought to converge to repair these lesions, with FA proteins garnering assistance from other repair machinery such as that involved in homologous recombination (HR) and nucleotide excision repair (4).
Eight of the 15 FA proteins (FANCA, B, C, E, F, G, L and M) form what is known as the FA core complex. All members of the core complex are essential for the monoubiquitylation of FANCD2 and FANCI after DNA damage or during S phase, and this event is considered the hallmark of FA pathway activation (5). Once monoubiquitylated, FANCD2 and FANCI are loaded onto chromatin (6), where they have been shown to co-localize in nuclear foci with three of the remaining FA proteins: FANCJ/BRIP1/BACH1, FANCN/PALB2 and FANCD1/BRCA2 (7–10). Recently identified as FA proteins, FANCO/RAD51C (11) and FANCP/SLX4 (12) are also involved in the later stages of ICL repair during HR. Owing to the increasing link between FA and familial breast cancer genes, this pathway is often referred to as the FA-BRCA pathway.
Mismatch repair (MMR) is a repair system highly conserved from Escherichia coli to humans for the correction of base substitutions and insertion–deletion loops (IDLs) that can arise in nascent DNA strands during replication (13). In humans, two protein complexes, MutSα and MutSβ, named for their homology with the E. coli protein MutS, exist for the recognition and binding of mismatches (14). MutSα, composed of MSH2 and MSH6, is primarily responsible for the detection of single-base mismatches and small IDLs, whereas MutSβ, composed of MSH2 and MSH3, is responsible for the detection of and repair of IDLs of up to 16 extra bases (15–17). Once detected, MutS complexes recruit the MutLα complex, composed of MLH1 and PMS2, which coordinates the remaining steps in MMR (18).
In addition to their role in MMR, MMR proteins have also been implicated in somatic hypermutation, VDJ recombination and the recognition of lesions caused both by the environment and chemotherapeutic agents (13). MutSα and MutLα have been shown to be required for the recruitment of ATR and ATRIP to O6-methylguanine adducts (19), and more recently, MSH2 was reported to be required for the recruitment of ATR after CDDP treatment (20). In addition, several previous reports suggest that MutS complexes may be involved in the detection and processing of ICLs. MutSα has been shown to bind ICLs produced by CDDP (21). Several groups have also reported that repair of psoralen ICLs is dependent on MutSβ (22–24). Taken together with other recent studies showing an interaction between FANCJ and MutLα (25), it seemed plausible that there might be a functional overlap between the MMR and FA-BRCA pathways.
In this study, we identify MSH2 and MLH1 as novel FANCD2-binding partners. We show by immunoprecipitation that the interaction between FANCD2 and MSH2 and MLH1 is induced upon treatment with DNA interstrand crosslinking agents. MSH2 specifically binds the monoubiquitylated form of FANCD2 (FANCD2-L), and this interaction requires ATR, but not ATM or BRCA1. MSH2-deficient cells show greatly diminished monoubiquitylation and chromatin loading of FANCD2 and FANCI and FANCD2 foci formation, whereas MLH1-deficient cells do not. Both MSH2- and MLH1-deficient cells display hypersensitivity and increased radial formation when exposed to DNA interstrand crosslinking agents. Studies in human cells and Drosophila mutants indicate an epistatic relationship between MSH2, MLH1 and FANCD2 with regard to ICL repair. These data suggest a significant role for MMR factors in ICL repair and in the activation of the FA pathway.
In addition, we provide the first evidence for a defect in MMR in FA cell lines. The interaction between MSH2 and MLH1 is reduced in both core complex and FANCD2 mutant cell lines. Using a plasmid-based assay, we also show that MMR is defective in several FA cell lines to a similar degree as MSH2-deficient cells. These data indicate a role for FA proteins in MMR and suggest significant crosstalk between the MMC and FA-BRCA pathways.
RESULTS
Identification of novel FANCD2-interacting proteins
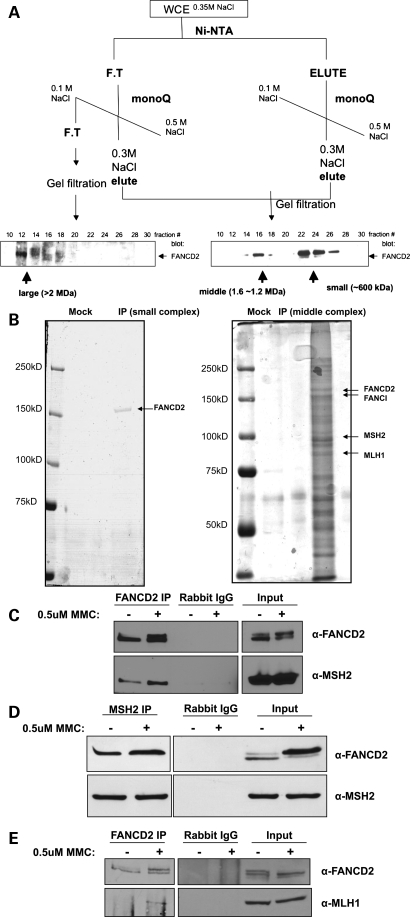
Given that the monoubiquitylation of FANCD2 and FANCI represents a central event in the FA pathway, we sought to purify FANCD2-containing complexes for the purpose of identifying novel FANCD2-interacting proteins. We have previously described a chromatography purification scheme that revealed the presence of three distinct FANCD2 subcomplexes [26 (permission granted)] (Fig. 1A). Using this scheme, followed by immunoprecipitation with anti-FLAG resin, we purified two FANCD2-containing protein complexes from whole-cell extracts of PD20 cells stably expressing FLAG-tagged FANCD2. The ‘small’ and ‘middle’ protein complexes were then eluted from the resin using FLAG peptide and subjected to SDS–polyacrylamide gel electrophoresis (PAGE), followed by silver-staining (Fig. 1B). Silver-stained bands were trypsin-digested and analyzed by liquid chromatography-mass spectrometry (LC-MS). Mass spectrometry results showed that the ‘small’ complex was composed solely of FANCD2, whereas the ‘middle’ complex contained many novel FANCD2-binding partners, including MSH2 and MLH1, in addition to known binding partners such as FANCI (27). In order to verify the interaction between FANCD2 and MMR proteins, endogenous FANCD2 was immunoprecipitated from whole-cell extracts of HeLa cells untreated or treated with MMC. Immunoblotting confirmed that MSH2 and MLH1 co-precipitate with endogenous FANCD2 and that this interaction is induced upon treatment with MMC (Fig. 1C and E). Reciprocally, FANCD2 co-precipitates with endogenous MSH2 (Fig. 1D).
Figure 1.
Identification of novel FANCD2-binding partners. (A) Whole-cell extracts from PD20 cells stably expressing FLAG-tagged FANCD2 were subjected to the chromatography scheme represented here. Three distinct FANCD2-containing protein complexes were identified by western blot and named ‘small’, ‘middle’ and ‘large’ complexes. (B) The ‘small’ and ‘middle’ protein complexes were immunoprecipitated using anti-FLAG resin, eluted using FLAG peptide and subjected to SDS–PAGE followed by silver-staining. The ‘small’ complex appeared as a single band, whereas silver-staining of the ‘middle’ complex revealed many protein bands, suggesting the presence of several different FANCD2-containing protein complexes. Bands were cut out, trypsin-digested and analyzed by mass spectrometry to reveal the novel FANCD2-binding partners, MSH2 and MLH1. (C–E) Immunoprecipitation of endogenous FANCD2 and MSH2 from HeLa whole-cell extracts confirms that FANCD2, MSH2 and MLH1 co-precipitate and this interaction is induced upon damage with MMC.
Characterization of the FANCD2–MSH2 and FANCD2–MLH1 interactions
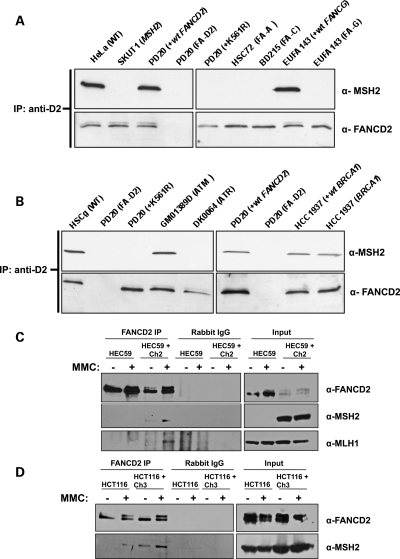
Since these proteins co-precipitated in an MMC-dependent manner, we next wanted to determine whether MSH2 and MLH1 interact with FANCD2-S or FANCD2-L. Endogenous FANCD2 was immunoprecipitated from whole-cell extracts of human cell lines deficient in several different FA core complex proteins or stably expressing wild-type FANCD2 or the ubiquitylation-resistant form of FANCD2 (K561R). Immunoblotting revealed that MSH2 co-precipitates with FANCD2 only in cells expressing wild-type FANCD2 and not in cells deficient in any core complex protein or in cells expressing only FANCD2-K561R (Fig. 2A). Immunoblotting was used to confirm that each cell line used failed to express detectable levels of the respective FA protein (data not shown; 28). These data demonstrate that MSH2 interacts with FANCD2-L, as FANCD2 cannot be ubiquitylated when any member of the core complex is missing or when cells express the FANCD2-K561R mutant protein.
Figure 2.
Characterization of the FANCD2–MSH2 and FANCD2–MLH1 interactions. (A) Endogenous FANCD2 was immunoprecipitated from whole-cell extracts of human cell lines deficient in FA core complex proteins or stably expressing wild-type FANCD2 or FANCD-K561R after an 18 h treatment with 50 nm MMC. MSH2 co-precipitates with FANCD2-L only. (B) Endogenous FANCD2 was immunoprecipitated from whole-cell extracts of cell lines deficient in ATR, ATM and BRCA1 after an 18 h treatment with 50 nm MMC. ATR is required for the interaction of FANCD2 and MSH2, but not ATM or BRCA1. (C) Endogenous FANCD2 was immunoprecipitated from extracts of MSH2-deficient and corrected cells that are untreated or treated with MMC for 24 h. MLH1 does not co-precipitate with FANCD2 in MSH2-deficient cells. (D) Endogenous FANCD2 was immunoprecipitated from MLH1-deficient cell extracts that are untreated or treated with MMC for 24 h. MSH2 co-precipitates with FANCD2 to a greater extent in MLH1-corrected cells, indicating that MLH1 enhances the interaction between FANCD2 and MSH2.
Since MSH2 has been shown to be involved in early signaling events following the recognition of a lesion, such as recruitment of ATR and ATRIP and activation of Chk1 (19), we next sought to determine whether the interaction between FANCD2 and MSH2 required other DNA repair proteins involved in both early and late signaling events during ICL repair. Immunoprecipitation of endogenous FANCD2 from extracts of human cell lines deficient in ATM, ATR, BRCA1 and their wild-type counterparts revealed that MSH2 co-precipitates with FANCD2 in ATM- and BRCA1-deficient cell lines (Fig. 2B). However, MSH2 did not co-precipitate with FANCD2 in ATR-deficient cells, indicating that ATR is required for this interaction. Immunoblotting was again used to confirm the identity of each cell line by measuring the expression level of the respective protein (data not shown). The interaction between FANCD2 and MSH2 was also intact in MSH3-, ERCC1- and XPF-deficient Chinese hamster cell lines (Ananth et al., data not shown), further emphasizing the importance of ATR in the binding of these proteins.
Given that MSH2 binds to MLH1 during its recruitment to DNA mismatches and IDLs (14), and FANCD2 binds to both MSH2 and MLH1, we were next interested in whether MSH2 was necessary for the interaction between FANCD2 and MLH1 and vice versa. Endogenous FANCD2 was immunoprecipitated from extracts from MMC-treated and untreated MSH2-deficient (HEC59) cells or cell lines corrected with wild-type MSH2 by chromosomal transfer (HEC59 + Ch2). Immunoblotting showed that MLH1 co-precipitates with FANCD2 only in the MSH2-corrected cell lines treated with MMC (Fig. 2C). This implies that MSH2 is either required to be physically present in complex with FANCD2 and MLH1 to stabilize the complex or is required for important upstream signaling events leading to the interaction between FANCD2 and MLH1. We then immunoprecipitated endogenous FANCD2 from MLH1-deficient (HCT116) cells or cell lines corrected with wild-type MLH1 by chromosomal transfer (HCT116 + Ch3). Interestingly, MSH2 co-precipitated with FANCD2 in MLH1-corrected cells, but this interaction was nearly undetectable in MLH1-deficient cells (Fig. 2D). These data show that MLH1, although largely considered to be a downstream effector protein in relation to MSH2, actually enhances the interaction between FANCD2 and MSH2.
MSH2 is required for normal kinetics of monoubiquitylation and chromatin loading of FANCD2 and FANCI
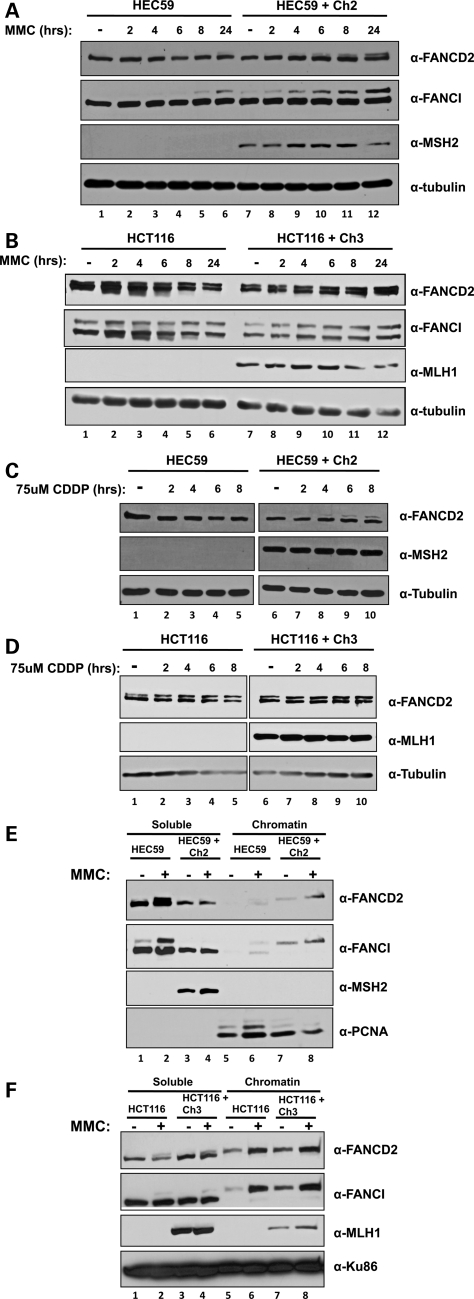
As MSH2 has been implicated in the detection and repair of ICLs (21–24), we next wanted to determine whether these FANCD2-binding partners were involved in the activation of the FA pathway. MSH2-deficient and corrected cells were treated with MMC at several time points over 24 h, collected and lysed, and the resulting cell lysates were analyzed by SDS–PAGE followed by immunoblotting. As shown in Figure 3A, FANCD2 monoubiquitylation is undetectable in MSH2-deficient cells except for a faint upper band corresponding to FANCD2-L at 24 h after treatment. FANCI monoubiquitylation follows suit, with FANCI-L becoming faintly detectable only by 8 h after treatment. However, both FANCD2 and FANCI become monoubiquitylated promptly after MMC treatment in corrected cell lines (compare lanes 1–6 and 7–12). In contrast, the same experiment performed in MLH1-deficient and corrected cell lines reveals that FANCD2 and FANCI are monoubiquitylated to similar extents in untreated cells and as soon as 2 h after MMC treatment (Fig. 3B, compare lanes 1–6 and 7–12). To confirm these results, we next treated the same cell lines over several time points with a second DNA interstrand crosslinking agent, CDDP. Immunoblotting shows that FANCD2 monoubiquitylation is undetectable in MSH2-deficient cells after up to 8 h of treatment with CDDP, whereas FANCD2-L is detectable in corrected cells as soon as 4 h after treatment (Fig. 3C, compare lanes 3–5 with 7–10). Again, MLH1-deficient and corrected cells display similar levels of FANCD2 monoubiquitylation after treatment with CDDP (Fig. 3D). Depletion of MSH2 and MLH1 in HeLa cells by siRNA transfection, followed by treatment with MMC for 4 and 24 h, showed a similar delay in FANCD2 monoubiquitylation after MSH2 depletion but not after MLH1 depletion (Supplementary Material, Fig. S1).
Figure 3.
MSH2 is required for normal kinetics of monoubiquitylation and chromatin loading of FANCD2 and FANCI. (A) MSH2-deficient and corrected cells or (B) MLH1-deficient and corrected cells were treated with 500 nm MMC over several time points for 24 h. Immunoblotting shows FANCD2 and FANCI monoubiquitylation is greatly diminished in MSH2-deficient cells, but not MLH1-deficient cells. (C) MSH2-deficient and corrected cells or (D) MLH1-deficient and corrected cells were treated with 75 μm CDDP over several time points for 24 h. Immunoblotting reveals a similar deficiency in FANCD2 monoubiquitylation in MSH2-deficient cells, but not MLH1-deficient cells. (E) MSH2-deficient and corrected cells or (F) MLH1-deficient and corrected cells were treated with 500 nm MMC for 24 h and then subjected to cellular fractionation. Immunoblotting reveals that although FANCD2 and FANCI chromatin loading is normal in MLH1-deficient cells, chromatin loading of both proteins is nearly undetectable in MSH2-deficient cells.
Monoubiquitylation of FANCD2 and FANCI leads to their chromatin localization, so we turned next to cellular fractionation experiments to determine whether chromatin loading was also impaired in MSH2-deficient cells. As expected, cellular fractionation of MSH2-deficient and corrected cells followed by immunoblotting revealed defective chromatin loading of FANCD2 and FANCI in both untreated and MMC-treated cells lacking MSH2. However, FANCD2 and FANCI were present in detectable levels the chromatin fraction of untreated MSH2-corrected cells, and increased upon treatment with MMC for 24 h (Fig. 3E, compare lanes 5–6 and 7–8). Since unmodified FANCD2 and FANCI are faintly detectable in the chromatin fraction of MSH2-deficient cells, we cannot rule out the possibility that FANCD2 and FANCI are loaded onto chromatin but are not monoubiquitylated in the absence of MSH2. As expected, MLH1-deficient cells are proficient in FANCD2 and FANCI chromatin loading, as both proteins were detectable at similar levels in mutant and corrected cell lines both before and after treatment with MMC (Fig. 3F, compare lanes 5–6 and 7–8). Importantly, mutant cell lines grew at similar rates and displayed similar cell-cycle profiles, so the phenotypes observed cannot be attributed to defects in growth or cell-cycle arrest (Supplementary Material, Fig. S1).
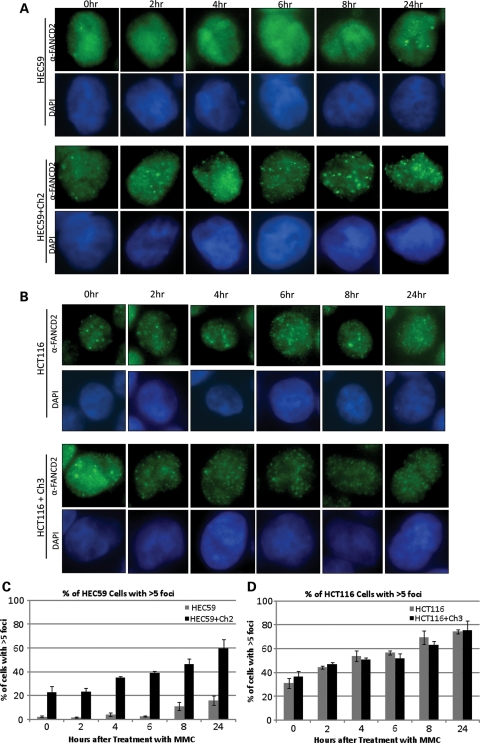
FANCD2 foci formation is impaired in MSH2-deficient cells
FANCD2 has been shown to form nuclear foci, thought to represent chromatin-bound FANCD2, in response to different types of DNA damage. Given that FANCD2 chromatin loading is impaired in MSH2-deficient cells, we next sought to examine FANCD2 foci formation in MMR cell lines. Cells were seeded into eight-well culture slides and then treated with MMC over several time points for 24 h. Immunostaining for FANCD2 reveals that FANCD2 foci formation is significantly delayed in MSH2-deficient cells compared with their corrected counterparts (Fig. 4A). Although foci were visible even in untreated cells and as early as 2 h after MMC treatment in corrected cells, FANCD2 foci did not appear in MSH2-deficient cells until 24 h after treatment with MMC. These results correlate well with the FANCD2 monoubiquitylation and chromatin-loading defects seen in the same cells. As expected, MLH1-deficient and corrected cells display efficient FANCD2 foci formation in both untreated and MMC-treated cells (Fig. 4B). Cells with greater than five foci were counted in each cell type and at each time point shown. As shown in Figure 4C and D, a clear delay in FANCD2 foci formation occurs only in MSH2-deficient cells, whereas all other cell types show an induction in this event following MMC treatment. Taken together, these data identify a significant role for MSH2 in the activation of the FA pathway, whereas MLH1 is functioning either downstream of FANCD2 and FANCI monoubiquitylation or in a separate pathway.
Figure 4.
FANCD2 foci formation is impaired in MSH2-deficient cells. (A) MSH2-deficient and corrected cells or (B) MLH1-deficient and corrected cells were treated with 500 nm MMC over several time points for 24 h. Immunostaining for FANCD2 shows that FANCD2 foci formation is greatly diminished in MSH2-deficient cells through all time points up to 24 h, but not in MLH1-deficient cells. At least 100 cells were counted to determine the percentage of cells with greater than five foci in both (C) MSH2-deficient and corrected cells and (D) MLH1-deficient and corrected cells. Graphs clearly show a significant delay and reduction in FANCD2 foci formation in MSH2-deficient cells.
MSH2 and MLH1 play a key role in ICL repair
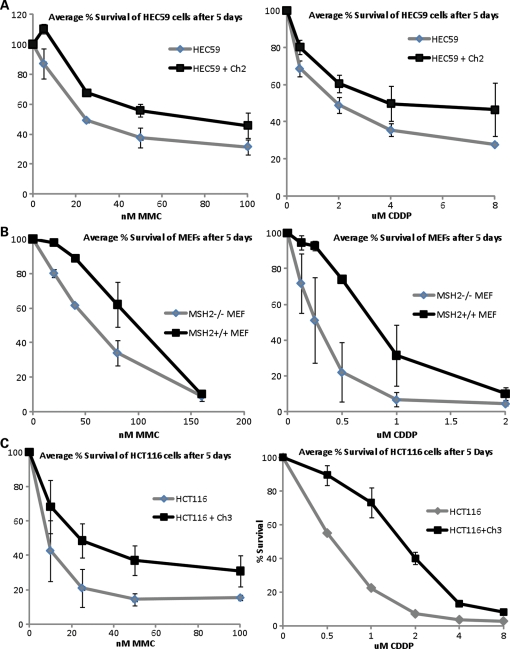
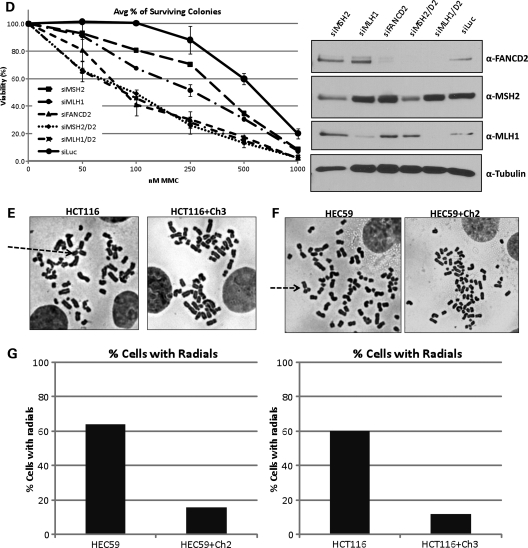
Both MSH2 and MLH1 interact with FANCD2 in response to treatment with DNA interstrand crosslinking agents; however, only MSH2 appears to be required for the activation of the FA pathway. To study the role of these proteins in ICL repair further, we decided to examine the MMR cell lines for two cellular phenotypes—sensitivity and radial formation after treatment with crosslinking agents. MSH2-deficient human and mouse cells and their wild-type counterparts were plated in six-well plates and treated with increasing concentrations of MMC and CDDP. After incubation for 5 days, cells were fixed and stained with crystal violet and then assessed for cell survival by dye extraction, followed by measurement of absorbance on a spectrophotometer. As shown in Figure 5A and B, both human and mouse cells deficient in MSH2 are hypersensitive to MMC and CDDP. Surprisingly, even though FA pathway activation is intact in MLH1-deficient cells, these cells are also clearly hypersensitive to both crosslinking agents (Fig. 5C). These results were also confirmed using colony-formation assays for both MSH2- and MLH1-deficient cells (data not shown). Clonal survival assays also show that MSH3-deficient Chinese hamster ovary (CHO) cells (29) are hypersensitive to MMC (Supplementary Material, Fig. S1). Depletion of MSH2 and MLH1 by siRNA transfection in HeLa cells results in hypersensitivity to MMC compared with control cells in colony-formation assays (Fig. 5D). In addition, the double knockdown of MSH2 and FANCD2 and MLH1 and FANCD2 results in comparable hypersensitivity to MMC as the single knockdown of FANCD2 alone, establishing an epistatic relationship between these three proteins with respect to ICL repair.
Figure 5.
MSH2 and MLH1 play a key role in ICL repair. (A) Human MSH2-deficient and corrected cells, (B) MSH2-deficient MEFs and their wild-type counterparts and (C) human MLH1-deficient and corrected cells were treated with increasing concentrations of MMC and CDDP and assessed for survival using crystal violet staining and extraction. Both MSH2- and MLH1-deficient cells are hypersensitive to DNA interstrand crosslinking agents. (D) Depletion of MSH2, MLH1, FANCD2, MSH2 and FANCD2, or MLH1 and FANCD2 by siRNA transfection followed by crystal violet survival assay shows that MSH2, MLH1 and FANCD2 are epistatic with regard to ICL repair. (E) MLH1-deficient and corrected cells and (F) MSH2-deficient and corrected cells were treated with MMC and dropped onto slides for chromosome breakage analysis. Increased radial formation is evident in both MSH2- and MLH1-deficient cells (marked by arrows). (G) Analysis of metaphase spreads shows a >3-fold increase in radial formation in MSH2- and MLH1-deficient cells versus their corrected counterparts.
We next examined metaphase spreads from the MMR mutant cell lines for increased radial formation in response to treatment with interstrand crosslinking agents, which is a diagnostic cellular phenotype of FA. Cells were treated with MMC for 24 h, collected, swollen in hypotonic buffer and fixed and dropped onto slides. After staining with Giemsa, slides were examined for chromosome breakage and scored for radial formation. As shown in Figure 5E and F, MSH2- and MLH1-deficient cells display increased radial formation in response to MMC compared with their corrected counterparts. MSH2-deficient cells displayed ∼4-fold more radial forms than corrected cells (P = 0.0005), whereas MLH1-deficient cells displayed ∼5-fold more radial forms than corrected cells (P = 0.0004) (Fig. 5G). Taken together, these data indicate a significant role for both MSH2 and MLH1 in ICL repair and an epistatic relationship between FANCD2, MSH2 and MLH1.
Mutation of MSH2 in Drosophila results in diexpoxybutane hypersensitivity and hypermutability
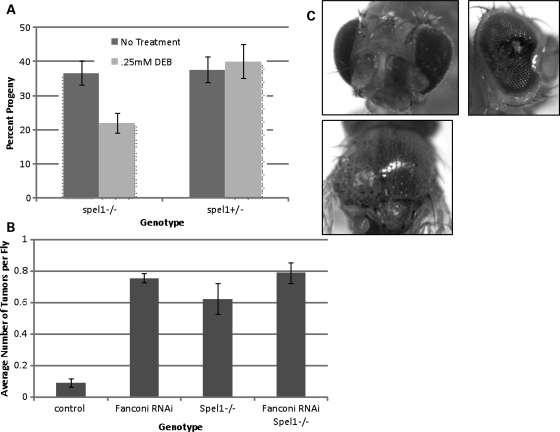
Several FA proteins in humans have been shown to harbor strong homology to Drosophila proteins, including FANCD2, FANCD1, FANCL and FANCM (30–34). Since MSH2 (spel1 in Drosophila) mutant flies have previously been constructed and have been shown to be deficient in MMR (35), we sought to characterize these flies with regard to ICL repair. The progeny from a cross between heterozygous Spel1 mutant flies were exposed to diexpoxybutane (DEB), and sensitivity to drug was assessed as the percent of surviving spel1−/− progeny. As shown in Figure 6A, Spel1 mutant flies are hypersensitive to DNA crosslinking agents, similar to the phenotype seen in human cell lines. FANCD2 RNAi flies have also been previously constructed and shown to exhibit this same hypersensitivity to crosslinking agents (34). Therefore, in order to examine whether the Drosophila homologues of MSH2 and FANCD2 show evidence of the epistatic relationship we see in human cell lines with regard to ICL repair, we assayed mutation frequency following DEB treatment in FANCD2 RNAi and spel−/− mutant flies. For this assay, we utilized flies with a heterozygous mutation in the tumor-suppressor gene, lats. Tumors arise with nearly 100% penetrance following mutation of the remaining functional copy of lats, allowing tumor number to serve as a measure of mutation frequency (36). Flies were crossed in vials, and parental flies were removed after 48 h of egg laying. Progeny were treated with DEB and then examined for tumor formation to determine mutation frequency in each fly genotype. As shown in Figure 6B, Fanconi RNAi, spel1−/− and Fanconi RNAi spel1−/− flies had significantly more tumors than controls. Fanconi RNAi spel1−/− flies did not have significantly more tumors than Fanconi RNAi alone (P = 0.65) or spel1 mutation alone (P = 0.16). These data demonstrate an epistatic relationship between the Drosophila homologues of FANCD2 and MSH2 with regard to ICL repair and suggest that the role for MSH2 in ICL repair may be conserved throughout many species.
Figure 6.
Mutation of MSH2 in Drosophila results in DEB hypersensitivity and increased mutagenesis. (A) spel1 mutant flies (MSH2-deficient) were assessed for sensitivity to DEB as percent of surviving progeny. The expected percentage of each genotype is 33% based on Mendelian ratios. Spel1 mutant flies are hypersensitive to DNA crosslinking agents. (B) Flies with heterozygous mutations in the tumor-suppressor gene lats were treated with 0.25 mm DEB and tumors were counted in resulting progeny to assess mutation frequency. Examples of tumors from Fanconi RNAi flies are shown in (C). Fanconi RNAi, spel1−/− and Fanconi RNAi spel1−/− flies had significantly more tumors than control flies, but Fanconi RNAi spel1−/− flies did not have significantly more tumors than Fanconi RNAi alone or spel1 mutation alone, indicating an epistatic relationship between Drosophila FANCD2 and MSH2 in ICL repair.
MMR is defective in FA cell lines
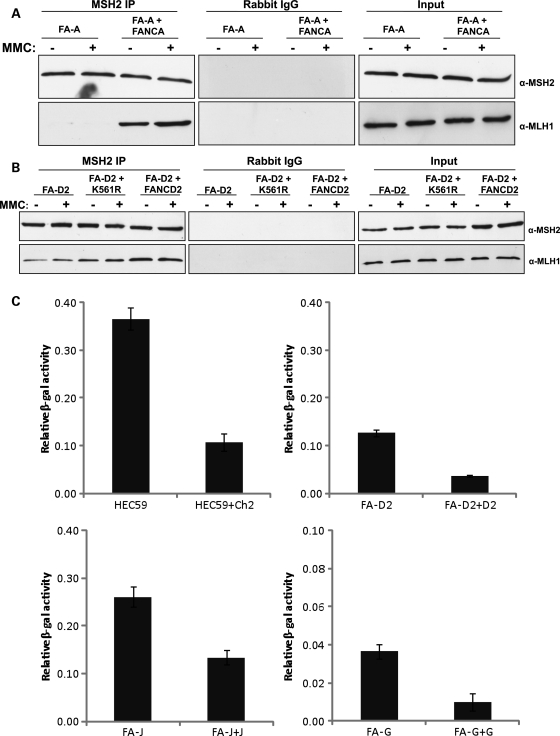
In view of the fact that FANCD2 interacts with both MSH2 and MLH1 and proteins in many DNA repair pathways have proven to be promiscuous in their repair functions, we were interested in whether FA proteins might be involved in the MMR pathway. To that end, we first immunoprecipitated endogenous MSH2 from whole-cell extracts of two FA mutant and corrected cell lines. Surprisingly, immunoblotting revealed that the interaction between MLH1 and MSH2 is compromised in FA mutant cell lines, but intact in corrected cell lines (Fig. 7A and B). Owing to this unexpected finding, we reasoned that FA cell lines may be deficient in MMR as well. MMR has previously been studied in many different contexts using pCAR reporter vectors (37,38). The pCAR-OF vector contains a β-galactosidase (β-gal) gene preceded by a 58 bp CA repeat region (CA29) that sets the reporter gene out of frame. MMR deficiency is characterized by instability of simple repeat sequences such as this one (39), so the reading frame can be restored if the replication machinery slips on the repeat sequence and defective MMR prevents correction of these errors. After transfection of the pCAR-OF reporter vector into mutant and corrected cells, an increase in β-gal expression would then indicate that MMR is deficient in these cells. We transfected this reporter vector into several FA mutant and corrected cell lines in addition to the MSH2-deficient cell line HEC59 and its corrected counterpart as a control. Significantly, both the MSH2-deficient cells and several different FA mutant cells displayed at least a 4-fold increase in β-gal expression compared with their corrected counterparts (Fig. 7C). For MSH2 mutants, P = 0.001. The FA cell lines used were deficient in FANCG (P = 0.001), FANCD2 (P = 0.001) and FANCJ mutant cells (P = 0.001), demonstrating that MMR is defective in cell lines corresponding to mutations in all three levels of the FA pathway subgroups—core complex, ID complex and downstream effectors, respectively. Notably, these data represent the first description of defective MMR in FA cell lines.
Figure 7.
MMR is defective in FA cell lines. (A) Endogenous MSH2 was immunoprecipitated from extracts of FANCA-deficient and corrected cells. MLH1 does not co-precipitate with MSH2 in FANCA-deficient cells. (B) Endogenous MSH2 was immunoprecipitated from extracts of FANCD2-deficient and corrected cells. The interaction between MSH2 and MLH1 is reduced in FANCD2-deficient cells. (C) Several FA mutant and corrected cell lines in addition to the MSH2-deficient cell lines HEC59 and its corrected counterpart were transfected with the pCAR-OF reporter vector to assess MMR activity. Both the MSH2-deficient cells and several different FA mutant cells displayed at least a 4-fold increase in β-gal expression compared with their corrected counterparts, indicating a defect in MMR in all FA cell lines tested on par with the defect observed in MSH2-deficient cells.
DISCUSSION
Although 15 FA proteins have been identified to date that participate in a common pathway to repair ICLs, much still remains unknown about the detection and processing of these deleterious lesions. In this study, we identify two novel FANCD2-binding partners, MSH2 and MLH1. MSH2 binds to FANCD2-L in response to crosslinking agents, and this interaction requires ATR and is enhanced by the presence of MLH1. MLH1 interacts with FANCD2 only in response to crosslinking agents and requires MSH2. In addition, we find that FANCD2 and FANCI monoubiquitylation and chromatin loading and FANCD2 foci formation are impaired in MSH2-deficient cells, but not in MLH1-deficient cells. However, both MSH2- and MLH1-deficient cells are hypersensitive to multiple crosslinking agents and display the characteristic increase in radial formation common to all FA mutant cells. Lastly, studies in both human cell lines and Drosophila mutants indicate an epistatic relationship between FANCD2, MSH2 and MLH1 in ICL repair. Although previous studies have demonstrated interactions between MMR and FA or FA-associated proteins (25,40,41), this is the first demonstration of a physical and functional interaction between the pivotal FA protein FANCD2 with MMR proteins (MSH2 and MLH1), and also the first report of an interaction between MSH2 and an FA protein.
Previous studies have implicated human MutS complexes in the recognition and early processing of ICLs. MutS complexes are able to bind ICLs in vitro (21), and the initial processing steps of an ICL require MSH2 (22), but not MLH1. Evidence has also been presented that MSH2 plays a role in ATR activation and recruitment to sites of DNA damage (19). Interestingly, MSH2 has also been shown to interact with ERCC1-XPF in response to ICLs induced by CDDP (42), and ERCC1-XPF has been proposed to perform the first incision of the crosslink in ICL repair (43). Taken together with our findings that MSH2 interacts with FANCD2 and is required for the monoubiquitylation and chromatin loading of FANCD2 and FANCI, we therefore suggest that MSH2 plays a crucial role in the detection of ICLs and the early signaling events leading to the activation of the FA pathway. Given that several FA core complex proteins are phosphorylated in an ATR-dependent manner, including FANCA, E, G and M, as well as FANCD2 in response to DNA damage (28,31,44–47), it is likely that MSH2 initiates a signaling cascade responsible for activation and chromatin loading of the FA core complex in addition to FANCD2 and FANCI as we have shown.
Our data show that MLH1-, MSH3- and MSH2-deficient cells are hypersensitive to DNA crosslinking agents. Previous reports have suggested that MMR-defective cells are resistant to CDDP and psoralen ICLs (48–50), but this result has not been consistent. MSH2-deficient cells have been shown by several groups to be hypersensitive to MMC and psoralen ICLs (51,52). In addition, PMS2-deficient HeLa cells and MLH1-defective Raji 10 cells have both been shown to be hypersensitive to MMC (52), supporting a role for the MutLα complex in ICL repair. Even more recently, a study now describes a clinical mutation in MLH1 that causes sensitivity to MMC (53), again lending credence to our data showing that MMR-defective cells are hypersensitive to DNA crosslinking agents. Lastly, other groups have shown that sensitivity to CDDP in some MMR-defective cells differs based on the p53 status of each subline (54,55). In this study, we have shown sensitivity to ICL agents in four different species deficient in one of three MMR proteins.
Other recent reports have shown that FANCJ interacts with the MutLα complex, and that disruption of this interaction results in hypersensitivity to interstrand crosslinking agents (25). It was therefore suggested that MLH1 may function to facilitate downstream ICL repair through the regulation of FANCJ helicase activity. Here we describe a damage-inducible interaction between FANCD2 and MLH1. As FANCD2 also binds to FANCJ (Chen and Kupfer, unpublished data, 2011), it is conceivable that FANCD2 escorts MLH1 to FANCJ to facilitate the unwinding of DNA and subsequent ICL repair. This would place MLH1 downstream of FANCD2 and FANCI monoubiquitylation and chromatin loading, and would explain the differences we observe between MSH2- and MLH1-deficient cells.
Mutations in MMR genes are most often associated with hereditary non-polyposis colorectal cancer (HNPCC) (56). Given that FA patients are generally not at a significantly increased risk for this type of cancer, it would be easy to dismiss the importance of the interaction between FA and MMR proteins. However, some cases of colon cancer have been reported in FA patients (57) and preliminary studies have suggested that some subsets of colorectal cancer patients may have alterations in FA genes (58). In addition, although monoallelic mutations in MMR genes result in HNPCC, biallelic mutations in MMR genes are now being described as the basis for constitutional mismatch-repair deficiency syndrome (CMMR-D) (59). Interestingly, similar to FA, CMMR-D is characterized by a predisposition to childhood cancers, mainly hematological malignancies or brain tumors, and café au lait spots (60). And like CMMR-D, monoallelic mutation of some FA genes results in familial breast cancer, whereas biallelic mutation of these same genes results in FA (61). CMMR-D has been attributed to biallelic mutations in MSH6, MSH2, MLH1 and PMS2 to date (62–65). The overlap between FA and CMMR-D as diseases is indicative of a likely functional overlap between the FA and MMR pathways.
A recent report by Xie et al. (66) also shows a possible link between FANCJ and MMR signaling. They demonstrate that a clinical mutation in MLH1, which ablates its ability to bind to FANCJ, causes sensitivity to MMC. However, they also find that FANCJ deficiency results in reduced MMR signaling as measured by a decrease in Chk1 phosphorylation after stimulation with methyl nitrosourea, an agent that induces O6-methylguanine formation and spurs an MMR response. These data suggest an overlap in function between MMR and FA proteins, and support our data showing that MMR is defective in multiple FA cell lines.
This is the first report of defective MMR in FA cells; however, other DNA repair proteins have been shown to be required for MMR that were previously unappreciated in this respect. Deficiency in MRE11, a protein first implicated in the repair of DNA double-strand breaks, has now been shown to lead to defective MMR and microsatellite instability (67). Similar to our studies, MRE11 interacts with MLH1, and depletion of MRE11 results in MMR deficiency as measured by an analogous plasmid-based assay. This suggests that perhaps even more DNA repair proteins are involved in the MMR pathway or other functions of MMR proteins remain to be discovered.
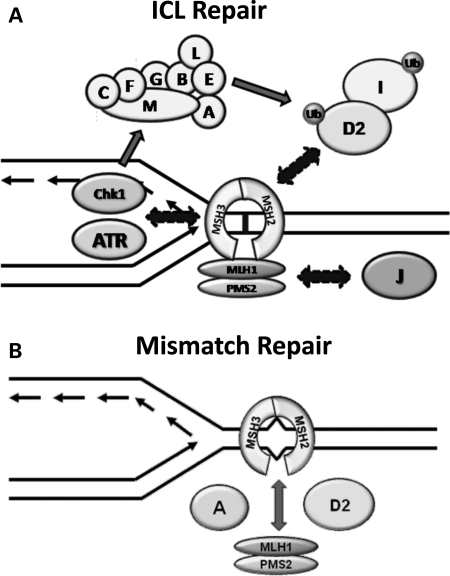
Our data suggest a significant physical and functional interaction between the FA and MMR pathways. We therefore propose a 2-fold model (Fig. 8) where MSH2 and MLH1 are required for ICL repair. MSH2 is responsible for the activation of the FA pathway, likely through the recognition of the lesion and activation and recruitment of ATR. This model is supported by a recent report showing a requirement for MSH2 in ATR recruitment following CDDP treatment (20). Downstream of FANCD2 and FANCI monoubiquitylation, MLH1 could be shuttled to FANCJ by FANCD2 in order to facilitate DNA unwinding and subsequent repair steps. Conversely, FA proteins are involved in MMR, where at least FANCA and FANCD2 enhance the interaction between MSH2 and MLH1.
Figure 8.
A model showing the overlapping functions of MMR and FA proteins in ICL and MMR. MSH2 is likely involved in the detection of ICLs and early signaling events leading to the monoubiquitylation and chromatin loading of FANCD2 and FANCI, such as recruitment of ATR. MLH1 may play a role in ICL repair downstream of FANCD2 and FANCI monoubiquitylation, dependent on its interaction with both FANCD2 and FANCJ. Conversely, FANCA and FANCD2, along with other members of the FA pathway, may be required for efficient binding between MSH2 and MLH1. This role in the MSH2–MLH1 interaction renders all FA cell lines tested defective in MMR.
MATERIALS AND METHODS
Antibodies
MSH2 polyclonal and monoclonal antibodies were obtained from Cell Signaling Technology and Calbiochem, respectively. MLH1 monoclonal and polyclonal antibodies were obtained from BD Pharmigen or Cell Signaling Technology, respectively. FANCD2 polyclonal and monoclonal antibodies were obtained from Abcam and Santa Cruz, respectively. PCNA antibody was obtained from Novus Biologicals. Topo II and α-tubulin antibodies were obtained from Calbiochem. Ku86 antibody was obtained from Santa Cruz. FANCI antibody was made as previously described. Secondary antibodies (ECL anti-rabbit IgG, HRP-linked; ECL anti-mouse IgG, HRP-linked; and Protein A, HRP-linked) were obtained from Amersham Health, Inc.
Cell culture
HeLa, H1299, FA-D2 mutant cells PD20, PD20 + FANCD2 K561R and PD20 + FlagFANCD2, MSH2++ MEFs and MSH2−/− MEFs (kindly provided by P.M.G., Yale University) and HCT116 cells (kindly provided by T. Kunkel, NIEHS) were cultured in DMEM (Invitrogen, Grand Island, NY, USA) containing 10% fetal bovine serum (Biowest, Miami, FL, USA) and 1% Pen-Strep (Invitrogen). HEC59 cells (kindly provided by T. Kunkel, NIEHS), EUFA30, EUFA30 + FANCJ, EUFA326 and EUFA326 + FANCG cells were cultured in DMEM (Invitrogen) containing 20% fetal bovine serum (Biowest) and 1% Pen-Strep (Invitrogen). HCT116 + Ch3 cells (kindly provided by T. Kunkel, NIEHS) were cultured in DMEM containing 10% fetal bovine serum, 1% Pen-Strep and 400 μg/ml G418 (Sigma, St Louis, MO, USA). HEC59 + Ch2 cells (kindly provided by T. Kunkel, NIEHS) were cultured in DMEM containing 20% fetal bovine serum, 1% Pen-Strep and 250 μg/ml G418 (Sigma). Cells were treated with either 500 nm MMC or 75 μm CDDP for the duration indicated for western blotting. Lymphoblastoid HSCg (GM02188; wild-type cells), GM01389D (ATM-deficient cells), DK0064 (Seckel syndrome cells with impaired ATR function), HSC72 (FA-A), BD215 (FA-C), EUFA143 (FA-G) were grown in RPMI1640 medium with 15% fetal calf serum (28). HSCg, GM01389D and DK0064 (68) were provided by Penny Jeggo (Sussex Centre for Genome Damage and Stability). SKUT1, an MSH2-deficient cell line (provided by Mark Meuth, University of Sheffield Institute of Cancer Research), HCC1937 (BRCA1−/−) and retrovirally cDNA-corrected HCC1937 + wt BRCA1 (provided by Helmut Hanenberg, University of Dusseldorf) and CHO cell lines Pro- and D35 provided by Larry Chasin (Columbia University) were all grown in DMEM with 10% fetal calf serum (29,69–71).
Purification of FANCD2 subcomplexes
FANCD2 subcomplexes were purified using methods previously described (26). Briefly, the ‘middle’ FANCD2 subcomplex was purified through several steps of chromatography, immunoprecipitated with anti-FLAG beads (Sigma), then subjected to SDS–PAGE, followed by silver-staining using SilverQuest Kit (Invitrogen, Carlsbad, CA, USA) and following manufacturer's instructions. Silver-stained bands were then trypsin-digested and analyzed by LC-MS for the identification of FANCD2-interacting proteins.
Immunoprecipitation
H1299 cells were cultured on 15 cm plates and treated with 500 nm MMC for 24 h before harvest by scraping. Following a PBS rinse, cells were lysed in non-denaturing lysis buffer (NDLB: 1% Triton X-100, 50 mm Tris–Cl, pH 7.4, 300 mm NaCl, 5 mm EDTA, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 2 mm sodium pyrophosphate, 2 mm sodium orthovanadate) and sonicated for 10 s. Extracts were cleared by centrifugation at 17 000g for 10 min at 4°C. The supernatant was removed and equal amounts of protein were used for each immunoprecipitation. One microgram of FANCD2 antibody or normal rabbit IgG (Santa Cruz, Santa Cruz, CA, USA) was added to samples and incubated overnight at 4°C on a rotator. Protein A sepharose beads (GE Healthcare, Waukesha, WI, USA) were then washed in NDLB, added to each sample and then left to rotate at 4°C for 1 h. Beads were then washed five times in 1 ml of wash buffer (0.1% Triton X-100, 50 mm Tris–Cl, pH 7.4, 300 mm NaCl, 5 mm EDTA), pelleted by centrifugation and then resuspended in equal amounts of wash buffer and SDS loading buffer. FANCD2–MSH2 co-immunoprecipitation in FA, ATM, ATR and BRCA1 cell lines (Fig. 2) was performed using Sigma's (Poole UK) EZview red protein gel affinity system as previously described (72,73), using FANCD2 antibody (Santa Cruz Biotechnology, USA) to immunoprecipitate and anti-human MSH2 (N-20, sc-494, Santa Cruz Biotechnology, UK) for western blotting. Total cell extracts were prepared from 1–2 × 107 exponentially growing cells treated with 50 nm MMC for 18 h.
Western blotting
Protein samples were suspended in SDS loading buffer, boiled for 5 min and then briefly centrifuged. Samples were then run on an 8% SDS–PAGE and transferred to nitrocellulose (Trans-Blot, Bio-Rad, Hercules, CA, USA). Membranes were blocked in 5% milk in PBS-T (PBS + 0.1% Tween-20) overnight at 4°C, and then incubated at room temperature for 1 h or overnight at 4°C in appropriate primary antibody diluted in PBS-T. After three 5 min washes in PBS-T, membranes were then incubated for 1 h at room temperature in appropriate secondary antibody at a dilution of 1:5000 in PBS-T + 0.5% milk. Finally, blots were washed five times for 5 min in PBS-T and developed by chemiluminescence (Supersignal West Pico Kit, Pierce, Rockford, IL, USA).
Cellular fractionation
Cellular fractionation protocol was adapted from published methods in Montes de Oca et al. (6). Cells were grown in 10 cm dishes, treated with 500 nm MMC for 24 h and collected by scraping. Soluble fractions were extracted by resuspending the pelleted cells in buffer A + 0.5% Triton X-100 (10 mm PIPES, pH 7, 100 mm NaCl, 3 mm MgCl2, 1 mm EGTA, 300 mm sucrose, 0.5 mm Na3VO4, 50 mm NaPO3, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mm PMSF) and incubating at room temperature for 3 min. Nuclei were pelleted by centrifugation for 3 min at 300g and were then washed three times in buffer A, followed by incubation in buffer A + DNase1 for 30 min at room temperature. The extract was centrifuged for 3 min at 300g and supernatant was set aside. The pellet was extracted again with non-denaturing lysis buffer (see immunoprecipitation methods) and the resulting supernatant was pooled with the buffer A + DNase 1 supernatant to form the chromatin fraction. Western blotting was used to analyze cellular fractions.
Cell-cycle analysis
Cell lines were harvested, washed and then fixed in 70% ethanol overnight at 4°C. Cells were then incubated in RNAse A solution (10 μg/ml) for 5 min at room temperature and stained with propidium iodide (Sigma). Flow cytometric analysis was then performed by the Yale FACs Core Facility, using a FACSCalibur flow cytometer.
Immunofluorescence staining
Cells were plated in eight-chamber slides, grown to 50% confluence and then treated with 500 nm MMC for the duration indicated. Slides were then rinsed in PBS, fixed in 4% paraformaldehyde for 5 min at room temperature, rinsed in PBS and then permeabilized in 0.3% Triton X-100 for 5 min at room temperature. After rinsing in PBS, slides were then blocked overnight at 4°C in PBS + 0.1% NP40 + 10% normal goat serum (Amersham Biosciences). Primary antibodies FANCD2 and normal rabbit IgG (Abcam and Santa Cruz, respectively) diluted 1:1000 in PBS + 0.1% NP40 + 0.5% BSA were then added and slides were incubated for 1 h at room temperature. Slides were washed three times for 5 min in PBS-T at room temperature, and then Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) secondary antibody was diluted 1:5000 in PBS + 0.1% NP40 + 0.5% BSA and added to slides for 1 h at room temperature. After washing three times for 5 min in PBS-T at room temperature and two times for 3 min in PBS at room temperature, slides were mounted using DAPI Vectashield Hard-Set (Vector Laboratories, Burlingame, CA, USA) and analyzed on a Nikon TE2000-E Eclipse inverted fluorescent microscope.
siRNA transfection
All siRNA duplex pools were obtained from Dharmacon Research and transfected using X-tremeGENE siRNA transfection reagent (Roche, Mannheim, Germany), OPTI-MEM (Invitrogen) and manufacturer's protocol. Cells were plated in six-well plates and transfected at 50% confluence. Cells transfected with siRNA directed towards MSH2 and FANCD2 were replated for survival assays and collected for western blotting 72 h after transfection.
Cell survival assays
Cells were seeded into six-well plates at 10 000 cells per well and treated with MMC or CDDP 24 h later. After incubation for 5 days, media were aspirated, and wells were rinsed once with PBS. Cells were then fixed in 10% methanol/10% acetic acid for 5 min at room temperature on a rocker. Fixing solution was aspirated and crystal violet solution (1% in methanol) was then added to cells and rocked for 5 min at room temperature. Plates were then rinsed in tap water to remove excess crystal violet and allowed to dry. Extraction of crystal violet dye was performed using extraction solution (methanol/SDS) and allowing plates to rock for 2 h. Absorbance was then measured at 595 nm. For colony survival assays in HeLa cells, cells were transfected as above, then replated in six-well plates at 500 cells per well 48 h after transfection. Twenty-four hours after plating, cells were treated with increasing concentrations of MMC for 1 h, rinsed with PBS and then fresh media were added and cells were incubated for 10 days. Colonies were fixed in 10% methanol/10% acetic acid, stained with crystal violet and counted. Clonal survival assays on CHO cell lines were performed as previously described (74).
Chromosome breakage analysis
Cells were treated with MMC for 24 h and 1 µm colcemid for 2–6 h and collected by trypsinization. Pellets were washed once in PBS, then swollen in hypotonic buffer (40 mm KCl, 25 mm sodium citrate) for 20 min at 37°C. Cells were pelleted and hypotonic solution was aspirated, followed by fixation in acetic acid:methanol (1:3) for 10 min at room temperature. Cells were then dropped onto slides and left to dry, stained with Giemsa stain and analyzed on a Nikon TE2000-E Eclipse inverted microscope. A minimum of 25 metaphase spreads of each cell type were analyzed for radial formation.
Drosophila mutagenesis
Fanconi RNAi lines and latsX1 flies have been previously described (34,36). Spel1 mutant lines were a gift from Carlos Flores (35). Pnr-Gal4 lines were obtained from the Bloomington Stock Center (75). Flies were crossed in vials containing 15 ml of standard Drosophila cornmeal-molasses media (www.flystocks.bio.indiana.edu/Fly_Work/media-recipes/media-recipes.htm). Following 48 h of egg laying, parental flies were removed. DEB (Sigma) was then diluted in 250 µl of ddH2O and added to vials containing progeny to obtain the final concentration indicated.
Statistical analysis
The frequencies are expressed as proportions of flies exhibiting mutations, and standard errors of these proportions are based on the binomial distribution. Significance for 2 × 2 tables were assessed using the χ2 test. Values were considered not significant if P > 0.05.
MMR assays
Cells plated in six-well plates were transfected with pCAR-OF and pCAR-IF constructs using Lipofectamine 2000, OPTI-MEM and manufacturer's protocol (Invitrogen). After incubation for 48 h, cells were collected and β-gal expression was measured using the Beta-Glo Assay System Kit (Promega, Madison, WI, USA) and a Lumat LB 9507 luminometer.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by NIH grant R01 HL063776 (G.M.K.) and NWCRF grant CR751 (N.J.J.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Kunkel (NIEHS) for HCT116 and HEC59 cell lines and C. Flores (University of Wisconsin) for spel1 mutant flies. We also thank Peggy Jeggo for HSCg, GM01389D and DK0064 cell lines, Mark Meuth for SKUT1 cells, Helmut Hanenberg for HCC1937 cell lines and Larry Chasin for CHO Pro- and D35 cell lines.
REFERENCES
- 1.Kennedy R.D., D'Andrea A.D. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. doi:10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 2.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. doi:10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 3.Collins N., Kupfer G.M. Molecular pathogenesis of Fanconi anemia. Int. J. Hematol. 2005;82:176–183. doi: 10.1532/IJH97.05108. doi:10.1532/IJH97.05108. [DOI] [PubMed] [Google Scholar]
- 4.McCabe K.M., Olson S.B., Moses R.E. DNA interstrand crosslink repair in mammalian cells. J. Cell. Physiol. 2009;220:569–573. doi: 10.1002/jcp.21811. doi:10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams S.A., Kupfer G.M. Ubiquitin and FANC stress responses. In: Bradshaw R.A., Dennis E.A., editors. Handbook of Cell Signaling. 2nd edn. Oxford: Oxford University Press; 2009. pp. 2265–2272. [Google Scholar]
- 6.Montes de Oca R., Andreassen P.R., Margossian S.P., Gregory R.C., Taniguchi T., Wang X., Houghtaling S., Grompe M., D'Andrea A.D. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. doi:10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. doi:10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Andreassen P.R., D'Andrea A.D. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. doi:10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain S., Wilson J.B., Medhurst A.L., Hejna J., Witt E., Ananth S., Davies A., Masson J.Y., Moses R., West S.C., et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. doi:10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F., Fan Q., Ren K., Auerbach A.D., Andreassen P.R. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma. 2010;119:637–649. doi: 10.1007/s00412-010-0285-6. doi:10.1007/s00412-010-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. doi:10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. doi:10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunz C., Saito Y., Schar P. DNA Repair in mammalian cells: mismatched repair: variations on a theme. Cell Mol. Life Sci. 2009;66:1021–1038. doi: 10.1007/s00018-009-8739-9. doi:10.1007/s00018-009-8739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. doi:10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 15.Genschel J., Littman S.J., Drummond J.T., Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. doi:10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 16.McCulloch S.D., Gu L., Li G.M. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J. Biol. Chem. 2003;278:3891–3896. doi: 10.1074/jbc.M210687200. doi:10.1074/jbc.M210687200. [DOI] [PubMed] [Google Scholar]
- 17.Tian L., Gu L., Li G.M. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSalpha and MutSbeta determine their differential mismatch binding activities. J. Biol. Chem. 2009;284:11557–11562. doi: 10.1074/jbc.M900908200. doi:10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. doi:10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 19.Yoshioka K., Yoshioka Y., Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. doi:10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pabla N., Ma Z., McIlhatton M.A., Fishel R., Dong Z. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J. Biol. Chem. 2011;286:10411–10418. doi: 10.1074/jbc.M110.210989. doi:10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada M., O'Regan E., Brown R., Karran P. Selective recognition of a cisplatin-DNA adduct by human mismatch repair proteins. Nucleic Acids Res. 1997;25:491–496. doi: 10.1093/nar/25.3.491. doi:10.1093/nar/25.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang N., Lu X., Zhang X., Peterson C.A., Legerski R.J. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. doi:10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J., Jain A., Iyer R.R., Modrich P.L., Vasquez K.M. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. doi:10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N., Liu X., Li L., Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst.) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. doi:10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng M., Litman R., Xie J., Sharma S., Brosh R.M., Jr, Cantor S.B. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. doi:10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhi G., Chen X., Newcomb W., Brown J., Semmes O.J., Kupfer G.M. Purification of FANCD2 sub-complexes. Br. J. Haematol. 2010;150:88–92. doi: 10.1111/j.1365-2141.2010.08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. doi:10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson J.B., Yamamoto K., Marriott A.S., Hussain S., Sung P., Hoatlin M.E., Mathew C.G., Takata M., Thompson L.H., Kupfer G.M., et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27:3641–3652. doi: 10.1038/sj.onc.1211034. doi:10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 29.Hinz J.M., Meuth M. MSH3 deficiency is not sufficient for a mutator phenotype in Chinese hamster ovary cells. Carcinogenesis. 1999;20:215–220. doi: 10.1093/carcin/20.2.215. doi:10.1093/carcin/20.2.215. [DOI] [PubMed] [Google Scholar]
- 30.Klovstad M., Abdu U., Schupbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. doi:10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. doi:10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E., et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. doi:10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 33.Castillo V., Cabre O., Marcos R., Surralles J. Molecular cloning of the Drosophila Fanconi anaemia gene FANCD2 cDNA. DNA Repair (Amst.) 2003;2:751–758. doi: 10.1016/s1568-7864(03)00046-6. doi:10.1016/S1568-7864(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 34.Marek L.R., Bale A.E. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst.) 2006;5:1317–1326. doi: 10.1016/j.dnarep.2006.05.044. doi:10.1016/j.dnarep.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 35.Flores C., Engels W. Microsatellite instability in Drosophila spellchecker1 (MutS homolog) mutants. Proc. Natl Acad. Sci. USA. 1999;96:2964–2969. doi: 10.1073/pnas.96.6.2964. doi:10.1073/pnas.96.6.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu T., Wang W., Zhang S., Stewart R.A., Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 37.Mihaylova V.T., Bindra R.S., Yuan J., Campisi D., Narayanan L., Jensen R., Giordano F., Johnson R.S., Rockwell S., Glazer P.M. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. doi:10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolaides N.C., Littman S.J., Modrich P., Kinzler K.W., Vogelstein B. A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol. Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strand M., Prolla T.A., Liskay R.M., Petes T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. doi:10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 40.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiácovo M.P., et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. doi:10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKay C., Declais A.C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T., et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. doi:10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan L., Hayashi T., Rabeya R.M., Nakajima S., Kanno S., Takao M., Matsunaga T., Yoshino M., Ichikawa M., Riele H., et al. Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells. DNA Repair (Amst.) 2004;3:135–143. doi: 10.1016/j.dnarep.2003.10.005. doi:10.1016/j.dnarep.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Fisher L.A., Bessho M., Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J Biol. Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. doi:10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 44.Collins N.B., Wilson J.B., Bush T., Thomashevski A., Roberts K.J., Jones N.J., Kupfer G.M. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009;113:2181–2190. doi: 10.1182/blood-2008-05-154294. doi:10.1182/blood-2008-05-154294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Kennedy R.D., Ray K., Stuckert P., Ellenberger T., D'Andrea A.D. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell Biol. 2007;27:3098–3108. doi: 10.1128/MCB.02357-06. doi:10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao F., Mi J., Wilson J.B., Zhi G., Bucheimer N.R., Jones N.J., Kupfer G.M. Phosphorylation of fanconi anemia (FA) complementation group G protein, FANCG, at serine 7 is important for function of the FA pathway. J Biol. Chem. 2004;279:46035–46045. doi: 10.1074/jbc.M408323200. doi:10.1074/jbc.M408323200. [DOI] [PubMed] [Google Scholar]
- 47.Ho G.P., Margossian S., Taniguchi T., D'Andrea A.D. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol. Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. doi:10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fink D., Nebel S., Aebi S., Zheng H., Cenni B., Nehmé A., Christen R.D., Howell S.B. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 49.Aebi S., Kurdi-Haidar B., Gordon R., Cenni B., Zheng H., Fink D., Christen R.D., Boland C.R., Koi M., Fishel R., et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 50.Wu Q., Vasquez K.M. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4:e1000189. doi: 10.1371/journal.pgen.1000189. doi:10.1371/journal.pgen.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Q., Christensen L.A., Legerski R.J., Vasquez K.M. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. doi:10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiumicino S., Martinelli S., Colussi C., Aquilina G., Leonetti C., Crescenzi M., Bignami M. Sensitivity to DNA cross-linking chemotherapeutic agents in mismatch repair-defective cells in vitro and in xenografts. Int. J. Cancer. 2000;85:590–596. doi: 10.1002/(sici)1097-0215(20000215)85:4<590::aid-ijc23>3.0.co;2-o. doi:10.1002/(SICI)1097-0215(20000215)85:4<590::AID-IJC23>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 53.Xie J., Guillemette S., Peng M., Gilbert C., Buermeyer A., Cantor S.B. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer Prev. Res. (Phila.) 2010;3:1409–1416. doi: 10.1158/1940-6207.CAPR-10-0118. doi:10.1158/1940-6207.CAPR-10-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin X., Ramamurthi K., Mishima M., Kondo A., Christen R.D., Howell S.B. P53 modulates the effect of loss of DNA mismatch repair on the sensitivity of human colon cancer cells to the cytotoxic and mutagenic effects of cisplatin. Cancer Res. 2001;61:1508–1516. [PubMed] [Google Scholar]
- 55.Vikhanskaya F., Colella G., Valenti M., Parodi S., D'Incalci M., Broggini M. Cooperation between p53 and hMLH1 in a human colocarcinoma cell line in response to DNA damage. Clin. Cancer Res. 1999;5:937–941. [PubMed] [Google Scholar]
- 56.Muller A., Fishel R. Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC) Cancer Invest. 2002;20:102–109. doi: 10.1081/cnv-120000371. doi:10.1081/CNV-120000371. [DOI] [PubMed] [Google Scholar]
- 57.Eiler M.E., Frohnmeyer D., Frohnmeyer L., Larsen K., Owen J., editors. Fanconi Anemia: Guidelines for Diagnosis and Management. 3rd edn. 2008. Fanconi Anemia Research Fund, Inc., Eugene, OR. [Google Scholar]
- 58.Palmieri G., Colombino M., Camboni M.G., Manca A., Baldinu P., Izzo F., Tatangelo F., Calemma R., Cossu A., Galimi F. Assessment of the role of Fanconi anemia (FA) genes in colorectal cancer: a new pathogenetic pathway? J. Clin. Oncol. 2006;24:3629. doi: 10.1200/JCO.2005.05.4882. doi:10.1200/JCO.2005.05.4882. [DOI] [PubMed] [Google Scholar]
- 59.Wimmer K., Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum. Genet. 2008;124:105–122. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
- 60.Rahman N., Scott R.H. Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum. Mol. Genet. 2007;16(Spec no. 1):R60–R66. doi: 10.1093/hmg/ddm026. doi:10.1093/hmg/ddm026. [DOI] [PubMed] [Google Scholar]
- 61.D'Andrea A.D. Susceptibility pathways in Fanconi's anemia and breast cancer. N. Engl. J. Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. doi:10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott R.H., Mansour S., Pritchard-Jones K., Kumar D., MacSweeney F., Rahman N. Medulloblastoma, acute myelocytic leukemia and colonic carcinomas in a child with biallelic MSH6 mutations. Nat. Clin. Pract. Oncol. 2007;4:130–134. doi: 10.1038/ncponc0719. doi:10.1038/ncponc0719. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q., Lasset C., Desseigne F., Frappaz D., Bergeron C., Navarro C., Ruano E., Puisieux A. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59:294–297. [PubMed] [Google Scholar]
- 64.De Rosa M., Fasano C., Panariello L., Scarano M.I., Belli G., Iannelli A., Ciciliano F., Izzo P. Evidence for a recessive inheritance of Turcot's syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. doi:10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 65.Whiteside D., McLeod R., Graham G., Steckley J.L., Booth K., Somerville M.J., Andrew S.E. A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe-au-lait spots. Cancer Res. 2002;62:359–362. [PubMed] [Google Scholar]
- 66.Xie J., Guillemette S., Peng M., Gilbert C., Buermeyer A., Cantor S.B. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer Prev. Res. (Phila.). 2010;3:1409–1416. doi: 10.1158/1940-6207.CAPR-10-0118. doi:10.1158/1940-6207.CAPR-10-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vo A.T., Zhu F., Wu X., Yuan F., Gao Y., Gu L., Li G.M., Lee T.H., Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438–444. doi: 10.1038/sj.embor.7400392. doi:10.1038/sj.embor.7400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alderton G.K., Joenje H., Varon R., Borglum A.D., Jeggo P.A., O'Driscoll M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. doi:10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- 69.Mohindra A., Hays L.E., Phillips E.N., Preston B.D., Helleday T., Meuth M. Defects in homologous recombination repair in mismatch-repair-deficient tumour cell lines. Hum. Mol. Genet. 2002;11:2189–2200. doi: 10.1093/hmg/11.18.2189. doi:10.1093/hmg/11.18.2189. [DOI] [PubMed] [Google Scholar]
- 70.Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D.M. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. doi:10.1016/S1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 71.Urlaub G., Kas E., Carothers A.M., Chasin L.A. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983;33:405–412. doi: 10.1016/0092-8674(83)90422-1. doi:10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- 72.Hussain S., Wilson J.B., Blom E., Thompson L.H., Sung P., Gordon S.M., Kupfer G.M., Joenje H., Mathew C.G., Jones N.J. Tetratricopeptide-motif-mediated interaction of FANCG with recombination proteins XRCC3 and BRCA2. DNA Repair (Amst.) 2006;5:629–640. doi: 10.1016/j.dnarep.2006.02.007. doi:10.1016/j.dnarep.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 73.Wilson J.B., Blom E., Cunningham R., Xiao Y., Kupfer G.M., Jones N.J. Several tetratricopeptide repeat (TPR) motifs of FANCG are required for assembly of the BRCA2/D1-D2-G-X3 complex, FANCD2 monoubiquitylation and phleomycin resistance. Mutat. Res. 2010;689:12–20. doi: 10.1016/j.mrfmmm.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones N.J. Genetic analysis of mitomycin C-hypersensitive Chinese hamster cell mutants. Mutagenesis. 1994;9:477–482. doi: 10.1093/mutage/9.5.477. doi:10.1093/mutage/9.5.477. [DOI] [PubMed] [Google Scholar]
- 75.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.