Abstract
Pigs are considered to be intermediate hosts and “mixing vessels,” facilitating the genesis of pandemic influenza viruses, as demonstrated by the emergence of the 2009 H1N1 pandemic (pdm/09) virus. The prevalence and repeated introduction of the pdm/09 virus into pigs raises the possibility of generating novel swine influenza viruses with the potential to infect humans. To address this, an active influenza surveillance program was conducted with slaughtered pigs in abattoirs in southern China. Over 50% of the pigs tested were found to be seropositive for one or more H1 influenza viruses, most commonly pdm/09-like viruses. Out of 36 virus isolates detected, one group of novel reassortants had Eurasian avian-like swine H1N1 surface genes and pdm/09 internal genes. Animal experiments showed that this virus transmitted effectively from pig to pig and from pig to ferret, and it could also replicate in ex vivo human lung tissue. Immunization against the 2009 pandemic virus gave only partial protection to ferrets. The continuing prevalence of the pdm/09 virus in pigs could lead to the genesis of novel swine reassortant viruses with the potential to infect humans.
INTRODUCTION
Pigs are considered to be “mixing vessels,” facilitating reassortment events among avian, swine, and human influenza viruses, which might allow the introduction of novel viruses into the human population (19, 20). The occurrence of the 2009 H1N1 influenza pandemic provided renewed evidence that pigs do play such a role in the influenza virus ecosystem (5, 11). Accumulated findings from epidemiological and genetic studies have revealed that each of the eight gene segments of the 2009 H1N1 pandemic virus (pdm/09) was generated through multiple reassortment events among viruses that had long been prevalent in and adapted to pigs (8, 24). However, due to a lack of systematic surveillance in North America, the direct precursor of the pdm/09 virus has not yet been recognized.
Another important lesson learned from the 2009 pandemic was that a virus of the same subtype as the prevailing human seasonal influenza virus could become pandemic if there were significant antigenic differences (8, 30). As such, any virus with the capacity to infect humans and with novel hemagglutinin (HA) genes that are antigenically distinct from those of circulating human strains should be considered to have pandemic potential.
Currently circulating swine influenza viruses are associated with the classical swine (CS) lineage, the North American triple reassortant (TR) lineage, or the Eurasian avian-like (EA) lineage (3, 13, 16, 27). Two influenza pandemics (those of 1918 and 2009) were caused by viruses with HA genes closely related to or directly belonging to the CS lineage (8, 27, 30). Sporadic human infection cases with different swine virus lineages are not rare events (4, 10, 15, 21).
Although the EA virus has been prevalent in Eurasian pig populations for more than 30 years, it is only occasionally detected in humans (1, 6, 9, 17). Currently, the majority of the human population is immunologically naive to EA-like viruses (27). Antibodies against pdm/09 are unlikely to confer substantial protection against EA-like viruses, since convalescent-phase human sera post-pdm/09 infection or human sera postvaccination did not cross-react with an EA H1N1 virus (26).
After the occurrence of the 2009 pandemic, the pdm/09-like virus was repeatedly introduced back into pigs in many countries (18, 26, 29). Recent influenza surveillance in Hong Kong showed that the CS, TR, and EA swine influenza virus lineages were cocirculating in pigs in southern China (24, 26, 27). Many of the contemporary swine virus isolates were reassortant variants of different swine influenza viruses (24, 26, 27), and the pdm/09 virus has also reassorted with other circulating swine influenza viruses (14, 25, 26). This raises concerns that pdm/09 reassortant variants within pigs may cause new threats to human health.
Since approximately half of the world's population of domestic pigs is farmed in China (7), this represents the largest localized collection of “mixing vessels” for influenza viruses in the world and therefore the greatest opportunity to generate reassortant viruses with the potential to infect humans. To understand the further development and impact of the pdm/09-like virus in pigs, active surveillance of influenza virus in pigs in southern China has been conducted since December 2009. Over 50% of pigs were seropositive for at least one H1 influenza virus (mostly pdm/09), and several viruses with novel genotypes were isolated. One isolate with EA-like surface genes and pdm/09 internal genes was tested for intra- and interspecies transmissibility in mammalian models. The findings emphasize the need for ongoing influenza virus surveillance of the pig population.
MATERIALS AND METHODS
Surveillance.
On an approximately weekly basis from December 2009 to June 2010, a total of 3,600 tracheal swabs and 1,020 serum samples were collected from slaughtered pigs at abattoirs in the Guangdong (GD) (swabs, n = 2,240; sera, n = 625; sampling occasions, n = 21) and Guangxi (GX) (swabs, n = 1,360; sera, n = 395; sampling occasions, n = 13) provinces of the People's Republic of China. The samples collected in Guangdong represent pigs introduced from many neighboring provinces, while those collected in Guangxi are mainly from local pigs.
HI assays.
Sera were pretreated with a receptor-destroying enzyme (RDE; Denka Seiken Co. Ltd., Tokyo, Japan) to destroy nonspecific inhibitors, followed by heat inactivation at 56°C for 30 min. RDE-treated sera were then absorbed with turkey red blood cells (TRBC) to remove nonspecific agglutination substances. Antibody titers were determined by testing serial 2-fold dilutions (1:10 to 1:2,560) of each serum in duplicate. Hemagglutination inhibition (HI) assays were performed in 96-well microtiter plates (Corning Costar Co.) with 0.5% turkey erythrocytes using four hemagglutination units of virus.
Serological survey.
HI assays were performed on each of the 1,020 serum samples collected, with a contemporary human H3N2 virus (A/Shantou/1328/2008) and five representative swine H1 influenza virus strains: A/Sw/HK/294/09 (CS H1N2), A/Sw/HK/915/04 (TR H1N2), A/Sw/HK/NS1583/09 (pdm/09 H1N1), A/Sw/HK/2433/09 (EA H1N1), and A/Sw/HK/1532/09 (EA-like H1N1 variant) (24, 26, 27).
Virus isolation and sequencing.
Isolation of viruses from tracheal swabs in Madin-Darby canine kidney (MDCK) cells, viral RNA extraction, cDNA synthesis, PCR, and sequencing were carried out as previously described (24, 26, 27). All 288 nucleotide sequences of the segments of the 36 influenza virus isolates detected in this study have been deposited in GenBank under accession numbers JN374994 to JN375281. The virus A/Swine/Guangdong/1361/2010 (H1N1) was plaque purified and resequenced to confirm its identity before use in subsequent experiments.
Phylogenetic analysis.
For each gene segment identified here and representative influenza virus sequences from GenBank, maximum-likelihood (ML) phylogenies were inferred using the heuristic tree search method Garli 1.0 (34). Phylogenetic support for branch points was estimated by bootstrap analysis with 100 replicates using the same ML method.
Animals.
All pigs (n = 16; local domestic hybrid pigs, Putian White × Nianbian variants) used were confirmed to be free of influenza virus by virus isolation in MDCK cells and to be seronegative against circulating swine and human influenza viruses (CS, TR, EA, pdm/09, and seasonal H1N1 and H3N2), as well as avian H5N1 and H9N2 viruses, by HI assays prior to the start of the study. Ferrets (n = 9) negative for both influenza virus and antibodies were obtained from the experimental ferret breeding program at Sangosho Pet Park Co., Ltd. Animal experiments were approved by the Shantou University Medical College in compliance with the University Policies “Animal Ethics and Welfare” and “Use of Animals in Research” and the guidelines of the World Health Organization and the International Council for Laboratory Animal Science.
Viral infectivity studies.
Infection and transmission studies were carried out in biosafety level 3 (BSL-3) containment laboratories at 20 to 21°C and 76.5% ± 2.1% relative humidity. All animals were moved to the BSL-3 lab at least 4 days prior to the experiment for acclimatization. A microchip (implantable programmable temperature transponder IPTT-300; BioMedic Data Systems) was implanted in the skin of each animal, between the shoulder blades, to measure subcutaneous temperatures.
Immunization of ferrets with the CA4 virus.
Twenty-two-week-old male ferrets (n = 3) were inoculated with 106 PFU of A/California/04/2009 (CA4, the prototype pdm/09 virus) and gained high titers of antibody (HI > 1,280) to this virus on the 14th day post-primary infection (dpi). These immunized ferrets were used for the transmission experiment at 80 dpi.
Infection and transmission experiments with Sw/GD1361 virus.
Four-week-old piglets (n = 7) were intranasally inoculated with 2 × 106 PFU, i.e., 2.56 × 106 TCID50 (median tissue culture infective dose) of Sw/GD1361 virus in 2 ml of minimum essential medium (MEM; Gibco), delivered with a mucosal atomization device (MAD nasal drug delivery device; Wolfe Tory Medical, Inc.) to mimic aerogenous infection (2). Two naive pigs and three naive ferrets inoculated with phosphate-buffered saline (PBS) were used as negative-control animals.
At 24 h post-inoculation (hpi) of the pigs, immunologically naive pigs (n = 7; 4 weeks old) and ferrets (naive ferrets, n = 3; ferrets with antibodies against CA4, n = 3; 33.5 weeks old) were introduced into the animal infection and transmission facility holding the inoculated pigs (n = 7) (see Fig. S3 in the supplemental material). Four immunologically naive pigs were used as physical contact animals, and three were used for aerosol contact. The aerosol contact pigs and ferrets were housed in adjacent double-layer steel wired cages with a distance of at least 10 cm from those holding the directly inoculated pigs. Airflow inside and between each pair of cages was <0.1 m/s. To avoid inadvertent physical contact and artificial transmission, aerosol contact animals were always handled first; drinking water and food stock, gloves, and any other items in contact with the animals or their bedding were kept sterile by decontamination or reserved for the exclusive use of each individual animal.
Animal monitoring.
Body weights and temperatures were recorded daily around the same time (9:30 to 10:30 a.m.) for each animal. Clinical signs were observed twice daily. Nasal swabs from each piglet were collected daily and placed into 0.6 ml of cold sterile PBS with antibiotics. Nasal washes from each ferret were collected daily into 1 ml of PBS with antibiotics. The endpoint infectivity titration (TCID50) was determined for all swabs and nasal washes in MDCK cells.
Animal serology and histology.
Blood was collected via venipuncture of the anterior vena cava of the pigs or from the ferret tail artery. Seroconversion was monitored by determining the HI titers of pre- and postexposure sera.
Four directly inoculated pigs were euthanized for postmortem examination (2 pigs at 4 dpi and 2 pigs at 6 dpi) by intracardiac injection of pentobarbital sodium (100 to 200 mg/kg of body weight). One physical contact pig was similarly euthanized at 5 days postcontact (dpc). Freshly excised tissues of the trachea, lungs, nasal turbinates, and other major organs of euthanized animals were fixed in 10% phosphate-buffered formalin, dehydrated, embedded in paraffin, and cut into 5-μm-thick sections. Standard hematoxylin and eosin (H&E) (Sigma) staining, as well as immunohistochemistry (IHC) assays with a mouse anti-NP (nucleoprotein) monoclonal antibody, were performed as described previously (32).
Infection of human lung tissue in ex vivo culture.
Similar to the method described in an earlier report (31), fresh lung tissues were surgically removed from patients with lung carcinoma, in accordance with a protocol approved by the Ethical Review Board of Shantou University Medical College. Only normal nonmalignant tissue fragments that were in excess of the requirements of clinical diagnosis were used. Tissues were cut into ∼3-mm by 4-mm by 2-mm cubes and placed in an F-12K nutrient mixture (Gibco) with l-glutamine and antibiotics. Three viruses were used for inoculation: CA4 (pdm/09), Sw/GD1361 (the novel reassortant; this work), and A/Wuhan/359/1995 (seasonal H3N2). For each virus, infections were performed in triplicate, with 7 lung tissue cubes per replicate inoculated with 0.5 × 106 PFU of virus in 0.5-ml inocula in 6-well plates, and allowed to absorb for 1 h at 37°C and 5% CO2. The tissue cubes were then washed three times with culture medium and incubated with 0.5 ml of F-12K medium supplemented with 0.2% TPCK (N-tosyl-l-phenylalanine chloromethyl ketone)-trypsin, 1% BSA (bovine serum albumin), and antibiotics. At 18 and 36 hpi, lung tissue cubes (n = 5 each per virus) were individually rinsed with medium, homogenized in 0.5 ml of cold PBS, and then clarified by centrifugation. Viral titers of the homogenates were determined by TCID50 assays with MDCK cells. The remaining cubes (n = 11 per virus) were fixed and examined for viral NP expression by immunohistochemical staining (31, 32).
RESULTS
From December 2009 to June 2010, active surveillance of pigs in the Guangdong (GD) and Guangxi (GX) provinces of southern China was undertaken for evidence of influenza infection and virus isolation.
Seroprevalence of influenza virus in pigs of southern China.
In the study period, based on hemagglutination inhibition (HI) assays, about 51% (521/1,020) of the sera were positive (HI titer ≥ 1:80) against at least one of five reference H1 strains, while all were negative to the contemporary human H3N2 influenza virus (HI titer ≤ 1:20) (Table 1; see also Fig. S1 in the supplemental material).
Table 1.
Seroprevalence of antibodies against different swine influenza virus lineagesa
| Seropositivity | No. (%) of sera positive |
||
|---|---|---|---|
| Guangdong | Guangxi | Subtotal | |
| None | 306 (49.1) | 193 (48.9) | 499 (48.9) |
| pdm | 104 (16.6) | 35 (8.9) | 139 (13.6) |
| TR | 2 (0.3) | 3 (0.8) | 5 (0.5) |
| CS | 8 (1.3) | 6 (1.5) | 14 (1.4) |
| EA | 35 (5.6) | 19 (4.8) | 54 (5.3) |
| pdm, TR | 15 (2.4) | 11 (2.8) | 26 (2.5) |
| pdm, CS | 25 (4.0) | 24 (6.1) | 49 (4.8) |
| pdm, EA | 13 (2.1) | 8 (2.0) | 21 (2.1) |
| TR, CS | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| TR, EA | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| CS, EA | 3 (0.5) | 7 (1.8) | 10 (1.0) |
| pdm, TR, CS | 25 (4.0) | 23 (5.8) | 48 (4.7) |
| pdm, TR, EA | 4 (0.6) | 0 (0.0) | 4 (0.4) |
| pdm, CS, EA | 15 (2.4) | 18 (4.6) | 33 (3.2) |
| TR, CS, EA | 1 (0.2) | 3 (0.8) | 4 (0.4) |
| pdm, TR, CS, EA | 69 (11.0) | 45 (11.4) | 114 (11.2) |
| Total sera positive | 319 (51.0) | 202 (51.1) | 521 (51.1) |
| Total sera tested | 625 | 395 | 1,020 |
Abbreviations of representative viruses: pdm, A/Sw/HK/NS1583/09 (H1N1); TR, A/Sw/HK/915/04 (H1N2); CS, A/Sw/HK/294/09 (H1N2); EA, A/Sw/HK/2433/09 (H1N1) and A/Sw/HK/1532/09 (H1N1). Viruses against which the swine sera showed an HI titer of ≥80 are given in the table.
Of the 625 sera collected in GD (over the entire surveillance period), 104 (16.6%) were positive solely for the pdm/09-like virus, while 35 (8.9%) of the sera from GX were positive solely for this virus. Additionally, 295 (28.9%) sera (GD, n = 166; GX, n = 129) were positive for pdm/09 and one or more other viruses (Table 1), indicating a high frequency of coinfection or multiple virus exposure in the pigs from southern China, although cross-reaction cannot be completely excluded. Some of the sera had extremely high HI titers (≥1:2,560) for different H1 reference viruses (see Fig. S1), suggesting a recent infection.
Genetic characterization of swine influenza virus isolates.
Thirty-six H1N1 and H1N2 influenza viruses were isolated from tracheal swabs obtained in Guangdong, but none were isolated from those taken in Guangxi. Full-length sequences were obtained for each of the eight gene segments of all 36 swine virus isolates. Phylogenetic analyses of the H1 hemagglutinin (HA) gene revealed that these viruses clustered into three different lineages: the pdm/09, CS, and EA virus lineages (see Fig. S2 in the supplemental material).
For each of the remaining genes, phylogenetic analyses revealed that these viruses belonged to four distinct genotypes: pdm/09-like H1N1 (n = 12, four sampling occasions), a previously undescribed EA-like H1N2 variant with its HA gene from the CS lineage and NA (N2) gene from the human lineage (n = 5, single sampling occasion), a novel reassortant with EA-like H1N1 surface genes and pdm/09-like internal genes (n = 10, single sampling occasion), and an EA-like H1N1 variant with the nonstructural (NS) gene from the TR lineage (n = 9, single sampling occasion) (Fig. 1; see also Fig. S2 in the supplemental material).
Fig. 1.
Genotypes of the swine influenza viruses identified. The name of a representative virus and the numbers of each variant isolated are given (left). Dates of sampling occasions A to G are as follows: 25 December 2009 (A), 31 December 2009 (B), 8 January 2010 (C), 29 January 2010 (D), 27 February 2010 (E), 7 May 2010 (F), and 28 May 2010 (G).
It was noted that viruses isolated from the same sampling occasion were very closely related to each other and always clustered together in the phylogenetic trees (see Fig. S2 in the supplemental material). All pdm/09-like viruses were detected from late December 2009 to the end of January 2010 (from four sampling occasions), while the remaining viruses were isolated in February and May 2010 (Fig. 1).
To see if there is any evidence of early adaptation of pdm/09-like viruses in pigs, the sequences of the 22 viruses containing pdm/09-like genes were compared with all of the pdm/09 sequences in GenBank (as of 5 March 2011). Nineteen of these viruses contained a total of 11 substitutions in 6 internal genes that were absent in all human pdm/09 virus sequences (Table 2). Ten of the novel reassortant viruses, represented by A/swine/Guangdong/1361/2010 (Sw/GD1361), had 6 of these substitutions (in the PB2, PA, M1, and NP genes), while Sw/GD/286/2010 had 2 unique substitutions (in the PB2 and PA genes). The other 8 viruses contained only one substitution, in the PB2 gene (5 viruses, represented by Sw/GD/275/2010) and the NS1/NS2 gene (3 viruses, represented by Sw/GD/94/2009). Only the substitution in the NS1/NS2 gene was observed on more than one sampling occasion.
Table 2.
Unique positions in novel isolates versus human and swine pdm/09 virusesa
| Protein | Substitution | No. of isolates | Sampling occasion | Genotype | Representative strain |
|---|---|---|---|---|---|
| PB2 | T81A | 10 | F | EA-pdm/09 | Sw/GD/1361/2010 |
| Q300K | 5 | D | pdm/09-like | Sw/GD/286/2010 | |
| N348D | 1 | B | pdm/09-like | Sw/GD/213/2009 | |
| I615V | 9 | F | EA-pdm/09 | Sw/GD/1361/2010 | |
| PA | I38T | 1 | D | pdm/09-like | Sw/GD/286/2010 |
| S140T | 10 | F | EA-pdm/09 | Sw/GD/1361/2010 | |
| V521I | 1 | F | EA-pdm/09 | Sw/GD/1425/2010 | |
| NP | H289Y | 10 | F | EA-pdm/09 | Sw/GD/1361/2010 |
| S344L | 10 | F | EA-pdm/09 | Sw/GD/1361/2010 | |
| NS1/NS2 | T5N | 3 | A, B | pdm/09-like | Sw/GD/94/2009 |
| M1 | V31I | 10 | F | EA-pdm/09 | Sw/GD/1361/2010 |
Substitutions for the residues in the novel isolates are relative to the human pdm/09 numbering and consensus sequences. For dates of sampling occasions, see Fig. 1.
Assessment of a novel H1N1 EA-pdm/09 reassortant virus.
The infectivity, transmissibility, and pathogenicity of a representative isolate with EA-like HA and NA genes and pdm/09-like internal genes (Sw/GD1361) were tested with experimental animals. Interspecies transmissibility and the potential for cross-protection from prior pdm/09 infection were investigated.
Infectivity and transmissibility in pigs.
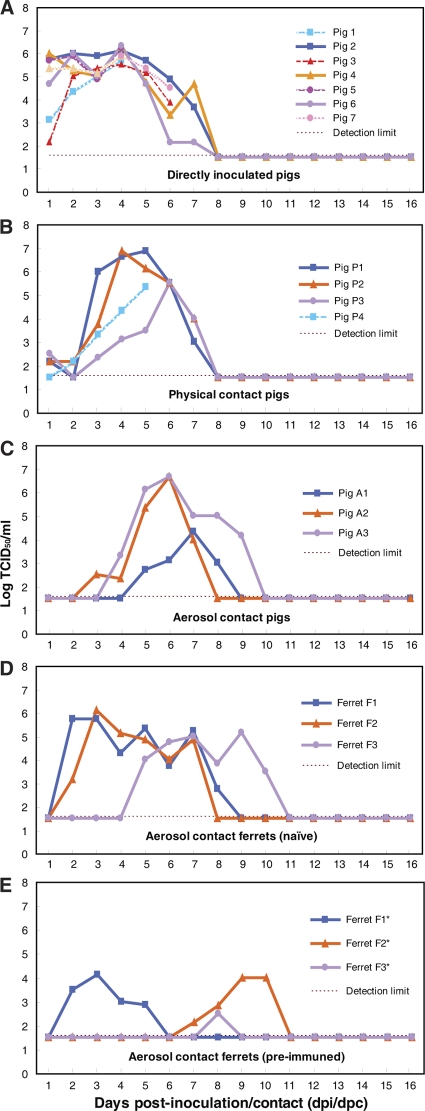
Seven uninfected pigs were cohoused, either in physical or aerosol contact, with seven pigs experimentally inoculated with the Sw/GD1361 virus (see Fig. S3 in the supplemental material). Virus shedding from each of the inoculated pigs was detected from the first day postinoculation (dpi) till the 7th day, with a peak at 4 dpi (6.0 ± 0.3 log TCID50/ml swab material) (Fig. 2A). In the physical-contact pigs, virus replication in the nasal cavity lasted from 1 to 7 days postcontact (dpc), with peak titers of 6.0 ± 0.7 log TCID50/ml (Fig. 2B). In the aerosol-contact group, pigs started to shed virus from the nasal route between 3 and 5 dpc, and virus was detected for at least 4 to 6 days, with peak titers of 5.9 ± 1.3 log TCID50/ml (Fig. 2C). There were no statistically significant differences in viral shedding among the inoculated, physical-contact or aerosol-contact pigs (Fig. 2A to C), and the peak virus shedding titers were comparable to those of prototype EA-like (27) and pdm/09 viruses (2, 12, 28, 29, 33), as previously reported. Lethargy, lower activity levels, and reduced interest in food occurred in the pigs, but no major clinical symptoms of infection were observed. On 15 dpi or 14 dpc, all experimental pigs, either inoculated or exposed by physical or aerosol contact, had seroconverted, with HI titers ranging from 160 to 1,280 (Table 3), which were comparable to those of EA-like viruses (27) and higher than those of pdm/09-like viruses (33). Cross-reaction of swine sera was observed only for the EA viruses. Naive pigs inoculated with PBS did not show virus shedding or seroconversion (data not shown).
Fig. 2.
Virus shedding of the infected pigs and contact animals from the nasal route. TCID50 values in MDCK cells from daily nasal swabs (pigs) or washes (ferrets) are shown. Codes for each animal are given in Fig. S3 in the supplemental material.
Table 3.
HI titer of sera from preexposure (3 days prior to Sw/GD1361 inoculation) and postexposure (15 dpi or 14 dpc) animals infected with Sw/GD1361a
| Animal group | Serum sampleb | HI titer forc: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CS, Sw/HK4167 |
pdm/09 |
Reassortant Sw/GD1361 |
EA |
||||||||||
| CA4 |
Sw/GD106 |
Sw/HK29 |
Sw/HK2433 |
||||||||||
| Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | ||
| Sw/GD1361, direct inoculation, pig | 1 | <10 | ND | <20 | ND | <20 | ND | <10 | ND | <10 | ND | <10 | ND |
| 2 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 320 | <10 | 320 | <10 | 320 | |
| 3 | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | |
| 4 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 320 | <10 | 320 | <10 | 80 | |
| 5 | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | |
| 6 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 160 | <10 | 160 | <10 | 80 | |
| 7 | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | |
| Sw/GD1361, physical contact, pig | P1 | <10 | 10 | <10 | <10 | <10 | <10 | <10 | 640 | <10 | 640 | <10 | 160 |
| P2 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 320 | <10 | 320 | <10 | 320 | |
| P3 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 320 | <10 | 320 | <10 | 160 | |
| P4 | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | <10 | ND | |
| Sw/GD1361, aerosol contact, pig | A1 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 1,280 | <10 | 1,280 | <10 | 320 |
| A2 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 320 | <10 | 320 | <10 | 80 | |
| A3 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 640 | <10 | 640 | <10 | 160 | |
| Sw/GD1361, aerosol contact, ferret | F1 | <10 | 1,280 | <10 | 1,280 | <10 | >1,280 | <10 | >1,280 | <10 | >1,280 | <10 | >1,280 |
| F2 | <10 | 1,280 | <10 | 1,280 | <10 | >1,280 | <10 | 640 | <10 | 640 | <10 | >1,280 | |
| F3 | <10 | >1,280 | <10 | 1,280 | <10 | >1,280 | <10 | >1,280 | <10 | >1,280 | <10 | >1,280 | |
| F1* | 160 | >1,280 | 160 | 1,280 | 160 | >1,280 | 160 | >1,280 | 80 | >1,280 | 80 | >1,280 | |
| F2* | 40 | >1,280 | 80 | >1,280 | 80 | >1,280 | 80 | >1,280 | 40 | >1,280 | 40 | >1,280 | |
| F3* | 640 | 320 | 640 | 320 | 640 | 640 | 320 | 640 | 160 | 640 | 160 | 320 | |
| Sw/HK4167 infected | Ferret | ND | >1,280 | ND | >1,280 | ND | ND | ND | ND | ND | >1,280 | ND | ND |
| Pig | <10 | 80 | <10 | 10 | <10 | 20 | <10 | <10 | <10 | <10 | <10 | <10 | |
| Sw/GD106 infected | Ferret | <10 | 1,280 | <10 | >1,280 | <10 | >1,280 | <10 | >1,280 | <10 | 640 | <10 | 1,280 |
| Pig | <10 | <10 | <10 | 80 | <10 | 80 | <10 | <10 | <10 | <10 | <10 | <10 | |
| Sw/HK29 infected | Ferret | ND | 640 | ND | 80 | ND | ND | ND | ND | ND | >1,280 | ND | ND |
| Pig | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 160 | <10 | 320 | <10 | 320 | |
Virus abbreviations: Sw/HK4167, A/Swine/Hong Kong/4167/1999 (classical swine H1N1 virus, CS); CA4, A/California/04/2009 (prototype pandemic H1N1 2009, pdm/09); Sw/GD106, A/Swine/Guangdong/106/2009 (pandemic H1N1 2009-like swine isolate, pdm/09); Sw/GD1361, A/Swine/Guangdong/1361/2010 (novel H1N1 reassortant); Sw/HK29, A/Swine/Hong Kong/29/2009 (Eurasian avian-like swine H1N1 virus, EA); Sw/HK2433, A/Swine/Hong Kong/2433/2009 (Eurasian avian-like swine H1N1 virus, EA). Reference ferret and pig antisera against Sw/HK4167, Sw/GD106, and Sw/HK29 were obtained from previous studies (27, 33).
Ferrets F1*, F2* and F3* were preimmunized with CA4 virus 80 days prior to this study. The codes for each animal are given in Fig. S3 in the supplemental material.
ND, not determined. Pigs 1 and 5 were sacrificed at 4 dpi, pigs 3 and 7 at 6 dpi, and pig P4 at 5 dpc. Pre, prior to infection (inoculation or contact); Post, postinfection (inoculation or contact exposure). Underlined boldface values are titers for homologous antigens.
Interspecies transmissibility of Sw/GD1361 to ferrets.
Ferrets were used as sentinel animals for an aerosol contact, interspecies transmission test. Two of the three immunologically naive ferrets (F1 and F2), held in separate cages with a distance of 10 cm from the cage of the infected pigs, began to shed virus at 2 dpc, with the third (F3) shedding virus at 5 dpc. Large amounts of virus (peak titer >5 log TCID50/ml) were secreted from the nasal discharges of the ferrets, lasting for 5 to 6 days (Fig. 2D). Sneezing, nasal discharge, inactivity, and slight fever (1.3 to 1.4°C increase in body temperature) were observed in the ferrets. On 14 dpc, all ferrets had seroconverted, with an HI titer of at least 640 against the homologous virus (Table 3). In general, ferret sera showed broad cross-reactivity to all H1 viruses tested. Naive ferret controls did not show virus shedding, clinical signs, or seroconversion (data not shown).
Ferrets (F1* to F3*) with seroconversion against the prototype pandemic virus (A/California/04/09 [CA4]) developed signs of infection when cohoused with the other ferrets. Symptoms similar to those of its naive counterpart (F1) developed in ferret F1* at 2 dpc, with virus shedding for at least 4 days (2 to 5 dpc). The other seroconverted ferrets (F2* and F3*) also shed virus, although the onset was delayed 5 to 6 days (Fig. 2E). Comparison of the pre- and postexposure HI titers from these ferrets showed that two of them (F1* and F2*) had 4- to 8-fold increases in antibodies against the Sw/GD1361 virus, whereas the third one (F3*) had a 2-fold increase (Table 3). Even though ferrets seropositive to pdm/09 could be infected by Sw/GD1361, the amount of virus shed and the duration of shedding were reduced from those for the immunologically naive ferrets (Fig. 2D and E). In the viruses recovered from the nasal washes of all six contact ferrets, no amino acid substitutions that related to the antigenic sites (Ca1, Ca2, Cb, Sa, and Sb) of the HA protein were observed (data not shown).
Infectivity in the pig respiratory tract and ex vivo human lung tissue.
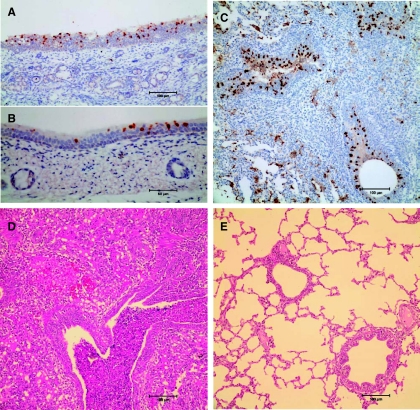
Euthanized pigs (four inoculated animals and one contact animal) showed similar lung lesions and clear evidence of virus replication in the nasal turbinate, trachea, and lower respiratory tract. Pulmonary consolidation and extensive bronchioalveolitis were also observed, characterized by multiple foci of severe inflammatory infiltrates and necrosis of bronchus epithelia (Fig. 3), which is similar to the effect of pdm/09-like virus infections (2, 12, 28, 29).
Fig. 3.
Representative pathology and virus replication in tissues from Sw/GD1361-infected pigs. Viral antigen reactions (with nucleoprotein shown as brown) in epithelial cells of a physical contact pig (pig P4, 5 dpc) are as follows: turbinate (A), trachea (B), and bronchiolar epithelial cells with intraluminal cellular debris (C). An H&E-stained lung section was taken from a physical contact pig (pig P4, 5 dpc) (D) and a naive control pig (mock infected with PBS) (E).
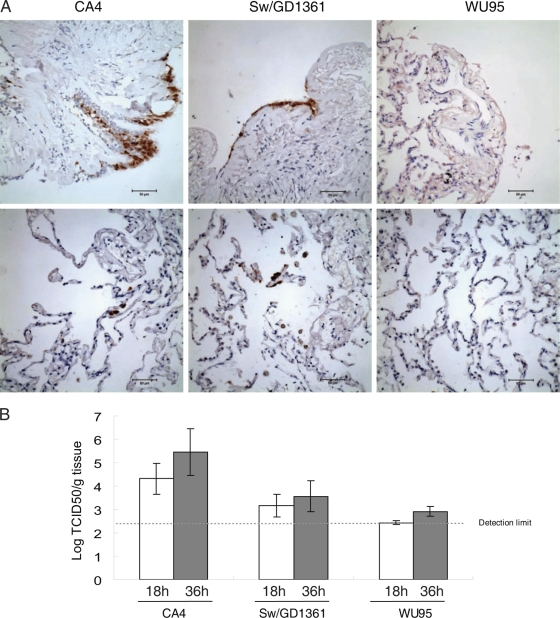
Immunostaining of viral proteins and TCID50 titers showed that Sw/GD1361 and the prototype pdm/09 virus (CA4) could infect human lung tissues, whereas only very limited infection was observed with human seasonal H3N2 influenza virus (A/Wuhan/359/1995) (Fig. 4).
Fig. 4.
Viral infectivity in ex vivo human lung tissue. Human ex vivo lung tissue cubes were inoculated with the virus CA4 (A/California/04/2009 pandemic H1N1 2009), Sw/GD1361 (A/Swine/Guangdong/1361/2010), or WU95 (A/Wuhan/359/1995 seasonal H3N2). (A) Immunohistochemical detection of viral antigen (nucleoprotein) in ex vivo human lung tissue sections (36 hpi). NP-positive cells are shown by brown staining. (B) Virus titers in the ex vivo human lung tissue cubes. The values are means (± standard deviation) for five replicate tissue cubes from three independent inoculations.
DISCUSSION
In the present study, virological surveillance revealed that pdm/09-like, TR-like, CS-like, and EA-like viruses were cocirculating in pigs in southern China with relatively high prevalences (Fig. 1 and Table 1; see also Fig. S1 in the supplemental material). Serological data from this surveillance suggested that infections with multiple different H1 viruses occur commonly in pigs (Table 1). Multiple infections, as observed here, highlight the possibility of further reassortment among these swine influenza virus lineages. Isolation of novel reassortant viruses with three differing genotypes during this surveillance demonstrated that such reassortment events do occur (Fig. 1).
Although the pdm/09-like virus has been repeatedly detected in pigs from different countries (18, 26, 29), whether it can become established in pigs remains unknown. Throughout this study, a high rate of seroconversion to the pdm/09 virus was observed in pig populations in southern China. Repeated detections of genetic reassortment between pdm/09-like and other swine viruses (14, 25, 26), in this study with EA-like viruses, suggest that the pdm/09-like virus might have been maintained in pigs for a period of time. It appears likely that the pdm/09-like virus will eventually become established in pigs.
Several unique substitutions were recognized in the pdm/09-like swine viruses isolated here that were absent in all human pdm/09-like viruses. Some of these viruses had multiple such substitutions over several genes. These changes implied that a process of adaptation of the current pandemic virus to swine might be in progress. This has parallels with the association of classical swine viruses with the 1918 pandemic virus and the emergence of the 1968 pandemic virus in pigs (22, 23).
Current H1 influenza viruses circulating in mammals fall into two major clades, the EA-like and the CS/human H1 clades (see Fig S2 in the supplemental material). All human H1 viruses established in the 20th century cluster with the CS lineage, from which the TR viruses and the pdm/09 virus were derived, and are distinct from the EA viruses. Viruses with EA-like HA genes rarely infect humans, and the human population would likely be immunologically naive to such a virus (1, 6, 9). Therefore, the ability of the EA-pdm/09-like reassortant detected here to cross the species barrier is relevant to the possibility of novel threats to human health arising from multiple infections of pigs with the pdm/09-like and other swine influenza viruses.
It was shown that the Sw/GD1361 virus not only could be transmitted efficiently from pig to pig but also could spread by aerosol from pig to ferret. These animal experiments and the replication of the virus in ex vivo human lung tissue suggest that the Sw/GD1361 EA-pdm/09-like reassortant virus could have the potential to cross the species barrier and infect humans.
Ferrets previously inoculated with and seroconverted to the pdm/09 virus could not avoid symptomatic infection with the Sw/GD1361 virus, indicating there was no substantial cross-protection between this EA-pdm/09 reassortant and the pdm/09 virus. As such, this reassortant, along with others like it that will be generated if pdm/09-like viruses become established in pigs, may represent a new threat to contemporary human populations. Prior exposure to currently circulating viruses is unlikely to provide protection from novel viruses of this type. Intensive surveillance of influenza viruses in pigs appears to be warranted to closely monitor their future evolution, the extent of reassortment, and their potential to impact public health.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge our colleagues from the International Institute of Infection and Immunity (Shantou) and State Key Laboratory of Emerging Infectious Diseases (Shenzhen and Hong Kong) for their excellent technical assistance.
This work was supported by the National Institutes of Health (National Institute of Allergy and Infectious Diseases contract HSN266200700005C), the Li Ka Shing Foundation, and the Area of Excellence Scheme of the University Grants Committee of the Hong Kong SAR (grant AoE/M-12/06).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Adiego Sancho B., et al. 2009. Human case of swine influenza A (H1N1), Aragon, Spain, November 2008. Euro Surveill. 14:pii=19120. [PubMed] [Google Scholar]
- 2. Brookes S. M., et al. 2010. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non-immune pigs. PLoS One 5:e9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74:29–46 [DOI] [PubMed] [Google Scholar]
- 4. Claas E. C., Kawaoka Y., de Jong J. C., Masurel N., Webster R. G. 1994. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology 204:453–457 [DOI] [PubMed] [Google Scholar]
- 5. Dawood F. S., et al. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605–2615 [DOI] [PubMed] [Google Scholar]
- 6. de Jong J. C., et al. 1986. Isolation of swine-influenza-like A(H1N1) viruses from man in Europe, 1986. Lancet ii:1329–1330 [DOI] [PubMed] [Google Scholar]
- 7. FAS/USDA March 2006, posting date Livestock and poultry: world markets and trade. Foreign Agricultural Service, USDA, Washington, DC: http://www.fas.usda.gov/dlp/circular/2006/06-03LP/swine_sum.pdf [Google Scholar]
- 8. Garten R. J., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gregory V., et al. 2003. Human infection by a swine influenza A (H1N1) virus in Switzerland. Arch. Virol. 148:793–802 [DOI] [PubMed] [Google Scholar]
- 10. Gregory V., et al. 2001. Infection of a child in Hong Kong by an influenza A H3N2 virus closely related to viruses circulating in European pigs. J. Gen. Virol. 82:1397–1406 [DOI] [PubMed] [Google Scholar]
- 11. Guan Y., et al. 2010. The emergence of pandemic influenza viruses. Protein Cell 1:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itoh Y., et al. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma W., et al. 2009. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 56:326–337 [DOI] [PubMed] [Google Scholar]
- 14. Moreno A., et al. 2011. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet. Microbiol. 149:472–477 [DOI] [PubMed] [Google Scholar]
- 15. Myers K. P., Olsen C. W., Gray G. C. 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 44:1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olsen C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199–210 [DOI] [PubMed] [Google Scholar]
- 17. Pensaert M., Ottis K., Vandeputte J., Kaplan M. M., Bachmann P. A. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull. World Health Organ. 59:75–78 [PMC free article] [PubMed] [Google Scholar]
- 18. Pereda A., et al. 2010. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg. Infect. Dis. 16:304–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scholtissek C. 1990. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med. Princ. Pract. 2:65–71 [Google Scholar]
- 20. Scholtissek C., Burger H., Kistner O., Shortridge K. F. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287–294 [DOI] [PubMed] [Google Scholar]
- 21. Shinde V., et al. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N. Engl. J. Med. 360:2616–2625 [DOI] [PubMed] [Google Scholar]
- 22. Shortridge K. F., Webster R. G., Butterfield W. K., Campbell C. H. 1977. Persistence of Hong Kong influenza virus variants in pigs. Science 196:1454–1455 [DOI] [PubMed] [Google Scholar]
- 23. Smith G. J., et al. 2009. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 106:11709–11712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 25. Starick E., et al. 2011. Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J. Gen. Virol. 92:1184–1188 [DOI] [PubMed] [Google Scholar]
- 26. Vijaykrishna D., et al. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vijaykrishna D., et al. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473:519–522 [DOI] [PubMed] [Google Scholar]
- 28. Vincent A. L., et al. 2010. Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross-reactivity with contemporary swine influenza virus antisera. Influenza Other Respi. Viruses 4:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weingartl H. M., et al. 2010. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 84:2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu R., et al. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J., et al. 2010. 2009 pandemic H1N1 influenza virus replicates in human lung tissues. J. Infect. Dis. 201:1522–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu H., et al. 2010. Substitution of lysine at 627 position in PB2 protein does not change virulence of the 2009 pandemic H1N1 virus in mice. Virology 401:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu H., et al. 2011. Pathogenicity and transmissibility of the pandemic H1N1 2009-related influenza viruses in mice, ferrets, and pigs. Influenza Other Respi. Viruses 5(Suppl. 1):82–84 [PubMed] [Google Scholar]
- 34. Zwickl D. 2006. Ph.D. thesis Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin, Austin, TX [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.