Abstract
Although human cancers have complex genotypes and are genomically unstable, they often remain dependent on the continued presence of single-driver mutations—a phenomenon dubbed “oncogene addiction.” Such dependencies have been demonstrated in mouse models, where conditional expression systems have revealed that oncogenes able to initiate cancer are often required for tumor maintenance and progression, thus validating the pathways they control as therapeutic targets. Here, we implement an integrative approach that combines genetically defined mouse models, transcriptional profiling, and a novel inducible RNAi platform to characterize cellular programs that underlie addiction to MLL-AF9—a fusion oncoprotein involved in aggressive forms of acute myeloid leukemia (AML). We show that MLL-AF9 contributes to leukemia maintenance by enforcing a Myb-coordinated program of aberrant self-renewal involving genes linked to leukemia stem cell potential and poor prognosis in human AML. Accordingly, partial and transient Myb suppression precisely phenocopies MLL-AF9 withdrawal and eradicates aggressive AML in vivo without preventing normal myelopoiesis, indicating that strategies to inhibit Myb-dependent aberrant self-renewal programs hold promise as effective and cancer-specific therapeutics. Together, our results identify Myb as a critical mediator of oncogene addiction in AML, delineate relevant Myb target genes that are amenable to pharmacologic inhibition, and establish a general approach for dissecting oncogene addiction in vivo.
Keywords: oncogene addiction, MLL-AF9, AML, Myb, Smyd2, RNAi, mouse models
Oncogene addiction describes the dependence of cancer cells on the continued expression of a driving oncogene, such that inactivation of the oncogene product triggers the selective cell death, senescence, or differentiation of cancer cells (Weinstein 2002). The concept was initially formulated based on studies using transgenic mice engineered to conditionally express particular oncogenes, where it was observed that withdrawal of oncogene expression could produce dramatic and complete tumor regressions, albeit with eventual disease relapse (Chin et al. 1999; Felsher and Bishop 1999; Gunther et al. 2003; Politi et al. 2006). This “addiction” of cancer cells to the driving oncogene was subsequently used to explain the remarkable efficacy and selectivity of certain molecularly targeted therapies; for example, those targeting Bcr-Abl in chronic myeloid leukemia and EGFR mutants in non-small-cell lung cancers (Druker et al. 2001; Lynch et al. 2004; Weinstein and Joe 2006). Subsequent studies have demonstrated that cancers can also become addicted to downstream mediators of driving oncogenes, providing additional entry points for therapeutic intervention (Soucek et al. 2008; Ehrenreiter et al. 2009; Hoelbl et al. 2010). Nonetheless, despite their potential importance, the precise molecular programs that mediate oncogene addiction remain poorly understood.
Acute myeloid leukemia (AML) exemplifies a genetically well-characterized cancer where specific lesions are linked to patient prognosis and therapy outcome (Estey and Dohner 2006). Leukemogenesis typically results from cooperating mutations that induce aberrant self-renewal in lineage-committed progenitors, often enforced by fusion proteins arising from chromosomal rearrangements, and others that increase proliferation, such as Ras pathway mutations. For example, translocations involving the MLL gene at 11q23 are among the most extensively characterized AML oncogenes, and are clinically associated with a dismal prognosis and poor therapy responses (Schoch et al. 2003). In patients, MLL rearrangements are often found in concert with activating mutations in Ras signaling pathways (Naoe et al. 1993; Liang et al. 2006; Radtke et al. 2009; Zuber et al. 2009; Chandra et al. 2010), which have been shown to cooperate with MLL fusion proteins to promote AML in mice (Ono et al. 2005; Stubbs et al. 2008; Zuber et al. 2009). Despite these advances in our understanding of the molecular mechanisms underlying AML pathogenesis, the majority of AML patients are not cured by the current chemotherapy regimens (Estey and Dohner 2006), underscoring the need for targeted therapeutics.
MLL encodes a histone H3K4 methyltransferase that activates transcriptional programs necessary for the establishment and maintenance of the hematopoietic system (Ernst et al. 2004). Leukemogenic fusion proteins aberrantly activate MLL targets, perhaps by recruiting alternative histone methyltransferases (Okada et al. 2005) and/or elongation factors (Lin et al. 2010; Yokoyama et al. 2010) to their promoters. The resulting inappropriate expression of canonical MLL target genes such as HoxA9 and Meis1 has been shown to suppress differentiation and establish aberrant self-renewal capabilities in lineage-committed progenitors, which is critical for the leukemogenic potential of MLL fusion proteins (Ayton and Cleary 2003; Cozzio et al. 2003; Krivtsov et al. 2006; Wong et al. 2007; Somervaille et al. 2009). In addition, transformation by MLL fusion proteins leads to the up-regulation of a self-renewal-associated transcriptional subprogram in “leukemia stem cells” (LSCs) that is similar to embryonic stem cells (Krivtsov et al. 2006; Somervaille et al. 2009) and overlaps with a transcriptional program controlled by the Myc oncoprotein (Kim et al. 2010). As self-renewal ability is likely critical for the maintenance of LSCs, targeting these programs presents a promising therapeutic opportunity in MLL fusion AML.
In this study, we set out to implement new RNAi technology to explore mechanisms of oncogene addiction in a manner that has not previously been possible in vivo. We chose to dissect the network's underlying addiction to an MLL fusion oncoprotein owing to the extensive body of work addressing its action in leukemia and the availability of tractable and well-characterized models of disease. Indeed, we demonstrated previously that a transplant-based mouse model of AML induced by the MLL-ENL fusion protein in cooperation with oncogenic Ras approximates the genetics, pathology, and treatment response observed in AML patients (Schoch et al. 2003; Neubauer et al. 2008; Zuber et al. 2009; Pardee et al. 2011). We thus reasoned that these systems would provide tractable models to interrogate mechanisms that underlie oncogene addiction in a relevant setting. Through iterative rounds of hypothesis generation and testing, we demonstrate that MLL-AF9 enforces a Myb-regulated program of aberrant self-renewal involving genes linked to LSC potential and poor prognosis in human AML, which is crucial to leukemia maintenance and is specific to cancer cells.
Results
An inducible mouse model of MLL fusion AML
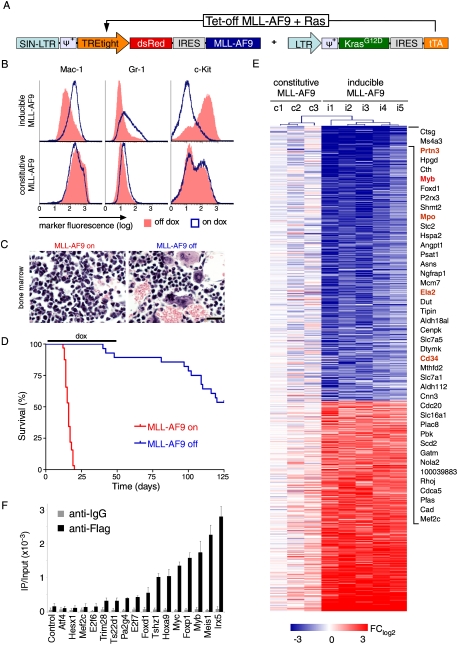
To explore oncogene addiction programs in a genetically defined leukemia context, we developed an AML mouse model driven by a conditional MLL-AF9 allele together with oncogenic Ras, which enabled us to examine the consequences of MLL-AF9 inhibition in an established disease. In order to produce a tightly regulated system that was easy to monitor, we constructed two retroviral vectors containing dsRed-linked MLL-AF9 under control of a tetracycline (tet) response element (TRE) promoter, and KrasG12D linked to the “Tet-off” tet transactivator (tTA), which activates TRE expression in a doxycycline (dox)-repressible manner (Fig. 1A). Leukemias were generated by retroviral cotransduction of both vectors into hematopoietic stem and progenitor cells (HSPCs), which were transplanted into syngeneic mice. Cells harboring both constructs (dsRed+) induced an aggressive myelomonocytic leukemia similar to existing mouse models based on constitutive MLL oncogene expression, which recapitulated key features of human MLL fusion-driven AML (Zuber et al. 2009).
Figure 1.
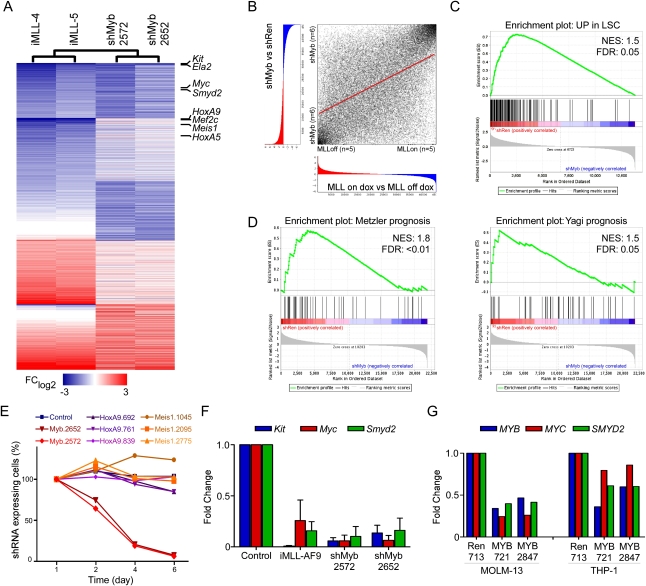
Withdrawal of MLL-AF9 induces differentiation and disease remission. (A) Retroviral constructs used to generate AML. (B) Representative immunophenotyping of primary Tet-off MLL-AF9;KrasG12D leukemia cells harvested from mice at terminal stage and briefly grown in vitro, with or without dox (4 d). Similar results were observed in three independent primary lines. (C) Representative bone marrow histology of syngeneic recipient mice transplanted with Tet-off MLL-AF9;KrasG12D AML, untreated and after dox treatment (9 d). Also see Supplemental Figure 3 for detailed histology. (D) Kaplan-Meier survival curves of mice transplanted with Tet-off MLL-AF9;KrasG12D AML, which were left untreated (MLL-AF9 on; n = 32) or treated with dox 5 d post transplantation for 45 d (MLL-AF9 off; n = 26). (E) Heat map of gene expression changes in primary AML lines with either Tet-off MLL-AF9 (i1–5) or constitutive MLL-AF9 (c1–3), 6 d after dox treatment to inactivate MLL-AF9 expression. Fold change relative to untreated leukemias rendered in a blue–white–red pseudo color scheme for all genes with P < 0.005 and FClog2 > 1.5 or FClog2 < −1.5. The most strongly down-regulated genes are indicated and include Myb and several of its known transcriptional target genes (marked in orange). (F) ChIP to assay MLL-AF9-Flag binding to promoters of down-regulated transcription factors. Bar graph represents enrichment of immunoprecipitation relative to input. Values represent the mean ± SE of three independent experiments.
The impact of withdrawing expression of MLL-AF9 on leukemia cell proliferation was dramatic. Dox treatment rapidly turned off dsRed, MLL-AF9, and HoxA9 expression (Supplemental Fig. 1) and triggered cell cycle arrest and terminal differentiation of leukemia cells (Fig. 1B; Supplemental Fig. 2). Withdrawal of MLL-AF9 in vivo led to disease regression and clearance of leukemia cells from all infiltrated organs (Fig. 1C; Supplemental Fig. 3) and prolonged the survival of leukemia-bearing mice (median, 16 vs. 127 d; P < 0.0001) (Fig. 1D). Most animals remained in remission upon discontinuing dox treatment after 45 d, and those with apparent “relapses” actually succumbed to a dsRed-negative host-derived lymphoid malignancy (Supplemental Fig. 4). These results, together with a previous report (Horton et al. 2008), demonstrate that leukemia cells require the continued expression of MLL-AF9.
Identifying critical mediators of leukemia maintenance
To identify molecular mechanisms underlying addiction to MLL-AF9, we performed genome-wide analysis of gene expression changes on leukemia cells following oncogene withdrawal. Six days following dox treatment, five independent Tet-off-regulatable MLL-AF9;NrasG12D leukemias showed a complex pattern of gene expression changes (Fig. 1E; Supplemental Table 1). Gene ontology analysis revealed a down-regulation of genes associated with cell cycle and mitosis, whereas those involved in mature myeloid cell functions were induced, suggesting that leukemia cells underwent a terminal differentiation program (Supplemental Table 2). In addition, gene set enrichment analysis (GSEA) (Subramanian et al. 2005) revealed a strong inverse correlation between global gene expression changes following MLL-AF9 withdrawal and a LSC-associated signature identified in MLL oncoprotein-expressing leukemias (normalized enrichment score [NES], 1.7 and −1.9; false discovery rate [FDR], 0.05 and 0) (Supplemental Fig. 5; Somervaille et al. 2009). Hence, the aberrant self-renewal and “stemness” programs induced by MLL-AF9 require continued oncogene expression.
To identify genes that might mediate these effects, we focused our subsequent analysis on differentially expressed transcription factors, as these molecules govern cell fate in normal hematopoiesis (Rosenbauer and Tenen 2007). Out of 17 strongly down-regulated transcription factors (P < 0.005, FClog2 < −1.5), 16 were validated by quantitative RT–PCR (qRT–PCR) analysis, including established MLL-AF9 targets such as HoxA9, HoxA5, and Meis1 (Supplemental Fig. 6). To distinguish between direct and indirect effects, we produced AMLs expressing a Flag-tagged MLL-AF9 transgene (Supplemental Fig. 7A,B) and analyzed MLL-AF9 promoter occupancy by chromatin immunoprecipitation (ChIP). In addition to binding the promoter regions of its canonical targets (HoxA9 and Meis1), MLL-AF9 was also enriched at the promoters of five additional transcription factors (Tshz1, Myc, FoxP1, Myb, and Irx5) (Fig. 1F; Supplemental Fig. 7C), suggesting that these also may be direct MLL-AF9 targets. The most differentially expressed direct MLL-AF9 target was the transcription factor c-Myb (Myb), which has been shown to be induced by MLL oncoproteins through an indirect mechanism (Hess et al. 2006). However, consistent with our results in murine leukemias, ChIP analysis of human THP1 AML cells expressing a Flag-tagged MLL-AF9 also showed strong occupancy of the fusion protein on the Myb promoter (Supplemental Fig. 7D). Besides Myb itself, several of its known transcriptional targets were among the most down-regulated genes following MLL-AF9 withdrawal (Fig. 1E; Supplemental Fig. 8; Ramsay and Gonda 2008), suggesting that Myb-dependent transcriptional programs may play a pivotal role in addiction to MLL-AF9.
Myb is required for the maintenance of leukemia
c-MYB was first identified as a proto-oncogene via its association with oncogenic retroviruses and as a target for insertional mutagenesis in mice (Klempnauer and Bishop 1984; Shen-Ong et al. 1986). More recent studies indicate that Myb is the target of somatic mutations and translocations in human cancers, and thus is an important human oncogene (Clappier et al. 2007; Persson et al. 2009). Functionally, Myb is a member of the basic helix–loop–helix family of transcription factors (Nomura et al. 1988), an important transcriptional regulator in normal stem and progenitor cell compartments (Emambokus et al. 2003; Malaterre et al. 2007), and is also a component of the LSC signature described above (Somervaille et al. 2009). Moreover, Myb inhibition is known to prevent the initiation of leukemogenesis by MLL-ENL (Hess et al. 2006) and has anti-proliferative effects in some leukemia cells (Luger et al. 2002; Somervaille et al. 2009). However, the role of Myb in leukemia maintenance has not been well characterized, in part because systems to efficiently inhibit gene expression in established disease in vivo were not available.
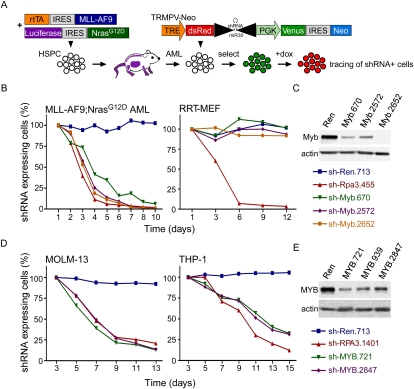
To study the role of Myb and other genes in leukemia maintenance, we took advantage of new methods to regulate gene expression using tet-responsive mir30-based shRNAs. We generated a “Tet-on RNAi-competent” model of MLL-AF9;NrasG12D AML that expresses an improved reverse tTA (rtTA3) linked to MLL-AF9, as well as NrasG12D-linked Luciferase to facilitate leukemia monitoring in vivo (Fig. 2A; Zuber et al. 2011a). These leukemia cells were then transduced with a new Tet-regulatable dual-color shRNA expression vector (TRMPV-Neo) (Zuber et al. 2011a). In this system, expression of the Venus fluorescent protein reports integration of the retrovirus, while dsRed fluorescence tracks expression of the shRNA, enabling precise fluorescent tracking and isolation of shRNA-expressing cells as the Venus+dsRed+ population.
Figure 2.
Expression of Myb is required for leukemia maintenance. (A) Schematic of the generation of Tet-on RNAi-competent MLL-AF9;NrasG12D AML and negative selection RNAi studies using the TRMPV system. Competitive proliferation assays in indicated mouse (B) and human (D) cell types. Graphs represent the percentage of shRNA-expressing (Venus+dsRed+) cells over time, normalized to initial measurement 1–2 d after dox treatment. (C) Immunoblot for Myb in Tet-on-competent MLL-AF9;NrasG12D leukemias transduced with indicated TRMPV-Neo-shRNAs, drug-selected and treated with dox for 3 d, and sorted for shRNA-expressing (Venus+dsRed+) cells to exclude cells that escape shRNA induction. (E) Immunoblot for MYB in Tet-on-competent human AML cell lines, dox-treated and sorted as described in C.
In competitive proliferation assays, TRMPV-Neo-transduced leukemia cells were mixed with uninfected leukemia cells and treated with dox to induce shRNA expression, and the percentage of shRNA-expressing (Venus+dsRed+) cells was monitored over time using flow cytometry. As expected, TRMPV-Neo-transduced leukemia cells expressing a neutral shRNA (targeting Renilla luciferase, Ren) were maintained in the cell population; in contrast, potent Myb shRNAs were rapidly depleted relative to non-shRNA-expressing cells (dsRed−) (Fig. 2B,C). These results mirrored the effects of suppressing replication protein A3 (Rpa3), an essential DNA replication factor required for proliferation (McJunkin et al. 2011; Zuber et al. 2011a). In contrast to the Rpa3 shRNAs, Myb shRNAs had no effect on immortalized rtTA-expressing mouse embryonic fibroblasts (RRT-MEFs) (Fig. 2B; Fellmann et al. 2011), demonstrating that Myb shRNAs are not generally toxic.
To extend these observations to human cancer cells, we evaluated the effects of Myb inhibition in 10 human leukemia cell lines. Suppression of Myb by two independent shRNAs impaired proliferation of all four MLL fusion-expressing cell lines tested, two of which coexpress MLL-AF9 with oncogenic Ras, as well as two other AML lines without MLL fusion proteins (Fig. 2D,E; Supplemental Fig. 9A). In four of six leukemia lines without MLL aberrations, Myb suppression had only minimal or no effect on cell proliferation, and these differences in sensitivity did not correlate with Myb expression levels (Supplemental Fig. 9B). Together, these results indicate that the sensitivity to Myb inhibition extends to human leukemia cells, particularly, but not exclusively, to MLL-rearranged contexts.
Myb suppression is tolerated in normal hematopoiesis
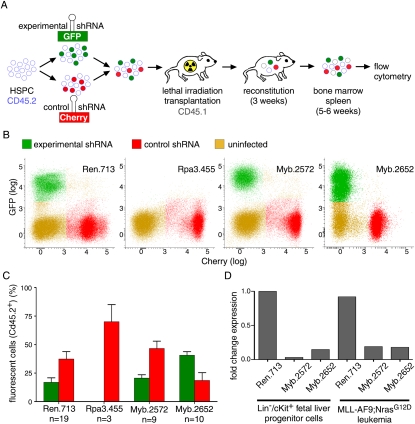
If Myb mediates cellular addiction to MLL-AF9, then myeloid leukemia cells should be more sensitive to Myb inhibition than their normal counterparts. To investigate the effects of RNAi-mediated Myb suppression in normal hematopoietic cells, we established a two-color competitive reconstitution assay in which HSPCs transduced with either a GFP-tagged experimental shRNA or an mCherry-tagged neutral internal control shRNA (Ren) are monitored for their ability to reconstitute the hematopoietic system of lethally irradiated recipient mice (Fig. 3A). As expected, 6 wk after transplantation, donor-derived bone marrow contained GFP- and mCherry-positive shRen-expressing cells, while cells expressing a GFP-tagged shRNA targeting Rpa3 were completely depleted (Fig. 3B,C).
Figure 3.
Suppression of Myb is tolerated in normal hematopoiesis. (A) Schematic of dual-color competitive reconstitution assay for evaluating effects of RNAi-mediated gene suppression in normal hematopoiesis. (B) Representative flow cytometry plots of donor-derived (CD45.2+) cells in bone marrow 4 wk after transplantation of HSPCs transduced with indicated experimental shRNAs (GFP+) and a neutral control shRNA (shRen, Cherry+). (C) Percentage of cells expressing indicated experimental shRNA (green) or the neutral control shRNA (red) in total bone marrow 4 wk after transplantation. Values represent the mean ± SE percentage of fluorescent cells within donor-derived (CD45.1+) full bone marrow of multiple mice. (D) qRT–PCR analysis for Myb mRNA in shRNA-expressing Tet-on-competent MLL-AF9;NrasG12D leukemias and fetal liver progenitor cells (FLPCs) (Lin−, cKit+). Values are normalized to actin and plotted relative to FLPC shRen.
In contrast, cells expressing Myb shRNAs were able to efficiently reconstitute recipient mice (Fig. 3B,C) even though Myb was suppressed to similar levels as in MLL-AF9;NrasG12D leukemia cells (Fig. 3D). These results differ from the consequences of complete Myb ablation observed in knockout mice (Mucenski et al. 1991; Lieu and Reddy 2009) and suggest that hypomorphic states produced by shRNA-mediated Myb suppression are better tolerated. Lineage analysis revealed that Myb inhibition does not impede normal erythropoiesis and myelopoiesis but, consistent with previous reports (Bender et al. 2004; Fahl et al. 2009), impairs normal lymphopoiesis (Supplemental Fig. 10). Interestingly, one of the shRNAs (shMyb.2652) showed enrichment in myeloid and progenitor compartments (Supplemental Fig. 10), which, together with a previous study (Emambokus et al. 2003), suggests that hypomorphic states of Myb induce expansion rather than depletion in the myeloid compartment. Of note, cells with Myb suppression are capable of supporting long-term myeloid reconstitution, and animals remained healthy and disease-free for >12 wk (data not shown). These results indicate that the consequences of suppressing Myb are dosage- and lineage-dependent and can result in highly cancer-specific sensitivities in the myeloid lineage. They also establish a rapid and general assay for exploring the impact of gene function during normal hematopoiesis.
Suppression of Myb eradicates AML in vivo
To explore the sensitivity to Myb inhibition in established AML in vivo, we transplanted a series of Tet-on-competent MLL-AF9;NrasG12D, TRMPV-Neo-transduced leukemias harboring shRNAs targeting Myb, the Ras effector Braf (another putative target in our model), or Ren, and induced shRNA expression with dox following leukemia detection in bioluminescence imaging (Supplemental Fig. 11A). Even using mixed populations of transduced cells, Myb suppression resulted in a delay in disease progression (Supplemental Fig. 11A) and a significant survival benefit (P < 0.005, Supplemental Fig. 11B). At terminal disease stage, bone marrow of mice transplanted with shRen and shBraf-transduced leukemia was completely infiltrated with shRNA-expressing cells (Venus+dsRed+), while leukemia cells expressing Myb-shRNAs were entirely gone (Supplemental Fig. 11C,D). Thus, in mice transplanted with shMyb-transduced MLL-AF9;NrasG12D leukemia, progressive disease resulted from the outgrowth of cells that failed to induce Myb shRNA expression (dsRed−).
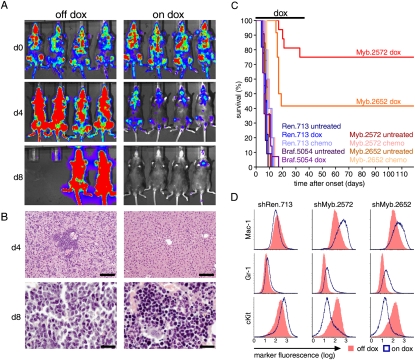
Even more dramatic results were observed when the experiment was repeated using single cell-derived clones that uniformly induce shRNA expression. While induction of Ren or Braf shRNAs had no effect on disease course, suppression of Myb induced the clearance of leukemia cells from all infiltrated organs, leading to complete remissions (Fig. 4A–C; Supplemental Fig. 11E). These remissions were durable, and most animals remained disease-free even after discontinuing dox treatment after 40 d (Fig. 4C). Again, those mice that subsequently developed disease harbored leukemia cells that were dsRed-negative, and thus did not express the Myb shRNA and no longer inhibited Myb expression (Supplemental Fig. 11F). This dramatic anti-leukemic response was in contrast to the minimal effects of a combined chemotherapy regimen mimicking clinical AML therapy (Ara-C + Doxorubicin) (Zuber et al. 2009) in mice bearing the same leukemias (Fig. 4C). Thus, transient inhibition of Myb invariably induces complete remission of MLL-AF9-driven AML in vivo, leading to eradication of an otherwise aggressive and treatment refractory disease.
Figure 4.
Suppression of Myb eradicates aggressive AML in vivo. (A) Bioluminescent imaging of recipient mice carrying clonal MLL-AF9;NrasG12D AML transduced with Myb shRNAs. Mice were treated with dox at disease onset (d0) and followed up after 4 and 8 d. (B) Liver (top) and bone marrow (bottom) histology from untreated or dox-treated mice at the indicated time point. Bars: for bone marrow, 20 μm; for liver, 100 μm. (C) Kaplan-Meier survival curves of recipient mice untreated, treated with dox, or treated with conventional chemotherapy (n > 4). Dox treatment was discontinued after 40 d. (D) Representative immunophenotyping of AML cells transduced with indicated TRMPV-Neo shRNAs with or without 3 d of dox treatment.
Myb regulates a LSC-associated program
We next performed another iteration of expression profiling and functional studies to explore the transcriptional networks and effector programs underlying the leukemia-specific dependency on Myb. Immunophenotyping and gene expression analysis revealed that suppression of Myb induces terminal differentiation into mature neutrophils as well as monocytes/macrophages (Fig. 4D; Supplemental Fig. 12), precisely phenocopying effects observed after inactivating the driving MLL-AF9 oncogene (Fig. 1B; Supplemental Fig. 12). The global gene expression changes following Myb suppression were remarkably similar to those observed after MLL-AF9 withdrawal (Spearman correlation, 0.50) (Fig. 5A,B; Supplemental Fig. 13A,B), as determined by a recently developed method to compare global expression signatures (Rank–Rank Hypergeometric Overlap [RRHO]) (Plaisier et al. 2010). Indeed, >40% of significantly altered genes in both signatures overlap (Fig. 5A; Supplemental Fig. 13C), and these influence the same biological processes (Supplemental Tables 3, 4). Such strong effects indicate that Myb is a dominant factor involved in aberrant transcription and impaired differentiation associated with MLL-AF9.
Figure 5.
Myb regulates a gene expression program overlapping with MLL-AF9 and associated self-renewal. (A) Heat map of fold change in gene expression for two representative Tet-off-inducible MLL-AF9;Ras leukemia cell lines (on dox relative to off dox) and Tet-on-competent MLL-AF9;Ras leukemia transduced with the indicated Myb shRNAs (shMyb relative to shRen) for all genes with >1.5-fold change in either comparison. (B) Overlap of MLL-AF9 and Myb-regulated gene expression signatures. Using the RRHO approach, genes were rank-ordered according to their differential expression between the indicated subclasses. Spearman rank correlation coefficient = 0.50 (P < 0.001). (C) GSEA plot evaluating LSC-associated genes (Somervaille et al. 2009) after expression of shRNAs targeting Myb (two independent shRNAs, n = 3) or controls (shRen or empty vector, n = 3). (D) GSEA plots evaluating expression of signatures associated with poor prognosis in AML patients in two different studies (Yagi et al. 2003; Metzeler et al. 2008) with or without suppression of Myb. (E) Competitive proliferation assay in MLL-AF9;NrasG12D leukemia. The graph represents the percentage of shRNA-expressing (Venus+dsRed+) cells over time, normalized to initial measurement after 1 d of dox treatment. (F) qRT–PCR analysis for Kit, Myc, and Smyd2 expression in Tet-off-inducible MLL-AF9;Ras leukemia treated with dox for 6 d (relative to off dox) and shRNA-expressing Tet-on-competent MLL-AF9;NrasG12D leukemia transduced with the indicated shRNA, treated with dox for 3 d, and sorted for shRNA-expressing (Venus+dsRed+) cells (relative to shRen). (G) qRT–PCR analysis of MYB, MYC, and SMYD2 in human AML cell lines expressing the indicated shRNAs. Cells were treated with dox for 4 d and sorted for shRNA-expressing (Venus+dsRed+) cells prior to RNA extraction.
The transcriptional module associated with Myb suppression also showed a strong inverse correlation with the LSC signature (NES, 1.5 and −1.7; FDR, 0.05 and 0.004) (Fig. 5C; Supplemental Fig. 13D), indicating that Myb is a master regulator of LSC-associated programs. Notably, the Myb-mediated expression profile was also significantly enriched for two gene sets associated with poor prognosis in pediatric and adult AML (NES, 1.8 and 1.5; FDR, <0.01 and 0.05) (Fig. 5D; Yagi et al. 2003; Metzeler et al. 2008), implying that this program also contributes to aggressive disease in humans. Interestingly, some canonical MLL target genes such as HoxA9 and Meis1 are excluded from the Myb-specific signatures (Fig. 5A; Supplemental Fig. 13C), indicating that Myb controls only a subset of MLL-AF9 targets and does not predominantly act as a cofactor in MLL complexes as proposed (Jin et al. 2010). These findings also suggest that HoxA9 and Meis1, despite their known role in leukemia initiation by MLL fusion proteins (Zeisig et al. 2004), may be less important for maintaining aberrant self-renewal. Supporting this notion, multiple HoxA9 and Meis1 shRNAs did not affect MLL-AF9;NrasG12D AML cells in competitive proliferation assays (Fig. 5E).
Components of the Myb-mediated self-renewal program are required for leukemia maintenance
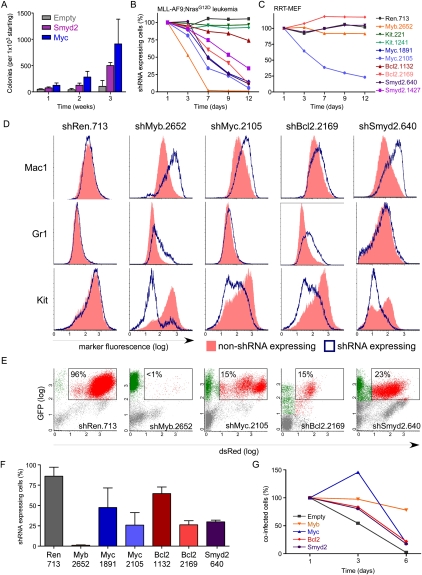
Several known Myb target genes have been associated with self-renewal in different contexts, and therefore might mediate Myb-dependent aberrant self-renewal in AML. For example, Kit, a target of somatic mutations in human AML (Estey and Dohner 2006), serves as a LSC marker in murine MLL-AF9-induced leukemia (Krivtsov et al. 2006). Myc contributes to self-renewal in normal stem cells (Smith et al. 2010) and may play a pivotal role in establishing the LSC signature described above (Kim et al. 2010). Bcl2 promotes the self-renewal of normal hematopoietic stem cells (Milyavsky et al. 2010), and its pharmacologic inhibition is purported to target LSCs (Konopleva et al. 2006). Finally, in different analyses integrating LSC-, Myb-, and MLL-AF9-associated signatures, we consistently identified the methyltransferase Smyd2 as a prominent LSC-associated gene suppressed upon Myb and MLL-AF9 withdrawal (Supplemental Tables 5, 6) and subsequently validated it as a Myb-regulated gene in mouse and human AML cells (Fig. 5F,G). Interestingly, Smyd2 is amplified in esophageal squamous cell carcinoma (Komatsu et al. 2009) and methylates and inhibits the p53 and Rb tumor suppressors (Huang et al. 2006; Saddic et al. 2010), two molecules that negatively regulate self-renewal (Galderisi et al. 2006; Zhao et al. 2010). Indeed, serial replating assays indicate that enforced Smyd2 expression enhances self-renewal of hematopoietic progenitors, similar to the known effects of Myc (Fig. 6A).
Figure 6.
Suppression of Myb-regulated genes partially phenocopies effects of Myb and MLL-AF9 withdrawal. (A) Serial replating assay of fetal liver cells transduced with the indicated cDNA expression vector. Bar graphs represent the number of GFP+ colonies normalized to the number seeded in Methocult. Values are the mean ± SEM of three independent experiments. Competitive proliferation assays in MLL-AF9;NrasG12D leukemia (B) or immortalized RRT-MEFs (C). (D) Representative immunophenotyping of MLL-AF9;NrasG12D leukemia cells transduced with the indicated TRMPV-Neo shRNAs 5 d after dox treatment, separated by shRNA-expressing (blue) and non-shRNA-expressing (red) cells within each sample. (E) Representative flow cytometry plots of donor-derived (CD45.2+) bone marrow cells in terminally diseased dox-treated mice. The percentage of shRNA-expressing cells (Venus+dsRed+) is indicated. (F) Percentage of shRNA-expressing cells in the bone marrow of moribund dox-treated mice in E. Values represent the mean ± SE (n > 4) percentage of Venus+dsRed+ cells among all donor-derived (CD45.2+) cells. (G) Competitive proliferation assay of MLL-AF9;NrasG12D leukemia cells cotransduced with TRN-shMyb.2572 and the indicated cDNA (tagged with GFP). See Supplemental Figure 15A for a schematic of the assay. Shown are the percentages of cells coexpressing the cDNA and shMyb.2572 (dsRed+GFP+) over time, normalized to the initial measurement after 1 d of dox treatment to induce shMyb expression.
Our system enables the rapid evaluation of candidate tumor maintenance genes without the production of new germline transgenic mice, simply by making potent shRNAs targeting a gene of interest and assessing their impact in our transplantable model. To evaluate a possible role of the Myb-regulated genes described above in maintaining aberrant self-renewal, we analyzed the impact of their TRMPV-shRNA-mediated suppression on AML proliferation and differentiation. Suppression of Kit by multiple potent shRNAs (Supplemental Fig. 14A) had no effect on MLL-AF9;NrasG12D AML (Fig. 6B). In contrast, leukemia cells expressing potent shRNAs targeting either Myc, Smyd2, or Bcl2 (Supplemental Fig. 14B–D) underwent terminal myeloid differentiation and were depleted from the population over time (Fig. 6B,D). Inhibition of Myc, but not Smyd2 or Bcl2, also impeded proliferation of RRT-MEF (Fig. 6C), indicating that strong RNAi-mediated suppression of Myc confers general anti-proliferative effects.
We next transplanted drug-selected TRMPV-Neo-transduced MLL-AF9;NrasG12D leukemias, induced shRNA expression, and evaluated the bone marrow for the presence of shRNA-expressing cells at terminal disease stage (Fig. 6E,F). Suppression of Myc, Bcl2, or Smyd2 led to the depletion of leukemia cells in vivo, although in no case were the effects as dramatic or complete as observed when targeting Myb. We also performed cDNA rescue experiments using a modified competition assay in which the relative frequency of shMyb-expressing cells is monitored over time in cells that ectopically express different cDNAs (Supplemental Fig. 15A). Consistent with RNAi-based phenotypes, overexpression of Myc, Bcl2, or Smyd2 delayed Myb-dependent depletion to varying degrees, but was unable to fully rescue the inhibitory and prodifferentiation effects of suppressing Myb (Fig. 6G; Supplemental Fig. 15B). While we cannot rule out that the less-dramatic effects observed after suppressing Myc, Smyd2, or Bcl2 are due to differences in shRNA knockdown efficiency, the inability of cDNAs encoding these genes to completely rescue the effects of Myb suppression suggests that none of them can completely replicate the effects of Myb. Instead, these results indicate that Myb exerts its central role in maintaining aberrant self-renewal through a coordinated program involving Myc, Bcl2, Smyd2, and, undoubtedly, additional target genes.
Discussion
Our study integrates mouse cancer models, transcriptional profiling, and inducible RNAi to systematically select and test candidate mediators of oncogene addiction in vivo. Building on a well-characterized and validated model, we implemented new and generalizable assays, enabling an unprecedented level of in vivo analysis of genetic dependencies in normal and cancer cells. These advances include a new Tet-on-inducible shRNA expression system optimized for target suppression in established tumors, as well as a new assay involving fluorescent-coupled shRNAs to study the consequences of suppressing the same genes in normal hematopoiesis. By combining these approaches, we identified Myb as a central mediator of oncogene addiction in AML and show that its suppression eradicates aggressive leukemia in vivo without impacting normal myelopoiesis. In addition, we identify key Myb effectors that coordinate this program, some of which are amenable to small molecule inhibition. Similar approaches can be used to systematically identify and validate tumor maintenance genes in other cancer models.
By comparing the expression profiles of AML cells produced by Myb suppression with those following MLL-AF9 withdrawal, we see that Myb is the dominant factor contributing to aberrant gene expression in MLL-AF9-expressing cells—with >40% overlap in differentially expressed genes. While down-regulation of Myb is required during hematopoietic differentiation (Emambokus et al. 2003), MLL-AF9 apparently “hijacks” Myb to enforce a program of aberrant self-renewal via transcriptional modules linked to LSC potential and poor prognosis in human AML. Moreover, while this Myb-dependent program is highly enriched for genes contained within the LSC signature, it excludes some canonical MLL targets such as HoxA9 and Meis1 that are known to contribute to leukemogenesis (Ayton and Cleary 2003; Wong et al. 2007). Interestingly, the LSC signature has recently been linked to Myc activity (Kim et al. 2010), which is a direct target of Myb (Evans et al. 1990) and depends on Myb expression. Accordingly, our data suggest that Myb can act as a dominant regulator of the LSC module in AML.
Previous studies suggest that Myb might act downstream from HoxA9 and Meis1 in transformation induced by MLL-AF9 or act more directly in MLL-mediated transcription as a component of the MLL-AF9 complex (Hess et al. 2006; Jin et al. 2010). Our finding that MLL-AF9 occupies the Myb promoter as strongly as some of its canonical targets raises the possibility that MLL-AF9 directly induces Myb transcription. While it remains to be determined which mechanisms predominate, the results are consistent with the possibility that Myb acts as part of a mutually reinforcing transcription program of self-renewal, as has been described for pluripotency factors in embryonic stem cells (Pan and Thomson 2007; Young 2011). In principle, such a situation could create a positive amplification loop; consequently, transient and partial Myb suppression collapses the LSC transcription module and eradicates aggressive AML without compromising normal myelopoiesis.
In addition to identifying mechanisms of oncogene addiction, our results provide compelling in vivo validation of Myb as a therapeutic target. We show that partial and transient suppression of Myb using RNAi precisely phenocopies MLL-AF9 withdrawal and eradicates aggressive AML in vivo by restoring a program of terminal myeloid differentiation. Suppression of Myb to levels similar to those that eliminate leukemia cells does not prevent normal myelopoiesis, indicating distinct requirements for Myb in cancer and normal cells and leading to a highly cancer-specific sensitivity. Indeed, Myb suppression using RNAi enhances myelopoiesis while impairing lymphopoiesis, which is consistent with studies using a Myb hypomorph mouse (Emambokus et al. 2003) and highlights the potential of RNAi to identify hypomorphic phenotypes of relevance to understanding human disease. Certainly, the impact of Myb inhibition on normal lymphopoiesis may present some “on-target” toxicities, but as transient Myb inhibition is sufficient to eradicate leukemia, such effects may prove manageable.
While the leukemia-specific dependency on Myb identified using a MLL-AF9;NrasG12D-expressing mouse model extends to human AML, this addiction program may not be strictly genotype-specific. For example, some cell lines without MLL fusion proteins were also sensitive to Myb suppression, suggesting that other lesions may engage the same Myb-dependent self-renewal program. Regardless, the remarkable sensitivity of leukemia cells expressing MLL fusion proteins to Myb inhibition may be unique, as we have evaluated hundreds of other potential AML maintenance genes using similar methods and, to date, none has as potent effects as Myb (Zuber et al. 2011b; J Zuber, CR Vakoc, J Shi, E Wang, C Chen, and SW Lowe, unpubl.). The potency of these effects is further underscored by the generally poor response of patients harboring MLL translocations to conventional therapy in the clinic.
Although targeting of Myb itself has proven elusive (Ratajczak et al. 1992; Luger et al. 2002), several key Myb effectors such as Bcl2 and Smyd2 are established or potential drug targets (Chonghaile and Letai 2008; Spannhoff et al. 2009). Still, our RNAi-based studies as well as preliminary drug trials using the BH3-mimetic ABT-737 (S Glaser and A Strasser, pers. comm.; data not shown) suggest that targeting Myb downstream effectors individually will not be as potent as inhibiting Myb. Nonetheless, improved drugs or adroit drug combinations hold promise for more effective and less toxic therapies. Moreover, in a parallel study, we identified the bromodomain protein Brd4 as a promising therapeutic target in AML (Zuber et al. 2011b). The dramatic responses to the Brd4 inhibitor JQ1 are in large part due to a role of Brd4 in sustaining Myc expression. Remarkably, expression changes following Brd4 suppression using RNAi or a small molecule inhibitor largely overlap with those seen after suppressing Myb (Supplemental Fig. 16), suggesting that Brd4 and Myb engage a common transcriptional network and providing another pharmacological entry point for targeting aberrant self-renewal programs. Together, our results demonstrate that suppressing a single mediator of aberrant self-renewal can re-establish pre-existing cell fate programs and eliminate aggressive AML, providing hope that targeting these pathways will lead to more effective therapies for leukemia and, potentially, other cancers.
Previous efforts to target mediators of oncogene addiction have predominantly focused on components of oncogenic signaling pathways; however, in many cases, the plasticity of these programs limits drug efficacy or facilitates drug resistance and, through various feedback mechanisms, can even enhance pro-oncogenic signaling (Poulikakos et al. 2010; Pratilas and Solit 2010). Our observation that suppressing a single key mediator of aberrant self-renewal can reverse this program, re-establish terminal differentiation, and thereby eliminate leukemia suggests that pre-existing cell fate programs remain hard-wired in cancer, and raises hope that agents inhibiting these programs will be less prone to resistance mechanisms. Such agents may also be effective at targeting so-called cancer stem cells, which require aberrant self-renewal programs to propagate the tumor and are considered more resistant to conventional therapies. More broadly, our study exemplifies how the combination of genetically defined mouse models and novel Tet-on RNAi technology can be used to systematically identify and evaluate new therapeutic targets for otherwise refractory cancers.
Materials and methods
Retroviral constructs
All constructs were derived from either the Murine Stem Cell Virus backbone (MSCV, Clontech) or the pQCXIX self-inactivating retroviral vector backbone (pSIN, Clontech) using standard cloning techniques. TRMPV vectors are available through Addgene (#27990, #27991, #27993, and #27996). shRNAs were designed using the Biopredsi algorithm and subcloned as described (for sequences, see Supplemental Table 7; Zuber et al. 2011a). See the Supplemental Material for more detailed vector information.
Generation and monitoring of AML
Primary leukemias were generated by cotransduction of retroviral constructs into fetal liver cells and transplantation into syngeneic recipient mice as described previously (Zuber et al. 2009). Leukemia cells were harvested from bone marrow and spleen and cultured in RPMI-1640 + 10%FBS. Peripheral blood smears and bone marrow cytospins were stained with Wright-Giemsa (Sigma); tissue specimens were fixed in 10% formalin and stained with Hematoxylin and eosin (Sigma). For immunostaining, cells were stained with specified fluorophore-conjugated antibodies (BioLegend) and analyzed by flow cytometry (LSRII, BD BioSciences). Mice were monitored and treated as described (Zuber et al. 2011a). Statistical evaluation of overall survival was based on the log-rank (Mantel-Cox) test for comparison of the Kaplan-Meier event time format. More detailed methods are provided in the Supplemental Material.
Gene expression analysis
Cultured and treated primary leukemia cells were sorted for shRNA-expressing cells when necessary using an Aria-II (BD Biosciences). Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using TaqMan reverse transcriptase reaction (Applied Biosystems). qRT–PCR analysis was performed on an iCycler mounted with an iQ5 multicolor real-time PCR detection system (Bio-Rad). All signals were quantified using the ΔΔCt method and normalized to β-actin levels. Microarray experiments were performed on Mouse Genome 430A 2.0 arrays (Affymetrix). All raw microarray data files are available from the Gene Expression Omnibus (GSE30747). For Western blot analysis, the following antibodies were used: c-Myb (clone1-1, 1:500; Millipore), Flag M2 (#1804, 1:1000; Sigma), Braf (F-7, 1:200; Santa Cruz Biotechnolgy), Bcl2 (BCL/10C4, 1:1000; BioLegend), Smyd2 (#4251, Cell Signalling), and actin (HRP-conjugated, 1:10000; Sigma). ChIP was performed as described (Steger et al. 2008). Detailed methods are provided in the Supplemental Material.
Acknowledgments
We thank J. Simon, E. Earl, and L. Bianco for mouse support; B. Ma for technical assistance; V. Thapar for assistance with gene expression analysis; C. Johns for performing microarrays; and K. McJunkin and L. Dow for comments and support. This work was supported by a Specialized Center of Research Grant from the Leukemia and Lymphoma Society, NIH grant R37 CA72614, and generous gifts from the Don Monti Memorial Research Foundation. J.Z. was supported by a research fellowship from the German Research Foundation (DFG) and the Andrew Seligson Memorial Fellowship at CSHL; A.R.R. was supported by an NIH traineeship and the Barbara McClintock fellowship; S.C.K. is the Leslie Rutherford Leukemia and Lymphoma Society Scholar; C.R.V. was supported by NIH training grant T32 HL007150 and by the CSHL President's Council; S.W.L. is a Howard Hughes Medical Institute investigator. J.Z. and A.R. designed research, performed experiments, analyzed data and cowrote the paper. W.L. performed array analysis. E.W., C.C., A.V.V., J.S., S.W., M.W., and M.T. performed experiments. T.G.G performed RRHO analysis. S.C.K. performed histological analysis. C.R.V designed research, performed experiments, and analyzed data. S.W.L. designed research, analyzed data, and cowrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17269211.
References
- Ayton PM, Cleary ML 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17: 2298–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K 2004. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5: 721–729 [DOI] [PubMed] [Google Scholar]
- Chandra P, Luthra R, Zuo Z, Yao H, Ravandi F, Reddy N, Garcia-Manero G, Kantarjian H, Jones D 2010. Acute myeloid leukemia with t(9;11)(p21-22;q23): common properties of dysregulated ras pathway signaling and genomic progression characterize de novo and therapy-related cases. Am J Clin Pathol 133: 686–693 [DOI] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, et al. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468–472 [DOI] [PubMed] [Google Scholar]
- Chonghaile TN, Letai A 2008. Mimicking the BH3 domain to kill cancer cells. Oncogene 27: S149–S157 doi: 10.1038/onc.2009.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, et al. 2007. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood 110: 1251–1261 [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev 17: 3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. 2001. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344: 1031–1037 [DOI] [PubMed] [Google Scholar]
- Ehrenreiter K, Kern F, Velamoor V, Meissl K, Galabova-Kovacs G, Sibilia M, Baccarini M 2009. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer Cell 16: 149–160 [DOI] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J 2003. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 22: 4478–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ 2004. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell 6: 437–443 [DOI] [PubMed] [Google Scholar]
- Estey E, Dohner H 2006. Acute myeloid leukaemia. Lancet 368: 1894–1907 [DOI] [PubMed] [Google Scholar]
- Evans JL, Moore TL, Kuehl WM, Bender T, Ting JP 1990. Functional analysis of c-Myb protein in T-lymphocytic cell lines shows that it trans-activates the c-myc promoter. Mol Cell Biol 10: 5747–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahl S, Crittenden R, Allman D, Bender T 2009. c-Myb is required for pro-B cell differentiation. J Immunol 183: 5582–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. 2011. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell 41: 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4: 199–207 [DOI] [PubMed] [Google Scholar]
- Galderisi U, Cipollaro M, Giordano A 2006. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene 25: 5250–5256 [DOI] [PubMed] [Google Scholar]
- Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, Cardiff RD, Chodosh LA 2003. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev 17: 488–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK 2006. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood 108: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, Fajmann S, Grebien F, Warsch W, Stengl G, et al. 2010. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med 2: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton SJ, Walf-Vorderwulbecke V, Chatters SJ, Sebire NJ, de Boer J, Williams O 2008. Acute myeloid leukemia induced by MLL-ENL is cured by oncogene ablation despite acquisition of complex genetic abnormalities. Blood 113: 4922–4929 [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM 2010. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest 120: 593–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer KH, Bishop JM 1984. Neoplastic transformation by E26 leukemia virus is mediated by a single protein containing domains of gag and myb genes. J Virol 50: 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, et al. 2009. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 30: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. 2006. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10: 375–388 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Liang DC, Shih LY, Fu JF, Li HY, Wang HI, Hung IJ, Yang CP, Jaing TH, Chen SH, Liu HC 2006. K-Ras mutations and N-Ras mutations in childhood acute leukemias with or without mixed-lineage leukemia gene rearrangements. Cancer 106: 950–956 [DOI] [PubMed] [Google Scholar]
- Lieu YK, Reddy EP 2009. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci 106: 21689–21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger SM, O'Brien SG, Ratajczak J, Ratajczak MZ, Mick R, Stadtmauer EA, Nowell PC, Goldman JM, Gewirtz AM 2002. Oligodeoxynucleotide-mediated inhibition of c-myb gene expression in autografted bone marrow: a pilot study. Blood 99: 1150–1158 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Malaterre J, Carpinelli M, Ernst M, Alexander W, Cooke M, Sutton S, Dworkin S, Heath JK, Frampton J, McArthur G, et al. 2007. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc Natl Acad Sci 104: 3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW 2011. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci 108: 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, et al. 2008. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 112: 4193–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, Lechman E, Hermans KG, Eppert K, Konovalova Z, et al. 2010. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell 7: 186–197 [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677–689 [DOI] [PubMed] [Google Scholar]
- Naoe T, Kubo K, Kiyoi H, Ohno R, Akao Y, Yoshida J, Kato K, Kojima S, Matsuyama T 1993. Involvement of the MLL/ALL-1 gene associated with multiple point mutations of the N-ras gene in acute myeloid leukemia with t(11;17)(q23;q25). Blood 82: 2260–2261 [PubMed] [Google Scholar]
- Neubauer A, Maharry K, Mrozek K, Thiede C, Marcucci G, Paschka P, Mayer RJ, Larson RA, Liu ET, Bloomfield CD 2008. Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a cancer and leukemia group B study. J Clin Oncol 26: 4603–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R 1988. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res 16: 11075–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121: 167–178 [DOI] [PubMed] [Google Scholar]
- Ono R, Nakajima H, Ozaki K, Kumagai H, Kawashima T, Taki T, Kitamura T, Hayashi Y, Nosaka T 2005. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J Clin Invest 115: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Thomson JA 2007. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res 17: 42–49 [DOI] [PubMed] [Google Scholar]
- Pardee TS, Zuber J, Lowe SW 2011. Flt3-ITD alters chemotherapy response in vitro and in vivo in a p53-dependent manner. Exp Hematol 39: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G 2009. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci 106: 18740–18744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG 2010. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res 38: e169 doi: 10.1093/nar/gkq636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE 2006. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 20: 1496–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Solit DB 2010. Targeting the mitogen-activated protein kinase pathway: physiological feedback and drug response. Clin Cancer Res 16: 3329–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J, Ganti R, Cai Z, Goorha S, Pounds SB, et al. 2009. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci 106: 12944–12949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ 2008. MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534 [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Hijiya N, Catani L, DeRiel K, Luger SM, McGlave P, Gewirtz AM 1992. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood 79: 1956–1961 [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7: 105–117 [DOI] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, Sage J 2010. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 285: 37733–37740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T 2003. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 102: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Shen-Ong GL, Morse HC 3rd, Potter M, Mushinski JF 1986. Two modes of c-myb activation in virus-induced mouse myeloid tumors. Mol Cell Biol 6: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S 2010. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML 2009. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 4: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M 2009. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4: 1568–1582 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, Kung AL, Armstrong SA 2008. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia 22: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB 2002. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK 2006. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 3: 448–457 [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, So CW, Cleary ML 2007. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 21: 2762–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, Mizutani S, Imashuku S, Ohki M, Ichikawa H 2003. Identification of a gene expression signature associated with pediatric AML prognosis. Blood 102: 1849–1856 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML 2010. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17: 198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA 2011. Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, et al. 2004. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 24: 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, Lowe SW 2010. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 24: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, McCurrach ME, Yang MM, Dolan ME, Kogan SC, et al. 2009. Mouse models of human AML accurately predict chemotherapy response. Genes Dev 23: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW 2011a. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011b. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukemia. Nature doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]