Summary
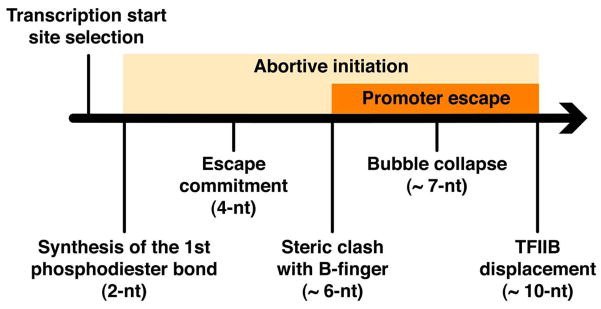
The initiation of transcription by RNA polymerase II is a multistage process. X-ray crystal structures of transcription complexes containing short RNAs reveal three structural states: one with 2- and 3-nucleotide RNAs, in which only the 3′-end of the RNA is detectable; a second state with 4- and 5-nucleotide RNAs, with an RNA-DNA hybrid in a grossly distorted conformation; and a third state with RNAs of 6 nucleotides and longer, essentially the same as a stable elongating complex. The transition from the first to the second state correlates with a markedly reduced frequency of abortive initiation. The transition from the second to the third state correlates with partial “bubble collapse” and promoter escape. Polymerase structure is permissive for abortive initiation, thereby setting a lower limit on polymerase-promoter complex lifetime and allowing the dissociation of nonspecific complexes. Abortive initiation may be viewed as promoter proofreading, and the structural transitions as checkpoints for promoter control.
The initiation of RNA polymerase II (Pol II) transcription is a focal point of cellular regulation. Initiation proceeds through multiple stages, each of which may be subject to intervention by regulatory factors. Stages so far recognized entail synthesis of transcripts with lengths of about 5, 10, and 25 nucleotides (nt) (1). Transcripts of less than 5 nt are unstable, resulting in frequent “abortive initiation.” At about 10 nt, interactions with general factors are disrupted, resulting in “promoter escape.” The initiation process concludes when, at a transcript length of about 25 nt, a transition is made to a stable “elongation complex.” Here, we report structures of Pol II with short transcripts that illuminate some of the earliest events of transcription initiation.
Previous biochemical studies showed that the synthesis of a 3- to 4-nt transcript confers a degree of stability, referred to as “escape commitment” (Fig. 1), revealed by a reduced incidence of abortive initiation and by the end of a requirement for adenosine triphosphate (ATP) hydrolysis for maintenance of a transcription “bubble” (2–8). A transcript length of about 7 nt induces partial collapse of the transcription bubble, which coincides with the start of promoter escape (9). An 8–base pair RNA-DNA hybrid is necessary and sufficient for the formation of a stable transcribing complex (10). Transcript “slipping,” in which a short RNA disassociates from the template DNA and reanneals with a repeating element upstream, correlates with hybrid strength as well (11, 12).
Fig. 1. Pol II initiation pathway.
Interactions with both Pol II and general transcription factors modulate transcript stability. Positively charged residues in the switch 2 region of Pol II are critical for the retention of a 5-nt transcript (13). The finger domain of the general transcription factor TFIIB exerts a similar stabilizing effect on the early transcribing complex, probably by direct contacts with both an RNA transcript of up to 5 nt and the DNA template strand (14). Accordingly, perturbation of TFIIB finger function alters the distribution of abortive transcripts (9). The mobile “clamp” domain of Pol II contacts both the hybrid and the downstream duplex DNA, thereby stabilizing early transcribing complexes (15). The general transcription factors TFIIE and TFIIF, both of which interact with the clamp, have been shown to stabilize short transcripts and facilitate escape commitment (5, 9, 16, 17).
Our initial attempts to crystallize transcribing complexes containing 4- and 5-nt RNAs were unsuccessful, resulting in crystals of Pol II alone with an open clamp (18), probably due to a low stability of these complexes. Attempts to stabilize the complexes by the addition of full-length TFIIB also yielded crystals of Pol II alone, probably because binding of the TFIIB zinc ribbon domain to the “dock” domain of the Rpb2 subunit of Pol II is incompatible with crystal packing. However, a TFIIB fragment (residues 50 to 217) from which the zinc ribbon was deleted, which has been shown to interact with a closed clamp (19), supported the crystallization of various early transcribing complexes, probably by maintaining a closed conformation of the clamp.
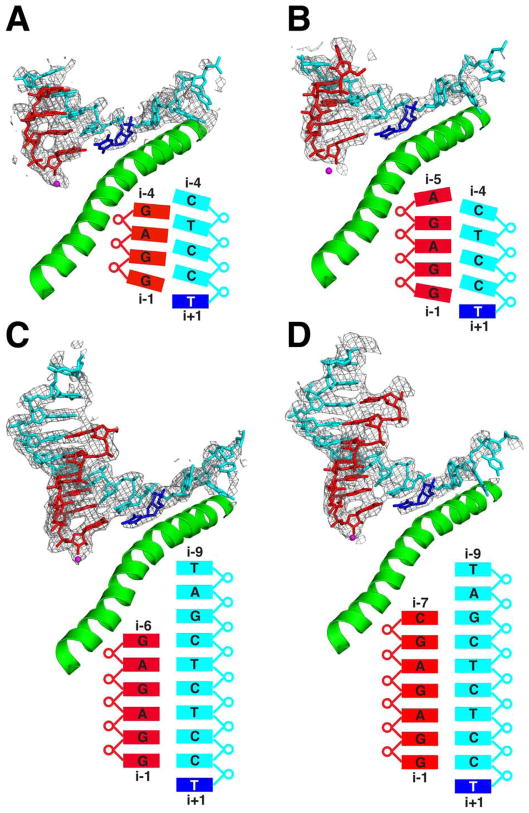
Structures of early transcribing complexes, formed from RNA oligonucleotides and a template DNA oligonucleotide with a downstream duplex region (table S1), were solved by molecular replacement with the structure of Pol II in the closed-clamp conformation. Models of the transcript RNA and template DNA strands were built into the unbiased Fobs – Fcalc electron density map (Fig. 2 and fig. S1). All complexes were in the posttranslocation state, with an empty nucleotide addition site opposite the i+1 base in the template strand. The downstream duplex DNA was largely disordered and was therefore omitted from model building. The clamp domain was not involved in crystal contacts and presumably remained mobile; this may be important for accommodating small conformational changes due to interaction with hybrids of different lengths.
Fig. 2. Structures of transcribing complexes with 4- to 7-nt RNAs.
Fobs – Fcalc electron density maps contoured at 2.0s are shown as gray mesh. Template DNA structure is cyan; transcript RNA is red. The i+1 nucleotide on the template DNA strand is blue. The active-site magnesium ion and the bridge helix are shown as a magenta sphere and a green ribbon, respectively. The sequence and conformation of the nucleic acids are diagrammed beneath the structures. (A) 4-nt RNA. (B) 5-nt RNA. (C) 6-nt RNA. (D) 7-nt RNA.
A series of individual nucleoside triphosphates (NTPs) and RNA oligonucleotides of 2 to 9 nt were used in the crystallization trials. To facilitate direct comparison, we ensured that the RNA and DNA oligonucleotides were the same in all complexes, except for additional nucleotides at the 5′-end of the RNA (Fig. 2, fig. S1, and tables S1 and S2). With DNA alone, or with DNA and ATP (matched to the coding base in the DNA at i+1), only the downstream DNA duplex could be seen in Fobs – Fcalc electron density maps, with no density for the upstream template strand. With a 2- or 3-nt RNA, scattered peaks for the RNA-DNA hybrid, most significant at the i–1 RNA position, could be seen in Fobs – Fcalc electron density maps (fig. S2); with RNAs of 4 nt or longer, clear connected density for the hybrid was observed. The 2- and 3-nt complexes showed no density for the i+1 nucleotide on the template strand (fig. S2), and modeling in the i+1 nucleotide resulted in strong negative electron density, indicating that the correct positioning of this nucleotide can only be achieved through the formation a stable hybrid (and not through nonspecific electrostatic interactions, as observed for nucleotides upstream on the template DNA strand). These findings are consistent with biochemical studies showing that 2- and 3-nt RNAs support expansion of the transcription bubble with ATP hydrolysis and nucleotide addition, whereas a transcript of at least 4 nt is required for formation of a stable initiation complex (2–8). The transition from a mobile state of the hybrid and i+1 nucleotide to one of strong interaction with the Pol II active center provides a molecular rationale for escape commitment.
Mobility of the i–3 ribonucleotide of the 3-nt complex may be explained by a deep cavity in the hybrid-binding pocket of Rpb2 facing i–3 and part of i–4 (fig. S3A). A molecular dynamics simulation supports this idea, showing that the i–3 ribonucleotide of the 3-nt complex, modeled on the basis of the 4-nt complex structure, is unusually mobile relative to the terminal ribonucleotide in complexes of 4 nt and longer (fig. S3B and movie S1).
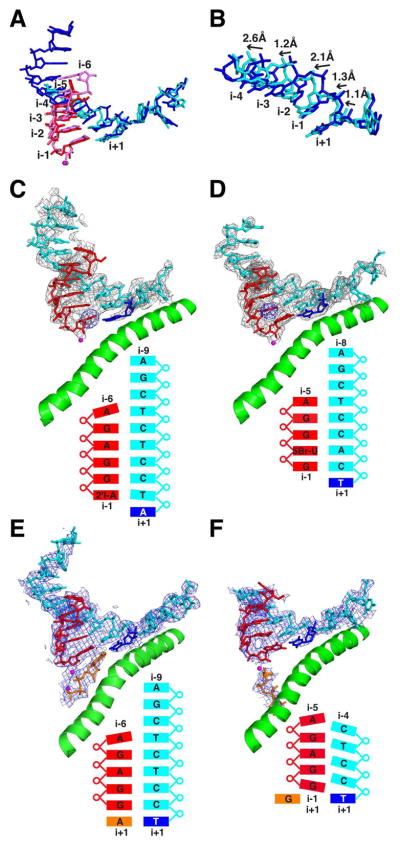
Comparison of the 4-, 5-, and 6-nt complexes reveals both an unusual hybrid conformation and a second structural transition. In the 4- and 5-nt structures, the i–1 and i–2 regions of the hybrid are most ordered. The densities for DNA bases at positions i–1 and i–2 are not separated from one another, and density for the template strand from i–5 to the end is missing altogether (Fig. 2, A and B). The 3′- and 5′-terminal ribonucleotides in both 4- and 5-nt structures are distinctly frayed (fig. S4), with ordering at 3′-end (i–1 position) apparently due to coordination of the phosphate by Rpb2 residues Lys979 and Lys987 (fig. S7). The 3′-terminal nucleotide partially overlaps the nucleotide addition site, opposite DNA nucleotide i+1. Superposition of the hybrid regions of the 5- and 6-nt structures reveals a shift of template nucleotides i+1 to i–4 in the 5-nt hybrid upstream by an average distance of 1.7 Å (Fig. 3, A and B). The conformations of RNA strands are similar in the two structures, except for the fraying of the 3′ and 5′ termini of the 5-nt hybrid. Thus, the i–2 RNA base appears to be paired with the i–1 DNA base, and so forth. This distortion of the 4- and 5-nt hybrids, and the mobility apparent from indistinct or absent electron density, indicate a degree of instability that would account for a high incidence of abortive initiation, which persists even after escape commitment (2).
Fig. 3. Conformational transitions during early initiation.
(A) Structures of 5- and 6-nt complexes are superimposed, with colors for the 5-nt complex as in Fig. 2 and the DNA and RNA of the 6-nt complex in blue and pink. (B) Sugar-phosphate backbones of the 5- and 6-nt complexes are superimposed and rotated 30° around the x-axis with respect to the view in (A) to display the shift between the two structures. (C) Anomalous diffraction peak from iodine incorporated by transcription in situ in the 5-nt complex is shown as blue mesh. Colors and other features in (C) through (F) are as in Fig. 2. (D) Anomalous diffraction peak from bromine at the i-2 position of 5-nt RNA is shown as blue mesh. (E) ATP soaked in crystals of the 5-nt complex, occupying the nucleotide addition (“A”) site, is shown in orange. (F) GTP soaked in crystals of the 5-nt complex, occupying the entry (“E”) site, is shown in orange.
By contrast, the 6-nt complex exhibits a canonical hybrid structure, and the template single strand upstream of the hybrid is also ordered (Fig. 2C). All hybrids from 6 to 9 nt are essentially indistinguishable, indicating that in the transition from the 5-nt complex a stable state has been achieved. This transition coincides with bubble collapse and promoter escape, shown to occur at around register 7 for human Pol II (9).
We next sought to determine whether the unusual hybrid structure of the 4- and 5-nt complexes was due to the particular sequence used. For example, the i–1 DNA base, a C, could pair with the i–2 RNA base, a G. Two sequence variants of the 5-nt complex were therefore investigated to eliminate the possibility of forming such a misaligned base pair. In one variant, the i–2 RNA base was changed to C, and the structure was the same as that of the original complex. In the second variant, the i–1 DNA base was changed to G, and the template strand no longer assumed a shifted conformation, probably because a shift would result in a steric clash between the G’s on the RNA and DNA strands (fig. S5). Distinctive features of the 5-nt complex nonetheless persisted: The densities due to neighboring bases were not separated, and the template DNA strand upstream of the hybrid region was not observed. Other sequence variants showed the transition to a canonical hybrid structure as early as 5 nt or as late as 7 nt (fig. S6 and table S1), but mobility and distortion of the hybrid helix and mobility of the upstream template DNA are apparently general, if not universal, features of the structure preceding the transition.
To determine whether the 5-nt complex with its distorted hybrid helix was active in transcription, we soaked crystals of the complex in a cryoprotectant solution containing 2′-iodo-ATP. An anomalous diffraction peak from the iodine atom demonstrated incorporation of the analog at the 3′-end of the RNA (Fig. 3C). As a test of the model-building of the nucleic acids, 5-bromouridine (5-BrU) was introduced at the second position from the 3′-end in the 5-nt RNA. An anomalous signal from the bromine atom was observed at the i–2 position, confirming the assignment of the U and the register of the hybrid (Fig. 3D). The 5-nt hybrid containing 5-BrU adopted the conformation of the 6–base pair hybrid (Fig. 3D), consistent with previous observations that 5-BrU confers higher stability of base pairing and that incorporation into RNA decreases the frequency of abortive initiation and transcript slippage (11, 12).
The effect of 5-BrU incorporation indicates that the 5-nt hybrid helix is on the verge of the transition to the canonical hybrid conformation. We investigated whether the presence of NTP in the nucleotide addition (i+1) site would tip the balance in favor of the canonical conformation. Crystals of a 5-nt complex bearing a 3′-deoxy terminus were soaked in cryoprotectant solution containing either ATP, matched to the DNA base at i+1, or guanosine triphosphate (GTP), mismatched to the base at i+1. ATP conferred upon the 5-nt complex many of the features of the 6-nt complex: alignment of the template DNA and RNA strands (i–1 RNA base pairing with i–1 DNA base and so forth); a canonical hybrid helix, except for the 5′-terminal nucleotide, which remained frayed; and partial ordering of the upstream template DNA strand beyond position i–5 (Fig. 3E). By contrast, the crystal soaked with GTP retained the original 5-nt complex structure, with the distorted conformation of the hybrid, shift of the template strand, and absence beyond i–5 (Fig. 3F). Evidently, the entry of matched NTP in the addition site directs the frayed RNA 3′-end into position opposite the i–1 DNA base and provides the number of base pairs required for the transition to a canonical hybrid helix.
Only two positively charged Pol II residues are located within 5 Å of the RNA in the 2- to 6-nt complex structures, and as mentioned above, both of these residues (Lys979 and Lys987 of the Rpb2 subunit) interact with the i–1 phosphate. The purpose is presumably the precise positioning of the 3′-end of the RNA for NTP addition, and consistent with such a critical role, these two residues are required for cell viability (20). In contrast, multiple lysine and arginine residues are located in close proximity to the template DNA strand, with one or two residues in contact with each of the nucleotides from i–1 to i–6 (fig. S7). In the 6-nt complex, the strictly conserved Rpb2 residues Arg857 and Arg942 contact the phosphate at i–6 (fig. S7), evidently critical for the 5- to 6-nt transition.
The lack of interaction of Pol II residues with short RNAs is counterintuitive. Especially in the case of 2- and 3-nt RNAs, interaction would be expected if the RNAs are to be retained in the absence of stable hybrid formation. Rather than contribute to hybrid stability, however, interactions with the template DNA have the opposite effect: They constrain a short hybrid in a distorted conformation. It is not impossible for a polymerase to promote helix formation with small RNAs. For example, crystals of bacteriophage T7 RNA polymerase complexes have been obtained that show stable retention of initiating NTP and canonical hybrid helix structure with 3-nt RNA (21, 22). Pol II structure is evidently designed to disfavor short hybrid formation, and thus to enhance the rate of abortive initiation. We suggest that this design enhances the specificity of transcription. DNA undergoes frequent helix openings, and the resulting incipient transcription bubbles may be captured by Pol II, leading to aberrant transcription. Reducing the lifetime of short hybrids increases the likelihood of Pol II dissociation from the DNA. Only at a promoter, where general transcription factors and sequence-specific interactions come into play, will the Pol II–DNA complex persist and transcription ensue. General factors may not influence abortive initiation, which is an inherent property of RNA polymerases, but they maintain polymerase-promoter association during abortive initiation and enhance the stability of complexes after escape commitment (14, 16).
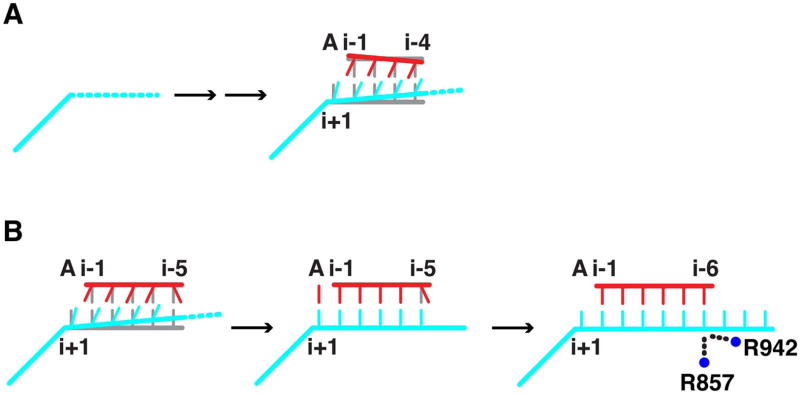
Abortive initiation is thus the basis of “promoter proofreading,” a form of kinetic proofreading (23, 24) supported by general transcription factors. The two structural transitions reported here, resulting in diminished abortive initiation and stable hybrid formation, may be viewed as checkpoints for promoter control (Fig. 4). Promoter proofreading is doubtless required in a cellular context. Nonspecific transcription by Pol II would likely have deleterious consequences for cell differentiation and development. By contrast, bacteriophage infection may be little affected by a high background of more or less random transcription, and a bacteriophage polymerase (such as the T7 enzyme) sacrifices specificity for speed of the initiation process.
Fig. 4. Schematic of structural transitions during initiation by pol II.
(A) The 3- to 4-nt transition is depicted with DNA in cyan, RNA in red, and a canonical hybrid helix in gray. Mobility of the upstream DNA template strand is symbolized by a dashed line. (B) The 5- to-6-nt transition is depicted as in (A). R857 and R942 mediating the transition in concert with base pair formation are symbolized by blue circles.
The 5- to 6-nt transition may make a further contribution to the transcription mechanism. Elsewhere we have proposed that 5-nt RNA makes a favorable interaction with the TFIIB finger region, resulting in a conformational switch of TFIIB and the release of its C-terminal region from contact with promoter DNA (14). Subsequent transcription beyond 5 nt creates a steric clash with the B finger, the complete release of TFIIB, rewinding of a part of the upstream DNA (“bubble collapse”), and promoter escape. The flexibility of the 5-nt complex, shown here by incomplete separation of electron density for the bases in the hybrid and a lack of density for the upstream template DNA, may facilitate the interaction with the B finger. Conversely, the rigidity of the 6-nt complex may accentuate the steric clash with the B finger and provide a driving force for promoter escape (Fig. 4B).
Supplementary Material
Acknowledgments
This research was supported by NIH grants GM049985 and AI21144 to R.D.K. X.L. was supported by the Jane Coffin Childs Memorial Fund fellowship. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Computing resources were provided by Dawning TC5000 supercomputing cluster, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences. Coordinates and structure factors have been deposited at the Protein Data Bank under accession code 3RZO, 3RZD, 3S14, 3S15, 3S16, 3S17, 3S1M, 3S1N, 3S2H, 3S2D, 3S1Q, and 3S1R (see Table S2).
References
- 1.Saunders A, Core LJ, Lis JT. Nat Rev Mol Cell Biol. 2006 Aug;7:557. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 2.Holstege FC, Fiedler U, Timmers HT. EMBO J. 1997 Dec 15;16:7468. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai H, Luse DS. J Biol Chem. 1987 Jan 5;262:298. [PubMed] [Google Scholar]
- 4.Luse DS, Kochel T, Kuempel ED, Coppola JA, Cai H. J Biol Chem. 1987 Jan 5;262:289. [PubMed] [Google Scholar]
- 5.Kugel JF, Goodrich JA. J Biol Chem. 2000 Dec 22;275:40483. doi: 10.1074/jbc.M006401200. [DOI] [PubMed] [Google Scholar]
- 6.Kugel JF, Goodrich JA. Mol Cell Biol. 2002 Feb;22:762. doi: 10.1128/MCB.22.3.762-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan M, Gralla JD. EMBO J. 1997 Dec 15;16:7457. doi: 10.1093/emboj/16.24.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan M, Gralla JD. J Biol Chem. 1999 Dec 3;274:34819. doi: 10.1074/jbc.274.49.34819. [DOI] [PubMed] [Google Scholar]
- 9.Pal M, Ponticelli AS, Luse DS. Mol Cell. 2005 Jul 1;19:101. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. J Biol Chem. 2000 Mar 3;275:6530. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 11.Pal M, Luse DS. Mol Cell Biol. 2002 Jan;22:30. doi: 10.1128/MCB.22.1.30-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene RG, Luse DS. J Biol Chem. 1999 Apr 23;274:11526. doi: 10.1074/jbc.274.17.11526. [DOI] [PubMed] [Google Scholar]
- 13.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. J Biol Chem. 2005 Oct 14;280:34917. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 14.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Science. 2004 Feb 13;303:983. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 15.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Science. 2001 Jun 8;292:1876. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 16.Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS. Mol Cell Biol. 2008 Jun;28:3757. doi: 10.1128/MCB.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen HT, Warfield L, Hahn S. Nat Struct Mol Biol. 2007 Aug;14:696. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cramer P, Bushnell DA, Kornberg RD. Science. 2001 Jun 8;292:1863. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Science. 2010 Jan 8;327:206. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domecq C, et al. Protein Expr Purif. 2009 Jan;69:83. doi: 10.1016/j.pep.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheetham GM, Steitz TA. Science. 1999 Dec 17;286:2305. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy WP, Momand JR, Yin YW. J Mol Biol. 2007 Jul 6;370:256. doi: 10.1016/j.jmb.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Ninio J. Biochimie. 1975;57:587. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 24.Hopfield JJ. Proc Natl Acad Sci U S A. 1974 Oct;71:4135. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.