Abstract
Mammalian target of rapamycin (mTOR) is a kinase that plays a key role in a wide array of cellular processes and exists in two distinct functional complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Although mTORC2 is primarily activated by growth factors, mTORC1 is regulated by numerous extracellular and intracellular signals such as nutrients, growth factors, and cellular redox. Previous study has shown that cysteine oxidants sufficiently activate mTORC1 activity under amino acid-depleted conditions and that a reducing agent effectively suppresses amino acid-induced mTORC1 activity, thereby raising the possibility that redox-sensitive mechanisms underlie amino acid-dependent mTORC1 regulation. However, the molecular mechanism by which redox regulates mTORC1 activity is not well understood. In this study, we show that the redox-sensitive regulation of mTORC1 occurs via Rheb but not the Rag small GTPase. Enhancing cellular redox potential with cysteine oxidants significantly increases Rheb GTP levels. Importantly, modulation of the cellular redox potential with a cysteine oxidant or reducing agent failed to alter mTORC1 activity in TSC1−/− or TSC2−/− mouse embryonic fibroblast cells. Furthermore, a cysteine oxidant has little effect on mTOR localization but sufficiently activates mTORC1 activity in both p18−/− and control mouse embryonic fibroblast cells, suggesting that the redox-sensitive regulation of mTORC1 occurs independent of the Ragulator·Rag complex. Taken together, our results suggest that the TSC complex plays an important role in redox-sensitive mTORC1 regulation and argues for the activation of mTORC1 in places other than the lysosome upon inhibition of the TSC complex.
Keywords: Amino Acid, Lysosomes, mTOR Complex (mTORC), Oxidation-Reduction, S6 Kinase, Signal Transduction, Tuberous Sclerosis (Tsc), Rapamycin, small GTPase
Introduction
The mammalian target of rapamycin (mTOR)2 belongs to the family of phosphatidylinositol 3-kinase (PI3K)-related kinases and shares high sequence similarity to PI3K despite possessing protein kinase activity (1). mTOR is an evolutionarily conserved protein kinase that forms two distinct functional complexes termed mTOR complex 1 and mTOR complex 2 (mTORC1 and mTORC2, respectively) (2–4). mTORC1 is known to be a rapamycin-sensitive complex that regulates a wide array of cellular processes such as cell growth and autophagy, whereas mTORC2 is known to be rapamycin-resistant and involved in the regulation of cell survival as well as cytoskeletal reorganization. Although mTORC2 activation is primarily mediated by growth factors, mTORC1 activation can be achieved by multiple inputs such as amino acids, growth factors, glucose, and oxidative stress (5, 6).
Recent studies have shown that two small GTPases, Rheb and Rag, play an essential role in the regulation of mTORC1 activation (7–10). Rheb interacts with and activates mTORC1, whereas Rag small GTPases function as essential spatial regulators of mTORC1 localization (8, 11–13). The Rag small GTPases function as heterodimers formed between RagA/B and RagC/D and bind to the MP1·p14·p18 complex (Ragulator), which is predominantly expressed on the lysosomal membrane (12, 14). Upon amino acid stimulation, the Rag heterodimer is activated and subsequently recruits mTORC1 to the lysosomal membrane, where it is activated by growth factor-regulated Rheb (8, 15). Rheb activity is tightly regulated by TSC2, a tumor suppressor protein containing a GTPase-activating protein (GAP) domain in its carboxyl terminus (15–20). TSC2 forms a physical and functional complex with TSC1 (21), and mutations in either TSC2 or TSC1 result in tuberous sclerosis complex (TSC), an inherited hamartoma syndrome characterized by the formation of benign tumors in multiple organs (22, 23). In response to growth factors, multiple kinases such as Akt and extracellular signal-regulated kinase (ERK) phosphorylate TSC2 and may inhibit its GAP activity toward Rheb, thereby activating mTORC1 (24–26). In contrast, suppression of mTORC1 activity occurs upon reduction of cellular energy levels via phosphorylation of multiple proteins such as TSC2, Raptor, and mTOR by the AMP-activated kinase (AMPK) (27–29) and Rheb by the p38-regulated/activated kinase (PRAK) (30).
Changes in intracellular energy levels as well as nutrient availability have been known to influence and modify the functionality of proteins regulated by redox potential (31, 32). Both redox sensing and redox signaling use sulfur switches, particularly Cys residues that are sensitive to reversible oxidation (31, 33). A previous study by Sarbassov and Sabatini (34) first demonstrated that cysteine oxidants such as phenylarsine oxide (PAO) and diamide activated mTORC1 activity in vivo and in vitro. Interestingly, cysteine oxidants were able to enhance S6K phosphorylation even in the absence of amino acids in culture, and a reducing agent effectively suppresses amino acid-induced mTORC1 activation (34). Furthermore, cysteine oxidants destabilized the mTOR-Raptor interaction but not the mTOR-Rictor interaction. These observations suggest that a redox-sensitive mechanism may underlie amino acid-dependent mTORC1 regulation, which is regulated by the Rag-Ragulator system (12, 34). Given the observation that cysteine oxidants enhance mTORC1 activity in vitro, it has been postulated that a component of mTORC1 or mTOR itself may sense redox potential to regulate mTORC1 function in response to extracellular amino acids (34–36). In this study, we show that cysteine oxidants specifically activate mTORC1 but not mTORC2. Moreover, our study indicates that the TSC-Rheb pathway, but not the Rag-Ragulator, plays an essential role in mTORC1 regulation in response to cellular redox potential. We also discuss redox-sensitive mTORC1 regulation by spatial activation of Rheb in places other than the lysosome.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Plasmids
PAO, British anti-Lewisite (BAL, also known as 2,3-dimercapto-1-propanol), diamide, 2-deoxyglucose (2-DG), U0126, and rapamycin were obtained from Sigma-Aldrich. HBSS was obtained from Invitrogen. No-Glucose Dulbecco's modified Eagle's medium (No-Glucose DMEM, Invitrogen) was used for glucose starvation treatment. [32P]Orthophosphate was obtained from MP Biomedicals. Mouse LAMP2 (H4B4) and rat LAMP2 antibodies (GL2A7) were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa and Abcam, respectively. HA and Myc antibodies were from Covance. AMPKα antibody was from Bethyl Laboratories. The p18 antibody was described previously (11). Other all antibodies used in this study were obtained from Cell Signaling. HA-S6K, Myc-Rheb, HA-TSC1, HA-TSC2, HA-RagA(QL), Myc-RagC(SN), HA-RagA(TN), and HA-RagC(QL) expression plasmids were described previously (7, 29). Lipofectamine (Invitrogen) was used for plasmid transfection according to the product manual.
Cell Culture and Treatments
HEK293T, HeLa, and mouse embryonic fibroblast cell lines (wild-type, TSC1−/−, TSC2−/−, p18−/−, and p18rev cells) were cultured in high glucose DMEM (Invitrogen) containing 10% fetal bovine serum (FBS, HyClone Laboratories) and penicillin/streptomycin (Invitrogen) (11). For amino acid stimulation assays, cells were treated with HBSS for 1 h and then cultured in low glucose DMEM for another 15 min. For cysteine oxidant or reducing agent treatment, cells were treated with HBSS for 1 h and then incubated in the reagent solution for 15 min under the following conditions: 5 μm PAO for HEK293T cells, 0.1 μm PAO for MEF cells, 250 μm diamide, and 0.5 mm BAL in HBSS. For the amino acid or glucose starvation treatments, cells were incubated in HBSS or No-Glucose DMEM for 1 h, respectively. 2-DG was added directly to the cell medium to a final concentration of 50 mm, and cells were incubated for 1 h. For rapamycin treatment, cells were preincubated with 20 nm rapamycin for 15 min before oxidant or reducing agent treatment.
Immunoblot and Immunoprecipitation
Cells were lysed on ice for 5 min in ice-cold lysis buffer (40 mm HEPES (ph 7.5), 120 mm NaCl, 1 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, 50 mm NaF, 1.5 mm Na3VO4, 0.3% CHAPS, and a mixture of protease inhibitors (Roche Applied Science)). After centrifugation at 13,000 × g for 15 min, the supernatant was mixed with 5× SDS sample buffer and boiled for 5 min. The samples were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. For immunoprecipitations, lysates were incubated with 1 μg of the indicated antibody for 2 h and precipitated with protein G/L-Sepharose.
In Vivo GTPase Assay
In vivo GTPase assays were performed as reported previously (17, 37). Briefly, cells were washed once with phosphate-free DMEM and incubated with 0.5 mCi/ml [32P]orthophosphate for 4 h. Cells were then lysed with lysis buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT, 0.5% Nonidet P-40, and protease inhibitors). Tagged small GTPase protein was immunoprecipitated with Myc or HA antibody. Immunoprecipitates were washed three times each with washing buffer I (50 mm Tris, pH 8.0, 0.5% Triton X-100, 500 mm NaCl, 5 mm MgCl2, 1 mm DTT) and buffer II (50 mm Tris, pH 8.0, 0.1% Triton X-100, 100 mm NaCl, 5 mm MgCl2, 1 mm DTT). The GTPase-bound nucleotides were eluted with elution buffer (2 mm EDTA, 0.2% SDS, 1 mm GDP, 1 mm GTP) at 68 °C for 15 min. Eluted nucleotides were separated on polyethyleneimine cellulose plates (Baker-Flex) in 0.75 m KH2PO4, pH 3.4.
siRNA Transfections
Effectene (Qiagen) was used to transfect 0.8 million HeLa cells in 6-cm dishes with siRNA oligonucleotides to TSC1 (SMARTpool, Dharmacon). 72 h after transfection, the cells were rinsed once with cold phosphate-buffered saline, lysed in ice-cold lysis buffer, and analyzed by immunoblotting as above.
Immunofluorescence Staining
Cells were fixed for 15 min with 4% paraformaldehyde. The fixed cells were rinsed three times with PBS and then permeabilized with 0.2% Triton X-100 in PBS for 10 min and blocked with 2% BSA-TBS for 30 min at room temperature. After rinsing three times with TBS containing 0.05% Tween (TBS-T), samples were incubated with primary antibodies in PBS for 1 h at room temperature, rinsed three times with TBS-T, incubated with secondary antibodies for 1 h at room temperature, washed two times with TBS-T, and rinsed two times with water. Samples were mounted on microscope slides using Prolong Gold anti-fade reagent (Invitrogen). Antibodies were used at the following dilutions: mTOR (1:400), RagC (1:200), LAMP2 (1:200), Alexa Fluor 488 anti-rat IgG (1:500), Alexa Fluor 594 anti-rabbit IgG (1:500), Alexa Fluor 488 anti-rabbit IgG (1:500), and Alexa Fluor 594 anti-mouse IgG (1:500). Images were obtained by the Fluoview 300 confocal microscope (Olympus) with 60× objective lenses. Observations were carried out three times with different samples prepared independently, and representative images were processed in the same manner using Adobe Photoshop CS3.
Statistical Analysis
All data were analyzed by analysis of variance with Scheffe's post hoc tests. Asterisks in Figs. 2 and 7 represent statistical significance (p value <0.05).
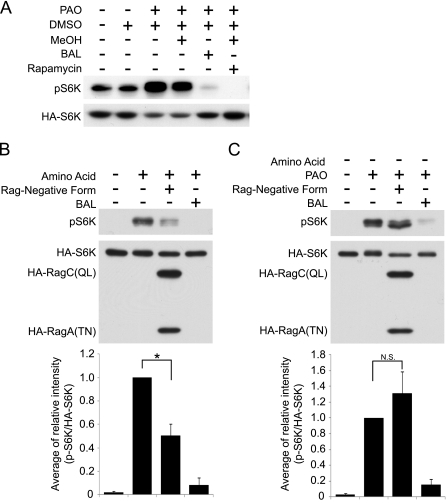
FIGURE 2.
PAO-induced S6K phosphorylation is resistant to the inactive Rag small GTPases. A, PAO induces phosphorylation of overexpressed HA-S6K. HEK293T cells were transfected with HA-S6K. After a 48-h transfection, the cells were treated with the combination of PAO, dimethyl sulfoxide (DMSO), MeOH, BAL, and rapamycin treatments as indicated. The cell lysates were analyzed by immunoblotting with phospho-S6K (pS6K) (Thr-389) and HA antibodies. B, amino acid-induced S6K phosphorylation was blocked by inactive Rag small GTPases (RagA(TN)·RagC(QL)). HA-S6K with or without HA-RagA(TN) and HA-RagC(QL) was transfected into HEK293T cells. Transfected cells were treated with HBSS for 1 h and then stimulated with amino acid in the presence or absence of BAL for 15 min. The cell lysates were analyzed by immunoblotting with phospho-S6K (Thr-389) and HA antibodies. Signal intensities (phospho-S6K and HA-S6K) from each immunoblot were quantified, and the relative ratio (phospho-S6K/HA-S6K) was shown. The data were expressed as mean ± S.E. (*, p < 0.05, n = 3). C, PAO-induced S6K phosphorylation was not blocked by inactive Rag small GTPases (RagA(TN)·RagC(QL)). HEK293T cells were transfected as in the same manner as B. After 1 h HBSS treatment, the cells were treated with PAO in the presence or absence of BAL for 15 min. N.S., not significant.
FIGURE 7.
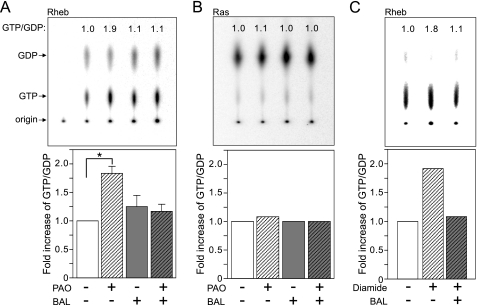
PAO activates Rheb. A and B, PAO specifically induces GTP-Rheb in vivo. Myc-Rheb (A) or HA-Ras (B) was transfected into HEK293T cells and labeled with [32P]orthophosphate. Cells were treated with PAO and/or BAL for 15 min as indicated. Radiolabeled Myc-Rheb or HA-Ras was immunoprecipitated with Myc or HA antibody, and bound nucleotides were eluted and separated on the polyethyleneimine cellulose plates. The ratio of GTP/GDP was calculated by the formula GTP counts/3 divided by GDP counts/2 and shown at top of each lane. In panel A, data were expressed as -fold increase of Rheb GTP/GDP ratio from three independent experiments (*, p < 0.05 versus other groups, mean ± S.E., n = 3). C, diamide also induces Rheb activity. The effect of diamide on GTP/GDP ratio of Rheb was examined in the same manner as in panel A.
RESULTS
mTORC1, but Not mTORC2, Is Activated by Cysteine Oxidant
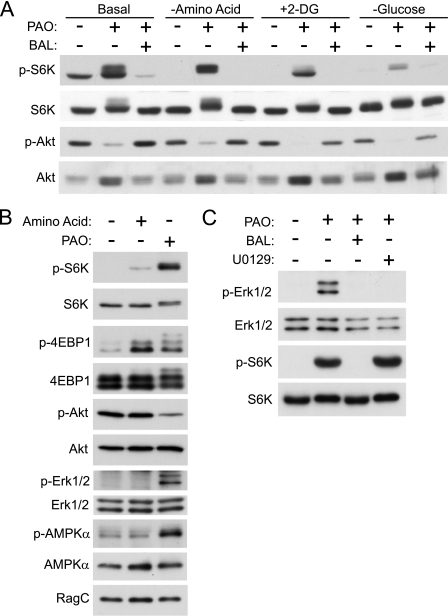
PAO is a cell-permeable oxidizing reagent that cross-links vicinal thiol groups and has been widely used as a cysteine oxidant (38–40). In contrast, BAL is a reducing agent that efficiently reverses oxidation reactions induced by PAO (41, 42). As reported previously, PAO treatment induced the phosphorylation of endogenous S6K (Thr-389), a substrate of mTORC1, in HEK293T cells (Fig. 1A) (34). BAL completely blocked the stimulatory effect of PAO on S6K phosphorylation, confirming that it is the oxidizing activity of PAO that is responsible for S6K phosphorylation. Interestingly, PAO decreased the phosphorylation of the AKT hydrophobic site (the site catalyzed by mTORC2), suggesting that PAO specifically activates mTORC1, but not mTORC2, and that cysteine oxidants may not directly enhance mTOR kinase activity. Furthermore, PAO induces the phosphorylation of S6K, the site catalyzed by mTORC1, in a manner independent of AKT activation. Amino acids are known to be required for insulin-mediated stimulation of TORC1 (43), but amino acids are not required for PAO to phosphorylate S6K (Fig. 1A). Pretreatment with 2-deoxyglucose, a compound that causes energy starvation by blocking glucose utilization, however, diminished PAO-induced S6K phosphorylation. Furthermore, in the absence of glucose, S6K activation by PAO was consistently compromised (Fig. 1A). These data suggest that glucose starvation may inhibit a step in the mTORC1 pathway downstream or parallel to PAO signaling. To validate the fact that PAO stimulates S6K phosphorylation via mTORC1 activation, we examined the effect of PAO on the phosphorylation of 4EBP1, another important TORC1 substrate under amino acid starvation conditions (44, 45). Consistent with the effect of PAO on S6K phosphorylation under amino acid starvation conditions, PAO treatment also induced 4EBP1 phosphorylation (Fig. 1B). To investigate the mechanism by which mTORC1 activity is enhanced by PAO, we also examined the activities of major upstream kinases such as AKT, ERK1/2, and AMPK under the same conditions. Consistent with the result shown in Fig. 1A, the phosphorylation of AKT, a major upstream positive regulator of mTORC1, is suppressed by PAO treatment under amino acid starvation conditions (Fig. 1B). We also found that the phosphorylation of both ERK and AMPK was enhanced by PAO treatment (Fig. 1B). Previous studies have determined that AMPK is a negative regulator for the mTORC1 pathway. Thus, the result suggests that PAO also acts downstream of AMPK to overcome its inhibitory effect on the mTORC1 activation. It has been reported that ERK stimulates mTORC1 activity by inhibiting the TSC2 tumor suppressor. To determine whether PAO-induced mTORC1 activation requires ERK1/2 activities, we examined the effect of the MEK inhibitor (U0126) on PAO-induced S6K phosphorylation. Interestingly, the MEK inhibitor had no effect on PAO-induced S6K phosphorylation, although it completely abolished PAO-induced Thr/Tyr phosphorylation on ERK1/2, the sites required for ERK1/2 activity (Fig. 1C). These results indicate that PAO does not require ERK1/2 activity to induce mTORC1 activation. Together, these data suggest that PAO does not require these regulatory kinases to stimulate mTORC1 activity.
FIGURE 1.
Redox specifically regulates mTORC1 but not mTORC2. A, PAO stimulates S6K phosphorylation, which is abolished by BAL treatment. HEK293T cells were pretreated with HBSS, 2-DG, or glucose-free DMEM for 1 h and then stimulated with PAO or BAL for 15 min before harvest. Lysates were analyzed by immunoblotting with phospho-S6K (p-S6K) (Thr-389), S6K, phospho-Akt (p-Akt) (Ser-473), and Akt antibodies. B, PAO treatment enhances both S6K1 and 4EBP1 phosphorylation under amino acid starvation conditions. HEK293T cells were treated with HBSS for 1 h and then stimulated with amino acid or PAO for 15 min. Cell lysates were prepared and analyzed by immunoblotting with the indicated antibodies. p-4EBP1, phospho-4EBP1; p-Erk1/2, phospho-Erk1/2; p-AMPKα, phospho-AMPKα. C, MEK inhibitor (U0126) has little effect on PAO-induced S6K phosphorylation. HEK293T cells were treated with HBSS for 1 h and then stimulated with PAO for 15 min in the absence or presence of BAL or U0126. 10 μm U0126 was added 30 min before PAO treatment.
PAO Stimulates mTORC1 in a Manner Independent of Rag Small GTPases
Cysteine oxidants are able to stimulate mTORC1 under amino acid starvation conditions. This raises the possibility that increased redox potential may activate Rag small GTPases because Rags are specific modulators for mTORC1 localization and because active Rags are able to induce mTORC1 activity under conditions of amino acid starvation (7, 8). To test this hypothesis, we examined the effect of a dominant negative form of the Rag complex (RagA(TN)·RagC(QL)) on PAO-induced S6K phosphorylation. Because transfection efficiency in HEK293T cells is ∼50% in our system, HA-tagged S6K1 (HA-S6K) was co-transfected with RagA(TN)·RagC(QL), and its phosphorylation was measured. For this aim, we first tested the effects of PAO and BAL on transfected HA-S6K. The results showed that transfected HA-S6K behaved similarly to the endogenous protein (Fig. 2A). PAO treatment enhanced the phosphorylation of HA-S6K, whereas BAL abolished PAO-induced S6K phosphorylation. If PAO enhances S6K phosphorylation by inhibiting a phosphatase, then rapamycin would not cause a dramatic decrease of S6K Thr-389 phosphorylation. In fact, as expected, rapamycin completely blocked S6K Thr-389 phosphorylation stimulated by PAO, indicating that PAO stimulates S6K phosphorylation by acting upstream of mTORC1. Consistent with previous observations, overexpression of dominant negative Rag components significantly reduced amino acid-induced S6K phosphorylation (Fig. 2B) (7, 8). However, similar levels of RagA(TN)·RagC(QL) expression failed to reduce PAO-induced S6K phosphorylation, suggesting that PAO may induce mTORC1 activation in a manner independent of Rag GTPases (Fig. 2C).
PAO-induced mTORC1 Activation Is Independent of Rag-Ragulator System
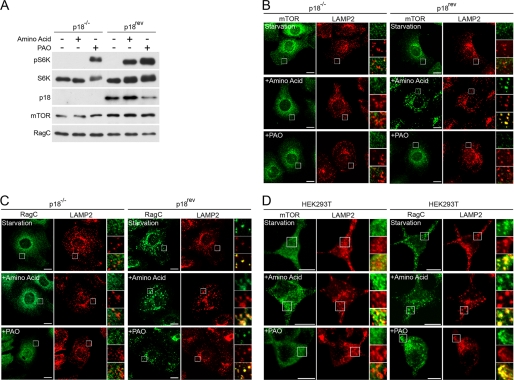
Translocation of mTORC1 to lysosomes is a critical step for amino acid-induced mTORC1 activation, where Ragulator (MP1·p14·p18 complex) plays a crucial role in anchoring Rag small GTPases on the lysosomal membrane (11, 12, 46). Sancak et al. (12) showed that amino acid stimulation failed to induce S6K phosphorylation and mTOR translocation to the lysosome in Ragulator-deficient cells. To further investigate the role of Rag in PAO-induced mTORC1 activation, we tested whether PAO stimulates S6K phosphorylation in Ragulator-deficient cells. p18−/− cells and p18−/− cells reconstituted with wild-type p18 (p18rev) cells were used as Ragulator-deficient and control cells, respectively (12). Consistent with previous observations, amino acid stimulation failed to induce S6K phosphorylation in p18−/− cells. However, PAO was able to stimulate S6K phosphorylation in p18−/− cells, further confirming a dispensable role of the Rag-Ragulator in PAO-induced mTORC1 activation (Fig. 3A). We also examined the effect of PAO and amino acid stimulation on mTOR and RagC localization in p18−/− and p18rev cells. As reported previously, mTOR was diffusely expressed around the nucleus without co-localizing with LAMP2 (late endosome or lysosome marker) in both p18−/− and p18rev cells under amino acid starvation conditions (Fig. 3B, upper panels) (12). Upon amino acid stimulation, mTOR perfectly co-localized with LAMP2 in p18rev cells but not in p18−/− cells (Fig. 3B, middle panels). However, we found that PAO failed to stimulate mTOR-LAMP2 co-localization in p18rev cells (Fig. 3B, bottom panels), indicating that PAO-induced mTORC1 activation is not associated with mTORC1 localization at the lysosome. RagC was always co-localized with LAMP2 in p18rev cells but was expressed diffusely throughout the cytoplasm in p18−/− cells regardless of the presence or absence of amino acids or PAO stimulation (Fig. 3C). Consistently, similar observations were seen in HEK293T cells (Fig. 3D).
FIGURE 3.
PAO induces S6K phosphorylation in a manner independent of Rag-Ragulator function. A, PAO induces S6K phosphorylation in p18−/− cells. p18−/− cells and p18rev cells were used as Ragulator-deficient and control cells, respectively. The cells were treated with HBSS for 1 h and then stimulated with amino acid or PAO for 15 min. The cell lysates were analyzed by immunoblotting with the indicated antibodies. B, amino acids but not PAO stimulate co-localization of mTOR and LAMP2 in p18rev cells. p18−/− cells and p18rev cells were treated in the same manner as A. mTOR and LAMP2 co-localization was examined by immunocytochemistry. Scale bars represent 10 μm. C, co-localization of RagC and LAMP2 were examined as in the same manner as B. D, amino acids but not PAO stimulate co-localization of mTOR and LAMP2 in HEK293T cells. Similar experiments were performed in the same manner as B in HEK293T cells.
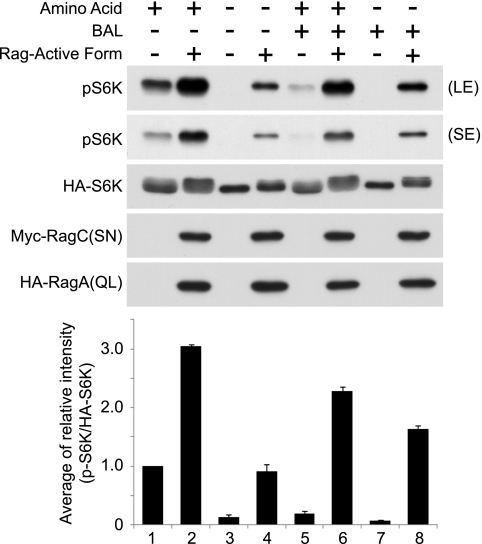
To further examine the relationship between redox potential and Rag-induced mTORC1 activation, we tested the effect of BAL treatment on S6K phosphorylation induced by the active Rag complex (RagA(QL)·RagC(SN)). Consistent with results shown in Figs. 1A and 2A, the levels of S6K phosphorylation were significantly reduced by BAL treatment under growth conditions (Fig. 4, lane 1 versus lane 5). However, BAL-induced reduction of S6K phosphorylation was attenuated when the active Rag complex was expressed (Fig. 4, lane 2 versus lane 6 and lane 4 versus lane 8). Together, these results support our hypothesis that cysteine oxidants induce mTORC1 activation in a manner independent of Rag-Ragulator function.
FIGURE 4.
Active Rag small GTPase-induced S6K phosphorylation is not inhibited by BAL treatment. HA-S6K with or without HA-RagA(QL) and Myc-RagC(SN) were transfected into HEK293T cells. Cells were incubated with or without amino acids for 1 h and treated with or without BAL for another 15 min. Cell lysates were analyzed by immunoblotting with phospho-S6K (pS6K) (Thr-389), HA, and Myc antibodies. Signal intensities (phospho-S6K and HA-S6K) from each immunoblot were quantified, and the relative ratio (phospho-S6K/HA-S6K) was determined. The data were expressed as mean ± S.E. (n = 3). (LE) and (SE) indicate long and short exposure, respectively.
TSC1 and TSC2 Are Required for Redox Regulation of mTORC1
A question that arises from the above observations is how mTORC1 activity is up-regulated by cysteine oxidants under conditions of amino acid depletion in a Rag GTPase-independent manner. To address this question, we investigated the role of TSC-Rheb signaling in redox-dependent mTORC1 regulation. We first monitored the effects of PAO and BAL on S6K phosphorylation under conditions of Rheb or TSC1·TSC2 overexpression (Fig. 5A). As expected, Rheb overexpression caused a strong enhancement of S6K phosphorylation. Interestingly, we found that Rheb-induced S6K phosphorylation was insensitive to both PAO and BAL treatment (Fig. 5A). Moreover, PAO-induced S6K phosphorylation was diminished by overexpression of TSC1·TSC2. These data suggest that PAO and BAL may act upstream of TSC1·TSC2 and Rheb.
FIGURE 5.
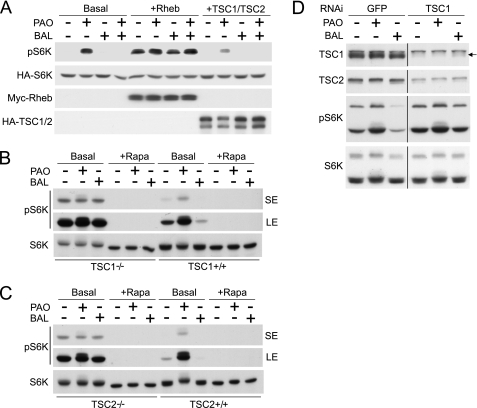
TSC1 and TSC2 are essential for redox-sensitive mTORC1 regulation. A, redox-sensitive S6K phosphorylation is disordered by the overexpression of Rheb or TSC1·TSC2. HA-S6K was transfected into HEK293T cells with or without Rheb (Myc-Rheb) or TSC1·TSC2 (HA-TSC1 and HA-TSC2). Cells were treated with or without PAO or BAL for 15 min. Cell lysates were analyzed by immunoblotting with phospho-S6K (pS6K) (Thr-389), HA, and Myc antibodies. B and C, loss of redox-sensitive mTORC1 regulation in TSC1−/− or TSC2−/− MEF cells. TSC1−/− and TSC1+/+ MEF cells (B) or TSC2−/− and TSC2+/+ MEF cells (C) were treated with PAO or BAL with or without rapamycin (+Rapa) for 15 min. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (LE) and (SE) indicate long and short exposure, respectively. D, transient knockdown of TSC1 reduces redox effect on S6K phosphorylation. HeLa cells with siRNA against human TSC1 or GFP1 (as control) were treated with or without PAO or BAL. Cells lysates were analyzed by immunoblotting with indicated antibodies. Arrow indicates TSC1.
To further test the function of the TSC complex in redox-induced mTORC1 regulation, we examined the effects of PAO and BAL in TSC1−/− and TSC1+/+ MEF cells. Consistent with previous observations, TSC1−/− cells showed a higher level of S6K phosphorylation than TSC1+/+ cells. As seen in HEK293T cells, PAO strongly enhanced S6K phosphorylation, whereas BAL abolished it in TSC1+/+ cells (Fig. 5B). However, we found that enhanced S6K phosphorylation in TSC1−/− cells was resistant to both PAO and BAL treatments but completely inhibited by rapamycin treatment (Fig. 5B). We also performed similar experiments in TSC2−/− cells. Results identical to those in TSC1−/− cells were also observed in TSC2−/− cells (Fig. 5C). PAO did not further increase S6K phosphorylation in the TSC2−/− cells, whereas it enhanced S6K phosphorylation in the control TSC2+/+ cells. Consistently, BAL did not inhibit S6K phosphorylation in the TSC2−/− cells, whereas it inhibited S6K phosphorylation in the TSC2+/+ cells, further supporting a role of TSC1·TSC2 in the redox regulation of mTORC1.
The functional importance of the TSC complex in redox regulation was further examined in cells with transient TSC1 knockdown (Fig. 5D). Expression of TSC1 was efficiently down-regulated by siRNA targeting TSC1 in HeLa cells and confirmed by anti-TSC1 immunoblotting. As expected, TSC1 knockdown simultaneously decreased TSC2 protein levels because TSC2 is stabilized in a complex with TSC1. In control siRNA-treated cells, PAO and BAL treatments resulted in an increase and a decrease of S6K phosphorylation, respectively. Importantly, knockdown of TSC1 in HeLa cells significantly diminished the effect of PAO or BAL on S6K phosphorylation. Together, these data suggest a model in which redox acts upstream of TSC1·TSC2 to regulate the mTORC1 pathway.
Effects of Redox on the Integrity of mTOR Complexes
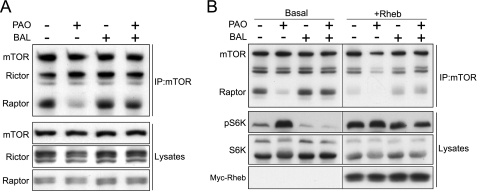
It has been reported that nutrients, such as amino acids, and cysteine oxidants affect the association between mTOR and Raptor under specific lysis conditions (9, 34, 47). We also observed that PAO treatment reduced the affinity between mTOR and Raptor and that this effect was attenuated by BAL (Fig. 6A). In contrast, PAO and/or BAL had no effect on the interaction between mTOR and Rictor, supporting a specific role of redox regulation in mTORC1. Subsequently, we examined the effect of Rheb on the interaction between mTOR and Raptor. Overexpression of Rheb also reduced the interaction between endogenous mTOR and Raptor with a concomitant increase in S6K phosphorylation (Fig. 6B). PAO treatment further diminished the interaction between mTOR and Raptor. These results suggest that PAO may activate Rheb, thereby decreasing the affinity between mTOR and Raptor.
FIGURE 6.
Effect of redox and Rheb on the integrity of mTOR complexes. A, redox modulates the Raptor-mTOR interaction. HEK293T cells were treated with or without PAO and/or BAL for 15 min. Cell lysates were prepared, and immunoprecipitation (IP) with mTOR antibody was performed. Lysates and immunoprecipitates were analyzed by immunoblotting with mTOR, Rictor, and Raptor antibodies. B, overexpression of Rheb also reduced the interaction between endogenous mTOR and Raptor. HEK293T cells transfected with or without Myc-Rheb were treated with PAO and/or BAL for 15 min as indicated. Cell lysates were analyzed in the same manner as in panel A. pS6K, phospho-S6K.
PAO Activates Rheb
The activity of Rheb in vivo can be assessed by measuring the ratio of GTP- to GDP-bound forms (17, 48). To test our assumption that the cysteine oxidant modulates the TSC-Rheb pathway, we examined the effects of PAO and/or BAL on the Rheb GTP/GDP ratio. We found that PAO treatment significantly increased Rheb in the GTP-bound state by ∼2-fold (Fig. 7A). BAL treatment alone did not decrease the overexpressed Rheb GTP level but did effectively attenuate the stimulating effect of PAO. This is consistent with the observation that BAL treatment fails to inhibit enhanced S6K phosphorylation under the condition of Rheb overexpression (Fig. 5A). The effect of PAO on guanine nucleotide loading was specific to Rheb as PAO did not affect the Ras GTP/GDP ratio (Fig. 7B). We also examined the effect of diamide, another cysteine oxidant. Diamide similarly enhanced the level of GTP-bound Rheb, and this effect was blocked by BAL (Fig. 7C). Therefore, our data indicate that PAO treatment specifically induces Rheb activity.
DISCUSSION
mTORC1 plays an essential role in a wide array of cellular processes such as translation and autophagy. The mechanism by which mTORC1 activity is regulated in response to various growth-promoting and inhibitory signals is an area of intense study. Multiple components such as kinases and small GTPases are implicated in temporal and spatial regulation of mTORC1 activity. Recent studies have highlighted that two small GTPases, Rheb and Rag, play crucial roles in growth factor- and amino acid-induced mTORC1 activation (49). Upon amino acid stimulation, Rag, anchored by Ragulators on the lysosome membrane, can be activated by an unknown mechanism and subsequently recruit mTORC1. On the lysosomal membrane, mTORC1 is activated by Rheb, whose activity is enhanced by growth factor signaling. This model nicely explains how amino acid and growth factor signals produce maximal activation of mTORC1 in a coordinate manner at the lysosome by active Rag and Rheb small GTPases. Because cysteine oxidants activate mTORC1 under conditions of amino acid depletion, we originally hypothesized that cysteine oxidants might activate Rag small GTPases because constitutively active Rag mutants are able to activate mTORC1 even in amino acid-depleted conditions (7, 8). Our study, however, has revealed that cysteine oxidants activate mTORC1 through Rheb activation. It is likely that the TSC complex itself may be a major target of cysteine oxidation to activate mTORC1 for the following reasons. Firstly, cysteine oxidants significantly increase the GTP/GDP ratio of Rheb. Secondly, the effects of both oxidant and reducing agents on mTORC1 activity are largely compromised in the absence of a functional TSC complex. Finally, the activity of major upstream regulators of the TSC complex does not account for cysteine oxidant-induced mTORC1 activation.
Blocking the glycolytic pathway with non-metabolized glucose (2-DG) treatment decreases cellular energy, thereby activating AMPK in HEK293 cells. AMPK phosphorylates multiple proteins in the mTORC1 signaling such as TSC2 and suppresses mTORC1 activity (29, 50). Besides AMPK activation, prolonged 2-DG treatment also activates p38-regulated/activated kinase that directly phosphorylates and inhibits Rheb activity by reducing its guanidine nucleotide interaction (30). Consistently, our data show that preincubation of 2-DG for 1 h significantly attenuated PAO-induced mTORC1 activation. Interestingly, Sarbassov and Sabatini (34) demonstrated that preincubation of PAO prevented 2-DG (15-min treatment)-induced mTORC1 inactivation. This result also supports our model that cysteine oxidants may inhibit the TSC complex because short term 2-DG treatment may not be able to block mTORC1 activation when the TSC complex is already under inhibition by PAO.
Given that PAO is known to react with the sulfhydryl groups of 2 adjacent cysteine residues, it might affect the structure of the TSC complex, resulting in the down-regulation of its GAP activity. The CXXC motif is utilized by many redox proteins for the formation, isomerization, and reduction of disulfide bonds (31). The C- or N-terminal cysteine in this motif can be replaced by serine or threonine (CXX(S/T) or (S/T)XXC), thereby modifying the functional repertoire of redox proteins (33). Indeed, mammalian TSC1 and TSC2 have 12 and 31 cysteine residues, respectively. In TSC1, 2 of these cysteines form a CXXS and TXXC motif. In TSC2, 9 cysteines form all CXXC, CXX(S/T), and (S/T)XXC motifs. It would be intriguing to test whether these motifs are indeed targets of cysteine oxidants and whether they have functional importance as regulatory residues for TSC2 GAP activity.
Although our study shows an important role of the TSC-Rheb pathway in the redox-sensitive mTORC1 regulation, the molecular mechanisms by which active endogenous Rheb induced by cysteine oxidants stimulates mTORC1 activity under amino acid-depleted conditions remain unclear. Two possible mechanisms can be proposed from our observations. Firstly, endogenous Rheb activated by the cysteine oxidant is also able to stimulate mTORC1 in places other than the lysosome. In this model, mTORC1 may move to the lysosome along with endogenous Rheb. Supporting this model, ectopic expression of Rheb is able to activate mTORC1 under conditions of amino acid starvation (17, 51, 52). Furthermore, mTORC1 interacts with not only active Rheb but also with inactive Rheb (52, 53).
Secondly, it is possible that a small portion of mTORC1 may still remain on the lysosomal membrane even under amino acid starvation conditions and that the remaining mTORC1 can be maximally stimulated by a cysteine oxidant through Rheb activation at the lysosome. If this model is correct, reduction of endogenous Rheb activity on the lysosome should inhibit active Rag-induced mTORC1 activation under amino acid starvation conditions. However, our data show that BAL has little effect on mTORC1 activation induced by active Rag under amino acid starvation conditions, indicating that redox-sensitive mTORC1 regulation does not occur on the lysosomal membrane. Although our study supports the former model in which redox-dependent mTORC1 regulation occurs at sites other than the lysosome, our data raise the possibility that the TSC complex is unable to suppress active Rheb once mTORC1 engages the lysosomal membrane.
Nutrient availability and oxygen pressure modulate the generation of both ATP and reactive oxygen species via oxidative phosphorylation in mitochondria as well as the glycolysis pathway (54). Although it has been documented that the levels of intracellular ATP or AMP are critical for the regulation of mTORC1 activity (29), the reactive oxygen species-mediated redox potential can also be a potential regulator of mTORC1. Especially in physiological and pathophysiological settings, it has been postulated that excess production of free radicals/reactive oxygen species may contribute to the progression of aging (from lower organisms to mammals) as well as the development of several cancers (55, 56). Given that rapamycin treatment extends life span and prevents cancer development, redox potential-mediated mTORC1 activation might play a role in the process of aging and cancer development.
In summary, we have demonstrated the role of redox potential in the regulation of the TSC-Rheb-mTORC1 pathway. Our data show that cysteine oxidants are able to activate mTORC1 via Rheb but not Rag activation in cells deprived of external amino acids. Furthermore, our study suggests that cysteine oxidant-induced GDP-GTP conversion of Rheb via TSC complex inactivation and concomitant activation of mTORC1 may occur at sites other than the lysosome. We note that the effects of cysteine oxidants and reducing agents used in this study may be beyond the physiological extent of cellular redox regulation in the mTOR signaling pathway. However, an understanding of such specificities toward Rheb and mTORC1 within mTOR signaling is a valuable tool to potently and transiently inhibit the function of the endogenous TSC complex in the context of temporal as well as spatial regulation of the mTORC1 pathway.
Acknowledgments
We appreciate the gift of TSC1 and TSC2 MEFs from Dr. David Kwiatkowski (Harvard University). We thank Tianqing Zhu for technical assistance. We also thank Dr. John Kim for a critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK083491 (to K. I.).
- mTOR
- mammalian target of rapamycin
- TSC
- tuberous sclerosis complex
- MEF
- mouse embryonic fibroblast
- GAP
- GTPase-activating protein
- AMPK
- AMP-activated protein kinase
- PAO
- phenylarsine oxide
- 2-DG
- 2-deoxyglucose
- BAL
- British anti-Lewisite
- HBSS
- Hanks' balanced salt solution
- S6K
- S6 protein kinase.
REFERENCES
- 1. Abraham R. T. (2004) DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 2. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 3. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 4. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 5. Hay N., Sonenberg N. (2004) Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 6. Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 7. Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 10. Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nada S., Hondo A., Kasai A., Koike M., Saito K., Uchiyama Y., Okada M. (2009) EMBO J. 28, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teis D., Taub N., Kurzbauer R., Hilber D., de Araujo M. E., Erlacher M., Offterdinger M., Villunger A., Geley S., Bohn G., Klein C., Hess M. W., Huber L. A. (2006) J. Cell Biol. 175, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sekiguchi T., Hirose E., Nakashima N., Ii M., Nishimoto T. (2001) J. Biol. Chem. 276, 7246–7257 [DOI] [PubMed] [Google Scholar]
- 15. Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. (2003) Mol. Cell 11, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 16. Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A. (2003) J. Biol. Chem. 278, 32493–32496 [DOI] [PubMed] [Google Scholar]
- 17. Inoki K., Li Y., Xu T., Guan K. L. (2003) Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saucedo L. J., Gao X., Chiarelli D. A., Li L., Pan D., Edgar B. A. (2003) Nat. Cell Biol. 5, 566–571 [DOI] [PubMed] [Google Scholar]
- 19. Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. (2003) Nat. Cell Biol. 5, 578–581 [DOI] [PubMed] [Google Scholar]
- 21. van Slegtenhorst M., Nellist M., Nagelkerken B., Cheadle J., Snell R., van den Ouweland A., Reuser A., Sampson J., Halley D., van der Sluijs P. (1998) Hum. Mol. Genet. 7, 1053–1057 [DOI] [PubMed] [Google Scholar]
- 22. Crino P. B., Nathanson K. L., Henske E. P. (2006) N. Engl. J. Med. 355, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 23. Kwiatkowski D. J. (2003) Ann. Hum. Genet. 67, 87–96 [DOI] [PubMed] [Google Scholar]
- 24. Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 25. Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 26. Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 27. Cheng S. W., Fryer L. G., Carling D., Shepherd P. R. (2004) J. Biol. Chem. 279, 15719–15722 [DOI] [PubMed] [Google Scholar]
- 28. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 30. Zheng M., Wang Y. H., Wu X. N., Wu S. Q., Lu B. J., Dong M. Q., Zhang H., Sun P., Lin S. C., Guan K. L., Han J. (2011) Nat. Cell Biol. 13, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchanan B. B., Balmer Y. (2005) Annu. Rev. Plant Biol. 56, 187–220 [DOI] [PubMed] [Google Scholar]
- 32. Poole L. B., Nelson K. J. (2008) Curr. Opin. Chem. Biol. 12, 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fomenko D. E., Gladyshev V. N. (2003) Biochemistry 42, 11214–11225 [DOI] [PubMed] [Google Scholar]
- 34. Sarbassov D. D., Sabatini D. M. (2005) J. Biol. Chem. 280, 39505–39509 [DOI] [PubMed] [Google Scholar]
- 35. Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S. (2005) J. Biol. Chem. 280, 20558–20564 [DOI] [PubMed] [Google Scholar]
- 36. Takahashi T., Hara K., Inoue H., Kawa Y., Tokunaga C., Hidayat S., Yoshino K., Kuroda Y., Yonezawa K. (2000) Genes Cells 5, 765–775 [DOI] [PubMed] [Google Scholar]
- 37. Li Y., Inoki K., Guan K. L. (2004) Mol. Cell. Biol. 24, 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoffman R. D., Lane M. D. (1992) J. Biol. Chem. 267, 14005–14011 [PubMed] [Google Scholar]
- 39. Kalef E., Gitler C. (1994) Methods Enzymol. 233, 395–403 [DOI] [PubMed] [Google Scholar]
- 40. Kosower E. M., Kosower N. S. (1995) Methods Enzymol. 251, 133–148 [DOI] [PubMed] [Google Scholar]
- 41. Bogumil R., Ullrich V. (2002) Methods Enzymol. 348, 271–280 [DOI] [PubMed] [Google Scholar]
- 42. Frost S. C., Schwalbe M. S. (1990) Biochem. J. 269, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., Avruch J. (1998) J. Biol. Chem. 273, 14484–14494 [DOI] [PubMed] [Google Scholar]
- 44. Beretta L., Gingras A. C., Svitkin Y. V., Hall M. N., Sonenberg N. (1996) EMBO J. 15, 658–664 [PMC free article] [PubMed] [Google Scholar]
- 45. von Manteuffel S. R., Gingras A. C., Ming X. F., Sonenberg N., Thomas G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Flinn R. J., Yan Y., Goswami S., Parker P. J., Backer J. M. (2010) Mol. Biol. Cell 21, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 48. Urano J., Ellis C., Clark G. J., Tamanoi F. (2001) Methods Enzymol. 333, 217–231 [DOI] [PubMed] [Google Scholar]
- 49. Zoncu R., Efeyan A., Sabatini D. M. (2011) Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarkar S., Korolchuk V. I., Renna M., Imarisio S., Fleming A., Williams A., Garcia-Arencibia M., Rose C., Luo S., Underwood B. R., Kroemer G., O'Kane C. J., Rubinsztein D. C. (2011) Mol Cell 43, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., Gulati P., Byfield M. P., Backer J. M., Natt F., Bos J. L., Zwartkruis F. J., Thomas G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. (2005) J. Biol. Chem. 280, 18717–18727 [DOI] [PubMed] [Google Scholar]
- 53. Long X., Ortiz-Vega S., Lin Y., Avruch J. (2005) J. Biol. Chem. 280, 23433–23436 [DOI] [PubMed] [Google Scholar]
- 54. Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., Chandel N. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dröge W. (2002) Physiol. Rev. 82, 47–95 [DOI] [PubMed] [Google Scholar]
- 56. Hipkiss A. R. (2006) Ann. N.Y. Acad. Sci. 1067, 361–368 [DOI] [PubMed] [Google Scholar]