Abstract
Amyotrophic Lateral Sclerosis (ALS) is a fatal motor neuron (MN) disease with astrocytes implicated as a significant contributor to MN death in familial ALS (fALS)1–5. However, these conclusions, in part, derive from rodent models of fALS based upon dominant mutations within the superoxide dismutase 1 (SOD1) gene which account for less than 2% of all ALS cases2, 4, 5. Here, we generated astrocytes from post-mortem tissue from both fALS and sporadic ALS (sALS) patients, and show that astrocytes derived from both patient groups are similarly toxic to MNs. In addition, we show that SOD1 is a viable target for sALS, as its knockdown significantly attenuates astrocyte-mediated toxicity towards MNs. Our data highlight astrocytes as a non-cell autonomous component in sALS and provide the first in vitro model system to investigate common disease mechanisms and evaluate potential therapies for sALS and fALS.
Amyotrophic lateral sclerosis (ALS), commonly referred to as Lou Gehrig’s disease, is a fatal neurodegenerative disease characterized by loss of motor neurons (MNs) in the motor cortex, brain stem, and spinal cord, resulting in muscle paralysis and ultimately death due to respiratory failure. Approximately 90% of all cases are classified as sporadic ALS (sALS), defined as having no family history of the disease6. The remaining cases are inherited in a dominant fashion termed familial (fALS) of which 20% are associated with mutations within the superoxide dismutase 1 (SOD1) gene6. Numerous hypotheses have been proposed to account for MN loss, however the precise molecular mechanisms leading to ALS remain unknown7. Studies performed in fALS mouse models have implicated non-neuronal cells such as microglia and astrocytes in the progression phase of fALS2, 8–11. In particular, in vitro co-culture systems have shown that MNs perish in the presence of astrocytes harboring SOD1 mutations1, 3–5. However, all of these in vitro and in vivo studies have been conducted using models that highly overexpress mutant SOD1, which may not fully mimic the actual disease. Despite significant knowledge gained from these studies, no efficacious drugs or therapies have been realized in human clinical trials to date. Indeed, models of ALS have come under scrutiny, given that drugs and therapies are tested in models of fALS, only accounting for a small fraction of ALS patients. Thus, efforts have been focused on developing in vitro cell based models for sALS, representing the majority of the patient population. Currently, there is a major impetus to develop in vitro cell-based models for ALS, utilizing induced pluripotent stem cell (iPSC) technology12, 13, although no reports have been published to date on sALS models. The generation of sALS disease models will be important to decipher whether astrocytes influence disease progression in sALS. There is precedent that glia are involved in the pathogenesis of sALS, since fALS and sALS patients show strikingly similar spinal cord pathologies with pronounced glial activation and upregulation of pro-inflammatory mediators and cytokines14. Indeed, in our analysis of ALS patient spinal cord samples, we observed increased glial cell numbers, which is indicative of astrocyte activation in the presence of atrophied MNs in both fALS and sALS patients (Supplementary Fig. 1,2). What remains to be determined is whether sALS astrocytes play an active role in the death of MNs, and whether this non-cell autonomous effect can be recapitulated in vitro. Here, we show that sALS and fALS astrocytes, derived from post-mortem spinal cord neural progenitor cells (NPCs), share a common non-cell autonomous toxicity, selectively killing MNs in a co-culture model system. Upon co-culture with MNs, we observed that inflammatory gene expression is upregulated within fALS and sALS astrocytes. In addition, we demonstrate that suppression of wild-type SOD1 in sALS astrocytes is neuroprotective in our model system, further implicating a role of wild-type SOD1 in sALS.
In order to test whether sALS astrocytes are similarly neurotoxic as fALS astrocytes, we derived astrocytes from adult NPCs isolated from post-mortem lumbar spinal cord tissue from fALS and sALS patients. We hypothesized that isolation of NPCs from these patients would provide a renewable source of multipotent cells that could be differentiated into astrocytes that were then used for co-culture studies with MNs, as previously described15. Sample collection was coordinated by The National Disease Research Interchange (NDRI) and all tissues were received within a time window of 48 to 72 hours post-mortem for immediate processing in our laboratory. Samples were non-identifiable with the exception of critical information regarding clinical diagnosis, gender, age at death, and length of time from diagnosis to death (Supplementary Table 1). To purify NPCs from these spinal cords, we utilized an enzymatic digestion protocol to dissociate the cells, followed by percoll gradient NPC enrichment, as previously described (Supplementary Fig. 3a)16, 17. Within 3–4 weeks, NPC cultures were established, and marker analysis showed robust levels of the NPC marker; NESTIN (Supplementary Fig. 3b,c). Furthermore, the NPCs proved to be tri-potent, capable of differentiating into neurons, oligodendrocytes, and astrocytes (Supplementary Fig. 3d–f). NPC cultures were successfully established from one fALS patient harboring a SOD1 A4V mutation (Supplementary Fig. 4), 7 sALS patients, and one non-ALS control. Further DNA sequencing verified that our cohort of sALS patients did not harbor mutations in common ALS gene loci (Supplementary Table 2). To increase the rigor of our analyses, we utilized two additional non-ALS controls: SCP-27; a fetal human NPC line, and 1800; a fetal human primary astrocyte cell line (Supplementary Table 1).
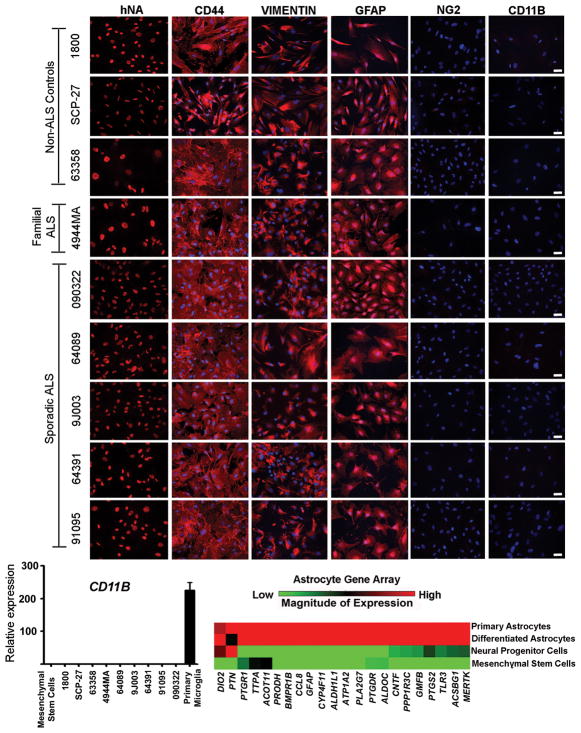
After initial expansion of all NPC lines, cells were differentiated into astrocytes by medium supplementation with 10% fetal bovine serum. Since NPCs can give rise to multiple cell types in these differentiation conditions, we performed an extensive marker analysis of our established astrocyte cell lines to evaluate the purity of our cultures. Indeed, we found high levels of expression of well-known astrocyte markers including VIMENTIN, GFAP, and the astrocyte precursor marker CD44 (Fig. 1a and Supplementary Table 3), without significant contamination of other cell types. We found no detectable oligodendroglial progenitors or microglia in our cultures as assessed by immunohistochemistry for NG2 and CD11B, respectively (Fig. 1a and Supplementary Table 3). The absence of microglia in the cultures was also confirmed by quantitative RT-PCR for CD11B and IBA1 (Fig. 1b and Supplementary Fig. 5). Additionally, we evaluated the maturity of astrocytes by measuring expression of a panel of genes known to be enriched in astrocytes in vivo18. NPC-derived astrocytes shared a similar gene profile to primary astrocytes isolated directly from the human spinal cord (Fig. 1c). All of these results suggest that highly enriched astrocyte lines can be derived from NPCs isolated from postmortem spinal cord tissues, which represent a renewable source of human fALS and sALS astrocytes for subsequent analysis.
Fig. 1.
NPCs can be differentiated into highly enriched astrocyte cultures which show a similar gene profile to spinal cord primary astrocytes. (A) Marker analysis of human NPC-derived astrocytes showing specific immunoreactivity against human nuclear antigen (hNA); high expression of the astrocyte markers CD44, Vimentin, and GFAP; absence of the glial progenitor marker NG2; and absence of the microglia marker CD11b. Scale bars, 50 μm. (B) Comparison of CD11b expression levels by quantitative RT-PCR in NPC-derived astrocyte cultures from control and ALS patients. Mesenchymal stem cells and human microglia were used as negative and positive controls, respectively. Error bars represent SEM. (C) Gene expression profile analysis of NPC-derived astrocytes. Comparisons were made to a human sample of mesenchymal stem cells, undifferentiated NPCs, and primary human astrocytes derived from spinal cord.
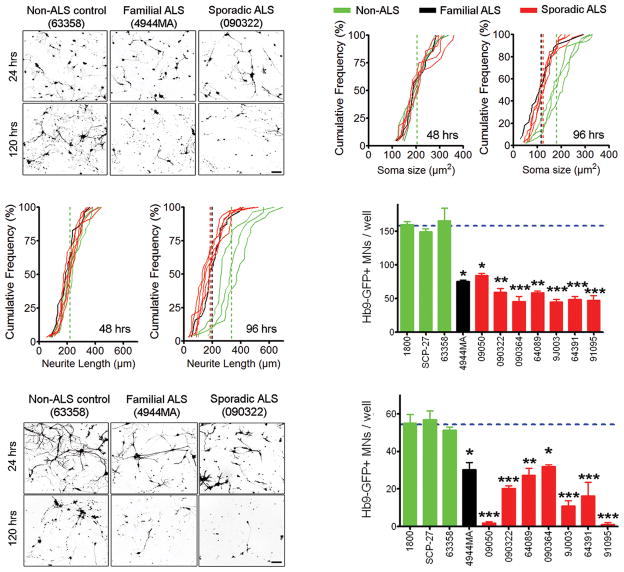
We next explored whether NPC-derived astrocytes from fALS and sALS patients were toxic to MNs by co-culturing mouse embryonic stem cell derived MNs with the differentiated astrocytes from each patient. We utilized an embryonic stem cell line containing an Hb9-eGFP reporter to allow for easy visualization of these MNs in co-culture with astrocytes19. For reproducibility of experiments, we enriched for Hb9-eGFP+ MNs by fluorescence activated cell sorting (Supplementary Fig. 6). Twenty-four hours following plating of the MNs onto a confluent layer of astrocytes, we found no significant difference in the number of MNs present between ALS and control astrocyte cultures (Fig. 2a), nor any significant difference in MN soma diameter or length of neuritic extensions up to 48 hours following co-culture initiation (Fig. 2b,c). However, after 96 hours in culture, MNs that were cultured on top of fALS astrocytes began to degenerate, evidenced by atrophy of the MN soma and shortened neurites (Fig. 2b, c). Indeed, quantification of MNs at 120 hours following co-culture showed that the number of MNs present in fALS astrocyte co-cultures was statistically reduced by 50% compared to non-ALS astrocyte control co-cultures (Fig. 2a, d). Similar results were observed when either SOD1 A4V or SOD1 G93A transgenes were overexpressed in non-ALS control astrocyte lines (Supplementary Fig. 7), supporting previous co-culture studies showing toxicity of astrocytes overexpressing mutant SOD1 1, 3–5, 15, 20.
Fig. 2.
Astrocytes derived from sALS and fALS patients cause MN death in co-culture. (A) Representative microscopic field showing the morphology of Hb9-GFP+ MNs in co-culture with human astrocytes at 24 and 120 hours. Scale bars, 100 μm. (B) Soma size and neurite length of Hb9-GFP+ MNs at 48 and 96 hours when co-cultured with astrocytes from fALS, sALS and non-ALS controls. (C) Counts of Hb9-GFP+ MNs per well after 120 hours of co-culture with astrocytes from fALS, sALS and non-ALS controls. *p<0.05, **p<0.01 and ***p<0.001 compared to 63358 (non-ALS control). Error bars represent SEM. (D) Representative microscopic field showing the morphology of Hb9-GFP+ MNs treated with astrocyte-conditioned media for 24 or 120 hrs. Scale bars, 100 μm. (E) Counts of Hb9-GFP+ MNs per well after 120 hours of treatment with astrocyte conditioned media. *p<0.05, **p<0.01 and ***p<0.001 compared to 63358 (non-ALS control). Error bars represent SEM.
Next, we shifted our focus to examine whether sALS astrocytes exhibit a similar non-cell autonomous toxicity towards MNs. Similar to our previous experiments utilizing fALS astrocytes, we noticed no significant changes in MNs co-cultured with sALS astrocytes for up to 48 hours. However, by 96 hours of co-culture, MNs co-cultured with all lines of sALS astrocytes began to degenerate (Fig. 2b). Quantification of MNs remaining after 120 hours of co-culture with sALS astrocytes showed a reduction of 45–70% compared to non ALS-controls (Fig. 2a, d). The MN damage elicited by the sALS patient-derived astrocytes was indistinguishable from fALS astrocytes, suggesting a shared mechanism for MN death in both types of co-cultures. To verify that the observed MN toxicity was caused by astrocyte-specific factors, we co-cultured MNs with ALS patient-derived fibroblasts and observed no significant decline in MN survival compared to MNs co-cultured with control fibroblasts (Supplementary Fig. 8a). To determine whether the fALS or sALS astrocytes affect survival of other neuronal cell types in vitro, we co-cultured GABAergic (gamma aminobutyric acid positive) neurons with ALS patient-derived astrocytes. Neither the fALS nor sALS astrocytes triggered death of GABAergic neurons, suggesting the ALS astrocyte-derived toxicity is specific toward MNs, thereby adding confidence to our assay (Supplementary Fig. 9a). Collectively, these results demonstrate that sALS astrocytes are also toxic towards MNs, and support the idea of a shared disease mechanism between fALS and sALS.
Previous work has shown that astrocytes isolated from the fALS SOD1 mouse model can cause MN death in vitro via secreted factors 1, 5. To determine whether our patient-derived fALS and sALS astrocytes also release neurotoxic factors, we treated MNs with astrocyte-conditioned media and measured MN survival. MNs cultured in conditioned media from sALS or fALS astrocytes died ~50% faster than MNs treated with conditioned media from non-ALS control astrocytes (Fig. 2e, f). Conditioned media collected from ALS patient-derived fibroblasts did not cause MN death, demonstrating the specificity of the astrocyte-derived toxicity (Supplementary Fig. 8b). Likewise, GABAergic neurons were unaffected by treatment with conditioned media prepared from fALS or sALS astrocytes indicating that MNs are selectively sensitive to factors present in the ALS astrocyte-conditioned media (Supplementary Fig. 9b). These results suggest that fALS and sALS astrocytes either secrete factors which are toxic to MNs, or alternatively do not provide proper MN supportive factors, resulting in cell death.
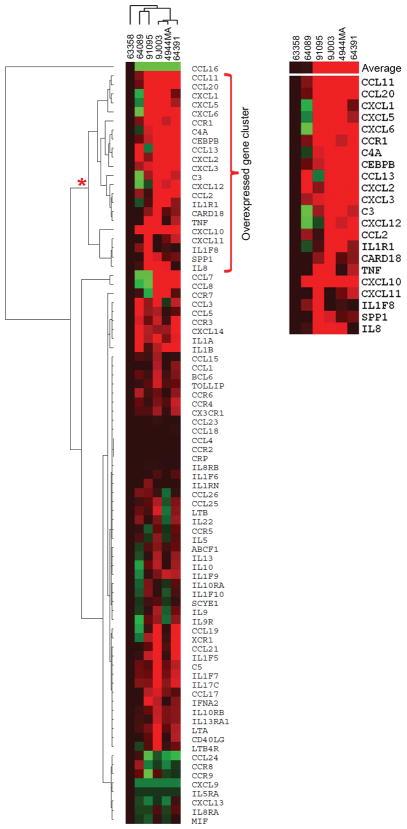
In order to investigate potential causes of the toxicity originating from fALS and sALS astrocytes, we examined genes involved in inflammatory signaling, since previous reports have documented heightened inflammatory gene activation in fALS astrocytes1, 3. We utilized quantitative Reverse Transcriptase polymerase chain reaction (qRT-PCR) array technologies to assay the expression of a functionally diverse set of 84 genes that includes both cytokine and chemokine ligands and receptors, and other intermediary signaling proteins all involved in inflammatory gene regulation. Gene expression analysis across several fALS and sALS astrocyte lines revealed that 35–60% of total inflammatory genes assayed were upregulated compared to non-ALS control astrocytes (Fig. 3 and Supplementary Table 4). We observed a degree of heterogeneity between our ALS astrocytes as expected with a varied disease etiology. However, clustering analysis revealed a set of 22 upregulated genes encompassing chemokines, proinflammatory cytokines, as well as components of the complement cascade, many of which have been previously implicated in ALS21 (Fig. 3b). We next performed a network-based pathway analysis using Ingenuity Pathway Analysis software, which identified 4 networks ranked by a statistical scoring method (Supplementary Table 5). The NF-κB signaling complex was identified as a major interactor within the highest ranked network (Score=33) based upon the high number of connections and its centralized location within the interactome (Supplementary Fig. 10). Additionally, IFN-α was also identified within this gene network interactome. Within the second ranked network (Score=12), pathways involving kinase signaling such as MAPK, JNK, and AKT, which establish a high number of interactions with the inflammatory genes in this cluster were identified (Supplementary Fig. 11). Many of these signaling networks have been previously implicated in fALS, and we now show evidence for their potential involvement in mediating inflammatory responses in sALS astrocytes22, 23.
Fig. 3.
Inflammatory gene activation in fALS and sALS astrocytes. (A) Hierarchical clustering analysis of inflammatory cytokine and receptor genes differentially expressed in ALS astrocytes compared to non-ALS control line (63358). (B) Cluster of inflammatory genes most upregulated in ALS astrocytes compared to non-ALS control. Shades of red correspond to the magnitude of expression increase, the intensity of green corresponds to the magnitude of the reduction in gene transcript. Display range was set at maximum of +/− 15 fold.
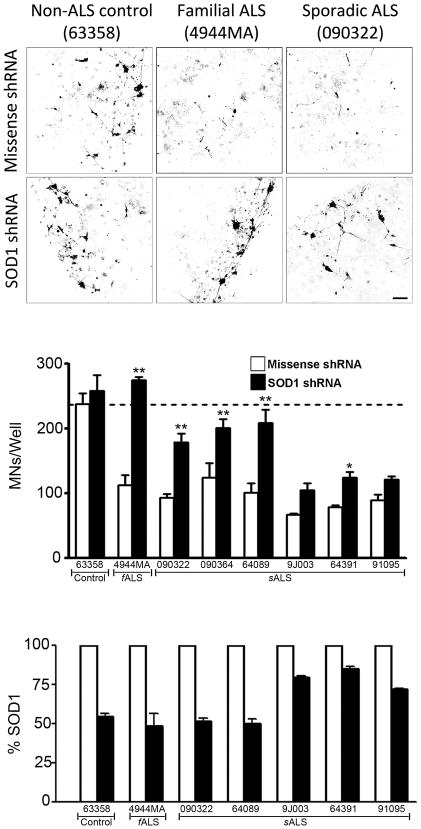
To increase the confidence of our assays, we determined whether suppression of mutant SOD1 in the fALS astrocytes would rescue MN death using a lentivirus encoding a shRNA 24 to specifically knockdown SOD1 levels. Suppression of mutant SOD1 expression in our fALS astrocytes resulted in a full rescue of astrocyte-mediated MN toxicity as compared to the non-ALS control line (Fig. 4a, b). As expected, the missense shRNA control did not alter the fALS astrocyte-mediated toxicity towards MNs (Fig. 4b). The indistinguishable pathological and clinical features of fALS and sALS have long insinuated a shared disease mechanism between these forms of ALS and recently SOD1 has been proposed as a common link. Several studies have suggested that wild-type SOD1 can adopt conformational changes in sALS patients that are similar to the known pathological forms of SOD1 mutated in fALS 25–27. These findings led us to test whether shRNA-based suppression of wild-type SOD1 in the sALS astrocytes would confer neuroprotection to MNs. In order to test this hypothesis, we performed experiments similar to our SOD1 knockdown in fALS astrocytes. All MNs co-cultured with sALS astrocytes expressing the missense shRNA control showed significant cell death, consistent with previous experiments. Remarkably, when we suppressed SOD1 in our sALS astrocytes, we observed statistically significant neuroprotective effects in 4 of the 6 sALS astrocyte lines (Fig. 4b). In the remaining 2 sALS astrocytes lines, we observed a trend of neuroprotection, although these results were not significant. This lower level of neuroprotection may reflect the observation that SOD1 was not efficiently suppressed in these lines, as quantified by ELISA analysis (Fig 4c), even though the level of lentiviral transduction efficiency was equivalent among all the astrocyte lines. Collectively, these results suggest that suppression of SOD1 in sALS astrocytes is beneficial and can confer significant MN protection from ALS astrocyte-derived toxicity, when greater than 50% of wild-type SOD1 knockdown is achieved. These results support previous findings 25–27 that wild-type SOD1 should be considered a possible therapeutic target for sALS.
Fig. 4.
Suppression of SOD1 in both fALS and sALS astrocytes is MN protective. (A) Representative microscopic fields showing the morphology of MNs stained with choline acetyl transferase (ChAT) after 120 hours of co-culture with human astrocytes transduced with either missense or SOD1 shRNA lentivirus. Scale bars, 100 μm. (B) Counts of MNs per well after 120 hours in co-culture with astrocytes expressing the missense shRNA (white bars) or the SOD1 shRNA (black bars). (C) Quantification of SOD1 levels in astrocyte cell lysate measured by ELISA. *p<0.05 and **p<0.01 compared to 63358 (non-ALS control). Error bars represent SEMs.
A major challenge in the field of ALS research has been to evaluate the similarities and differences between fALS and sALS. Here, we tested whether astrocytes derived from the more common form of the disease, sALS, showed a similar glial-mediated toxicity towards MNs as demonstrated in various fALS models. Our results indicate a shared mechanism leading to MN death between fALS and sALS through astrocyte-mediated toxicity. In addition, we provide evidence that wild-type SOD1 is involved in this toxicity, suggesting that therapies targeting SOD1 in astrocytes may be beneficial not only to fALS patients with SOD1 mutations, but also to sALS patients as well. Recently, novel vectors have been shown to efficiently target astrocytes of adult brain and spinal cord28 and constitute an attractive therapeutic option. Alternatively, astrocyte replacement has also been proposed as a potential therapy for slowing disease progression in ALS, and stem cell transplants are currently in human clinical trials29.
Although glia represents a potential therapeutic target in ALS, an ultimate goal in the ALS research field is to understand why MNs perish in the disease. Of note, our studies presented here investigated only ALS astrocyte-derived toxicity on non-ALS derived MNs. The derivation of patient-specific MNs will be important to further evaluate pathological abnormalities in these cell types. To aid in this endeavor, we recently demonstrated that human MNs can be efficiently and rapidly generated from pluripotent stem cells30. This novel strategy should be advantageous in generating MNs from patient-specific iPS cells. In sum, this work introduces the first model system to investigate molecular disease mechanisms and evaluate potential therapies for sALS, and highlights the great potential to use patient-specific samples for studying complex diseases.
Online Methods
Human tissue samples
Post-mortem spinal cord and skin samples were obtained from the National Disease Research Interchange (NDRI, Philadelphia, PA), and informed consents were obtained from all subjects before sample collections by the NDRI. Receipt of human tissues was granted through Nationwide Children's Hospital Institutional Review Board (IRB08-00402, Investigating the Role of Glia in Amyotrophic Lateral Sclerosis), and use of all human samples is in accordance with their approved protocols. Tissues samples were processed in approximately 24 to 72 hours postmortem.
Tissue processing for histological analysis
Spinal cord tissues were fixed in 4% PFA for 24 hours, followed by two rinses of 0.1M sodium phosphate buffer. Tissues were then transferred to 30% sucrose in 0.1M sodium phosphate buffer and allowed to incubate for two days at 4°C or until the spinal cords sank to the bottom of the solution. Spinal cord tissues were cut at 5–6 mm section and embedded in Tissue-Tek OCT compound (Sakura Finetek) and frozen with dry ice. Tissues were then sectioned at 10 microns with a cryostat and then stored at −20°C in an antifreezing solution prior to immunocytochemical analysis. For thionin staining, spinal cords were fixed in 4% PFA for 24 hours, paraffin embedded, and sectioned at 8μm. Sections were then stained for thionin for easy visualization of motor neurons.
Neural progenitor cell isolation and maintenance
All procedures were performed in sterile fashion in a class II biosafety cabinet. A representative portion (2.5 cm) of three regions (thoracic, cervical and lumbar) of the spinal cord was used for Neural Progenitor Cell (NPC) isolation as described previously16. Briefly, tissue was diced and a single cell suspension was obtained by enzymatic dissociation of the tissue at 37 °C for approximately 30–40 minutes with 2.5 U/ml papain (Worthington Biochem, Lakewood, NJ), 250 U/ml of DNase I (Worthington Biochem), and 1 U/ml neutral protease (Roche, Indianapolis, IN). After dissociation, the cell suspension was mixed with DMEM/F12 (Invitrogen, CarlsBad, CA) with 10% fetal bovine serum (FBS, Gibco, CarlsBad, CA), passed through a 70 μM filter, and centrifuged. The cell pellet was resuspended in DMEM/F12 with 10% FBS and combined 1:1 with percoll (GE Healthcare, Piscataway, NJ). The cell/percoll mixture was centrifuged at 20,000 g for 30 min at room temperature and the low buoyancy fraction (10 ml) above the red blood cell layer was collected. Cells were washed and resuspended in NPC medium containing DMEM/F12 (Invitrogen) supplemented with 10% FBS (Gibco), 10% BIT9500 (Stem Cell Technologies, Vancouver, BC), 1% N2 supplement (Invitrogen), 20 ng/ml of FGF-2 (Peprotech, Rocky Hill, NJ), 20 ng/ml of EGF (Peprotech), and 20 ng/ml of PDGF-AB (Peprotech). NPCs were cultured on fibronectin (Chemicon, Billerica, MA) coated plates and after 24 hours, the media was replaced with serum-free NPC medium. Half of the medium was subsequently replaced every 2 days. Cells were passed when 60–70% confluence was reached in about 3–4 weeks.
Neural progenitor cell differentiation
Neurons, astrocytes and oligodendrocytes were obtained from NPC cultures by removal of growth factors from the media and supplementation with differentiation factors. To differentiate NPCs into neurons, medium was supplemented with 2 uM all-trans retinoic acid (Sigma, St. Louis, MO) and 5 uM of forskolin (Sigma). To induce astrocytic differentiation, NPCs were supplemented with 10% FBS (Gibco). Oligodendrocyte differentiation was induced by culturing cells in 200 ng/ml of IGF-1. In all conditions, cells were allowed to differentiate for 1 week.
Astrocyte culture and maintenance
Once NPC cultures from ALS patients and controls were established, astrocytes were generated by media supplementation with 10% FBS. Astrocytes were cultured in laminin coated plates and media was changed every three days. Astrocytes were passaged when 80% confluent. Fetal human NPCs, isolated from the frontal brain cortex of a 28 week term fetus, (SCP-27, P1) were obtained from the National Human Neural Stem Cell Resource (NHNSCR, Orange, CA). Human fetal astrocyte cell line (1800) isolated from cerebral cortex was obtained from ScienCell research laboratories (Carlsbad, CA).
Skin fibroblast isolation and maintenance
In brief, a 2 cm2 skin sample was rinsed twice in PBS and then placed in a 10cm2 dish with the epidermal side facing down. The subcutaneous layer was then scraped off using a surgical scapel and cut into 0.5 cm2 strips. Skin tissue was then allowed to incubate with 25 ml of 0.05% trypsin/EDTA (Invitrogen, Carlsbad, CA) in a 50 ml conical tube at 37 °C for 45 minutes with gentle agitation every 15 minutes. After the incubation period, 20 ml of DMEM media containing 10% FBS was added to inactivate the trypsin and cells were pelleted at 350g for 4 minutes. Cells were then resuspended in DMEM media containing 10% FBS and plated to a 6 well dish, and within two weeks fibroblasts were usually confluent on a 10 cm2 tissue culture dish.
Immunocytochemistry
Cells in culture were fixed with 4% PFA for 10 minutes, followed by three rinses of TBS for 5 minutes, and then blocked for 1 hour with 10% donkey serum with 0.1% Triton X-100. Spinal cord tissue sections were blocked similarly prior to antibody addition. All primary antibodies were diluted in blocking solution and tissue sections or cells were incubated overnight at 4°C. Primary antibodies and dilutions utilized are shown in supplemental table 6. Nuclei were counterstained with DAPI. All images were captured using a Zeiss LSM510-META confocal laser-scanning microscope (Zeiss, Thornwood, NY).
DNA sequencing of ALS patient samples
The SOD1 gene was sequenced in all ALS samples. For SOD1 sequencing, primers were designed that flanked all 5 exons of the SOD1 within intronic regions. High fidelity accuprime polymerase (Invitrogen,Carlsbad CA) was utilized to amplify exonic regions and was subsequently sequenced at Eurofins MWG Operon (Huntsville, Al) utilizing standard Sanger sequencing protocols. In all ALS cases SOD1 coding regions were wild-type, except for one familial ALS case containing a C>T transition 14 nucleotides downstream of the start adenine nucleotide rendering an alanine to a valine amino acid substitution at position 4 in the protein (SOD1A4V).
RNA isolation and quantitative RT-PCR
RNA was harvested using the RT2 q-PCR grade RNA isolation kit (SABiosciences, Frederick, MD) and total RNA was reverse transcribed with RT2 First Strand Kit (SABiosciences) according to the manufacturer’s instructions. Real-time quantitative PCR reactions were performed using RT2 Real-Time SYBR Green/Rox PCR Master Mix (SABiosciences, Frederick, MD) and PCR arrays were run on an ABI Prism 7000.
Gene array analysis
Astrocyte-enriched gene expression was analyzed in proliferating NPCs, astrocytes, mesenchymal stem cells, and primary human astrocytes using a customized RT2 Profiler PCR Array (SA Biosciences). Inflammatory gene expression was measured in astrocytes co-cultured with MNs using a cataloged RT2 Profiler PCR Array for human Inflammatory Cytokines and Receptors (SA Biosciences). All analyses were performed using the web-based PCR Array Data Analysis Software available on the SA Biosciences website. Gene expression was normalized using a set of 5 housekeeping genes and fold regulation was used to analyze changes in gene expression. Cluster 3.0 was used to cluster the row of genes based on the Euclidean distance, and at the same time cluster the patients based on centered correlation. Unstandardized fold change values against control were used as input for Clustergram. Phylogenetic trees were displayed by TreeView. The overexpressed inflammatory cluster of genes (also called focus genes) common to all ALS patients was uploaded into IPA 9.0 (Ingenuity Systems Inc, Mountain View, CA, USA). The IPA web tool was utilized to identify molecular networks that involve these focus genes, which were then ranked by a statistical scoring algorithm with P<0.001. Interactomes based upon the top 2 statistically ranked networks were generated by IPA 9.0 by probing the ingenuity knowledge base, which is the largest annotated database of previously published data.
Virus production
SOD1 expression in astrocytes was knocked down by lentiviral transduction expressing siRNA sequences previously described24. In addition, lentiviruses were used to overexpress either human wild-type SOD1, SOD1 G93A or SOD1 A4V by the CMV promoter in astrocytes. Viruses were produced by transient transfection into HEK293 cells using calcium phosphate, followed by supernatant viral purification by ultracentrifugation.
ES motor neuron differentiation
Mouse embryonic stem cells that express GFP driven by the Hb9 promoter (HBG3 cells, kind gift from Tom Jessell) were cultured on primary mouse embryonic fibroblasts (Millipore, Billireca, MA,) and differentiated to motor neurons (MNs) in the presence of 2 μM retinoic acid (Sigma) and 2 μM purmorphamine (Calbiochem). After 5 days of differentiation, the embryoid bodies were dissociated and sorted for GFP on a BD FACSVantage/DiVa sorter.
Co-culture of motor neurons and astrocytes
Human astrocytes were plated in 96 well plates coated with laminin (5 ug/ml, Invitrogen) at a density of 10,000 per well. Two days after, GFP positive motor neurons (MN) were sorted by FACS and cultured on top of the astrocytes at a density of 10,000 per well in MN media [DMEM:F12 (Invitrogen), 5% horse serum, 2% N2 (Invitrogen), 2% B27 (Invitrogen) + GDNF (10 ng/ml, Invitrogen), BDNF (10 ng/ml, Invitrogen), CNTF (10 ng/ml, Invitrogen)]. After 24 hrs, cytosine arabinose (1 mM) was added for 48 hrs in order to eliminate any remaining dividing NPCs or embryonic stem cells. Media was changed every other day subsequently. For the astrocyte conditioned media experiments, media was collected from the co-culture every 2 days, supplemented with GDNF, CNTF, and BDNF, and added to MNs that were cultured on laminin coated plates.
Quantification of SOD levels
Quantification of SOD 1 levels in NPCs derived astrocytes lines was performed either by western blot or Elisa. For western blot, cell lysates were obtained using Tissue Protein Extracting Reagent (TPER, Pierce, Rockford, IL) and 20 ug of protein was loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen). The proteins were run at 150 V for 1 hour then transferred to an Invitrolon PVDF membrane (Invitrogen). The membrane was blocked in 5% non-fat milk, 0.1% Tween-20 in TBS for 1 hour and then incubated overnight in primary antibody to human SOD1 (#2770, Cell Signaling, 1:1000). Bound primary antibody was detected by horseradish peroxidase conjugated secondary antibody (Jackson) followed by chemiluminescence (ECLTM Western Blotting Detection Reagents, Amersham Biosciences). The blots were then stripped and re-probed with a β-actin antibody (#A5060, Sigma-Aldrich, 1:1000) to control for protein loading. To evaluate the levels of SOD1 knockdown in sALS, SOD1 isozyme was analyzed by ELISA (Abnova, Taipei City, Taiwan). Elisa was performed in 75 ng of cell lysate following manufacture recommendations.
GABAergic neuron differentiation
Mouse fetal neural progenitors were cultured on laminin-coated plates (5 ug/ml, Invitrogen) in Neural Stem Cell Media [DMEM:F12 (Invitrogen) and 5% N2 (Invitrogen)]. GABAergic neurons were differentiated from mouse fetal neural progenitors in DMEM:F12 (Invitrogen) with 0.1% FBS (Invitrogen), retinoic acid (1 μM, Sigma), and forskolin (5 μM, Sigma) for 7 days.
Statistical analyses
All statistical tests were performed by multiway analysis of variance followed by a Bonferroni post hoc analysis of mean differences between groups (GraphPad Prizm Software). Experiments were performed in triplicate or quadruplicate.
Supplementary Material
Acknowledgments
This work was funded by NIH R01 NS644912-1A1, RC2 NS69476-01, Project A.L.S. and Packard Center for ALS Research (P2ALS) and Helping Link Foundation to B.K.K. and an NIH NRSAF31NS058224 to A.M.H. K.M. is supported by a fellowship from the Swiss National Science Foundation (SNSF).
Footnotes
Author Contributions
Conceived and designed the experiments: M.E.H., A.M.H., C.J.M., A.H.M.B., J.R.M., & B.K.K. Performed the experiments: M.E.H., A.M.H., C.J.M., K.M., L.B., A.F., S.S., S.L., M.J.M., K.D.F., M.R., A.E., A.K., & A.C. Analyzed the data: B.K.K., M.E.H., A.M.H., & C.J.M. Wrote the manuscript: M.E.H., A.M.H., C.J.M., & B.K.K with input from the other co-authors.
References
- 1.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka K, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchetto MC, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown RH., Jr Amyotrophic lateral sclerosis. Insights from genetics. Arch Neurol. 1997;54:1246–1250. doi: 10.1001/archneur.1997.00550220050013. [DOI] [PubMed] [Google Scholar]
- 7.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Boillee S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 10.Clement AM, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 11.Beers DR, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulting GL, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 14.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodge JC, et al. Delivery of AAV-IGF-1 to the CNS extends survival in ALS mice through modification of aberrant glial cell activity. Mol Ther. 2008;16:1056–1064. doi: 10.1038/mt.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer TD, et al. Cell culture. Progenitor cells from human brain after death. Nature. 2001;411:42–43. doi: 10.1038/35075141. [DOI] [PubMed] [Google Scholar]
- 17.Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560–573. doi: 10.1016/j.mcn.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 20.Dodge JC, et al. AAV4-mediated expression of IGF-1 and VEGF within cellular components of the ventricular system improves survival outcome in familial ALS mice. Mol Ther. 2010;18:2075–2084. doi: 10.1038/mt.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobsiger CS, Boillee S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci U S A. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Yang B, Zhang D. Activation of interferon signaling pathways in spinal cord astrocytes from an ALS mouse model. Glia. 2011;59:946–958. doi: 10.1002/glia.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bendotti C, et al. Inter- and intracellular signaling in amyotrophic lateral sclerosis: role of p38 mitogen-activated protein kinase. Neurodegener Dis. 2005;2:128–134. doi: 10.1159/000089617. [DOI] [PubMed] [Google Scholar]
- 24.Miller TM, et al. Virus-delivered small RNA silencing sustains strength in amyotrophic lateral sclerosis. Ann Neurol. 2005;57:773–776. doi: 10.1002/ana.20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zetterstrom P, Graffmo KS, Andersen PM, Brannstrom T, Marklund SL. Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic ALS model mice. J Biol Chem. 2011 doi: 10.1074/jbc.M111.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepore AC, et al. Focal transplantation-based astrocyte replacement is neuroprotective in a model of motor neuron disease. Nat Neurosci. 2008;11:1294–1301. doi: 10.1038/nn.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hester ME, et al. Rapid and Efficient Generation of Functional Motor Neurons From Human Pluripotent Stem Cells Using Gene Delivered Transcription Factor Codes. Mol Ther. 2011 doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.