Abstract
To uncover shared pathogenic mechanisms among the highly heterogeneous autism spectrum disorders (ASDs), we developed a protein interaction network that identified hundreds of new interactions among proteins encoded by ASD-associated genes. We discovered unexpectedly high connectivity between SHANK and TSC1, previously implicated in syndromic autism, suggesting that common molecular pathways underlie autistic phenotypes in distinct syndromes. ASD patients were more likely to harbor CNVs that encompass network genes than control subjects. We also identified, in patients with idiopathic ASD, three de novo lesions (deletions in 16q23.3 and 15q22 and one duplication in Xq28) that involve three network genes (NECAB2, PKM2, and FLNA). The protein interaction network thus provides a framework for identifying causes of idiopathic autism and for understanding molecular pathways that underpin both syndromic and idiopathic ASDs.
INTRODUCTION
Autism spectrum disorders (ASDs) are a heterogeneous group of neuro-developmental disorders with three core features: impaired social skills (e.g., gaze avoidance), delayed language development, and repetitive or stereotyped behaviors (1–4). “Classic” or idiopathic autism principally involves these three features; “syndromic” ASDs are those disorders in which the autistic phenotype is one aspect of a much broader clinical syndrome. In Tuberous Sclerosis Complex (TSC), for example, which is caused by a single gene mutation in either TSC1 or TSC2, the core autistic phenotype is accompanied by seizures, developmental delay, cortical tubers, facial angiofibromas, and other skin lesions (5, 6). Similarly, Phelan-McDermid syndrome (PMS) is caused by microdeletions of chromosome 22q13.3 encompassing the SHANK3 gene (encoding SH3 and Ankyrin domain containing protein), and is characterized by general hypotonia, seizures, intellectual disability and ASD (7). Autistic features are commonly observed in Fragile X syndrome, Angelman symdome, phosphatase and tensin homolog (PTEN) hamartoma, Rett syndrome, Timothy syndrome, and in individuals with Neuroligin mutations (Table S1) (8). The majority (85–90%) of ASD cases do not show such clinically distinct phenotypes and the genetic causes remain largely unknown; in these cases the ASD is considered “non-syndromic”.
Although approximately 50 genes or genomic variants that either cause or predispose individuals to ASDs have been identified (1–4), each accounts for no more than 0.5–2% of total ASD cases (9–11), and many of these correspond to the syndromic ASDs. Together, these genes and genomic variants account for at most 30% of all ASD cases (1, 12, 13). For non-syndromic ASD, copy number variations (CNV) of submicroscopic DNA segments may prove more relevant; growing numbers of ASD-susceptibility loci have been reported based on genome-wide association studies.
To make the challenge of understanding the pathogenesis of ASDs even more daunting, the known ASD-related proteins (including those mapping within CNVs) span diverse categories, from transcription factors (4, 14) and RNA-binding proteins (15) to cell adhesion molecules (16) and enzymes involved in protein modification (2, 8, 13) and degradation (12, 17). Given the clinical heterogeneity of ASDs, it would not be surprising if mutations in hundreds or even thousands of genes cause ASD phenotypes—but might these genes converge on a few pathways?
We hypothesized that, if this were actually the case, it would best be revealed by a protein-protein interaction analysis of the existing autism-associated genes. Several years ago, when confronting a slightly different challenge with another group of neurological disorders, the inherited ataxias, we created an “ataxia interactome”. Using this interactome we identified interactors of ataxia-associated proteins (18) and mapped their inter-relationships, uncovering unexpected functional relationships (19–21). Lacking a unifying neuropathology for the ASDs, we decided for the present study to rely upon key phenotypic features to create a protein-protein interaction network that would reveal functional relationships between the gene products. Uncovering functional relationships among diverse ASD-related proteins is a first step towards the ultimate goal of developing therapies that might benefit multiple functionally or mechanistically related ASDs.
RESULTS
We began by identifying protein partners of ASD-associated proteins (1–3) and determining whether any of them interacted with other ASD proteins. We selected genes from three groups: those whose mutation results in syndromic ASDs (“syndromic ASD proteins”, Table S1), those whose mutation causes severe language delay, and those whose products are paralogs, known binding partners, or functionally related to syndromic ASD proteins. We refer to the second and third groups as “ASD-associated” genes in order to distinguish them from the first group, syndromic ASD proteins (Table S1). Although language delay can occur without accompanying autistic features, and the reasons for language delay may be as heterogeneous as the ASDs themselves, we reasoned that language development might depend on the same pathways as those involved in social communication deficits observed in ASDs.
We performed a yeast-two hybrid (Y2H) screen of a human cDNA library using 192 bait fragments for 35 gene products, each of which encoded either full-length or partial segments of coding sequences. After a series of stringent tests (18, 22), we obtained 7,933 interacting prey clones, which belonged to 783 unique proteins; 539 passed a second round of testing in yeast to demonstrate interactions held up in an independent reconstitution system (22). We considered only these 539 proteins as candidate binary interacting partners of the bait proteins. These 539 proteins comprised 848 interactions with 26 syndromic ASD proteins or ASD-associated proteins (Tables S1 and S2). Among them, only 32 interactions (4%) were previously reported (Tables S2 and S3). Baits for nine proteins failed to identify definite binding partners in this stringent Y2H screen (Table S1).
To validate the interaction data in a mammalian system, we performed glutathione-sepharose affinity co-purifications (GST-APs) in HEK293T cells for 52 randomly selected interactions (6% of the total) (Fig. S1). The mammalian cells recapitulated 44 out of 52 interactions (85%) (Fig. S1 and Table S2). In general, bona fide binary interactions do not validate at more than 50% (23–25) when using different assay systems; our unusually high validation rate thus supports the reliability of our stringent screening methods compared to prior interactomes (18, 22, 26). We did not remove candidate pairs from the screening data (Table S2) when they failed in the validation assays, because the Y2H screen is excellent at detecting transient and biologically relevant interactions that are often difficult to recapitulate in co-immunoprecipitation and affinity purification systems (18, 22–25).
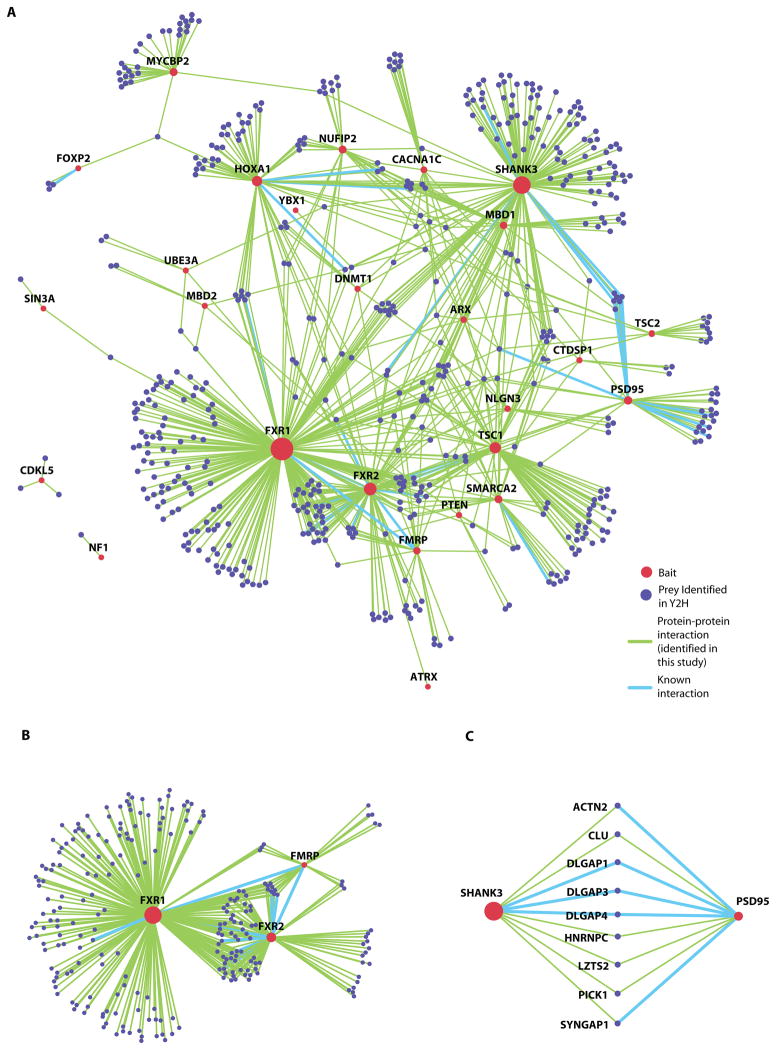
Using the interaction data from the Y2H screen (Table S2), we generated an ASD protein interaction network (Fig. 1A). Twenty-four out of the 26 syndromic ASD or associated proteins were interconnected in one major component (CDKL5 and NF1 were located outside) (Fig. 1A). The Fragile X-related proteins 1 and 2 (FXR1 and FXR2) showed the greatest connectivity, and were also connected to the Fragile X mental retardation protein (FMRP) (Fig. 1B). The ASD network thus recapitulates and expands previously described in vivo associations and functional relationships among the Fragile X-related proteins (27, 28). Similarly, we expanded previously established relationships among the tuberous sclerosis complex proteins 1 and 2 (TSC1 and TSC2) (29) by identifying four new partners (Fig. 1A and Table S2). We also confirmed that the post-synaptic proteins SHANK3 and PSD95 interact in vivo (30, 31) and identified nine shared partners between them (ACTN2, CLU, DLGAP1, DLGAP3, DLGAP4, HNRNPC, LZTS2, PICK1, and SYNGAP1 Fig. 1C).
Figure 1. Landscape of protein-protein interactions for syndromic ASDs and associated disorders.
(A) The protein interaction network derived from the Y2H screens; inset indicates color codes for baits, prey, and interactions. Twenty-six bait proteins are included.
(B) Fragile-X mental retardation protein FMRP and its paralogs (FXR1 and FXR2) are highly connected via shared interactors.
(C) The synaptic proteins SHANK3 and PSD95 share nine interactors.
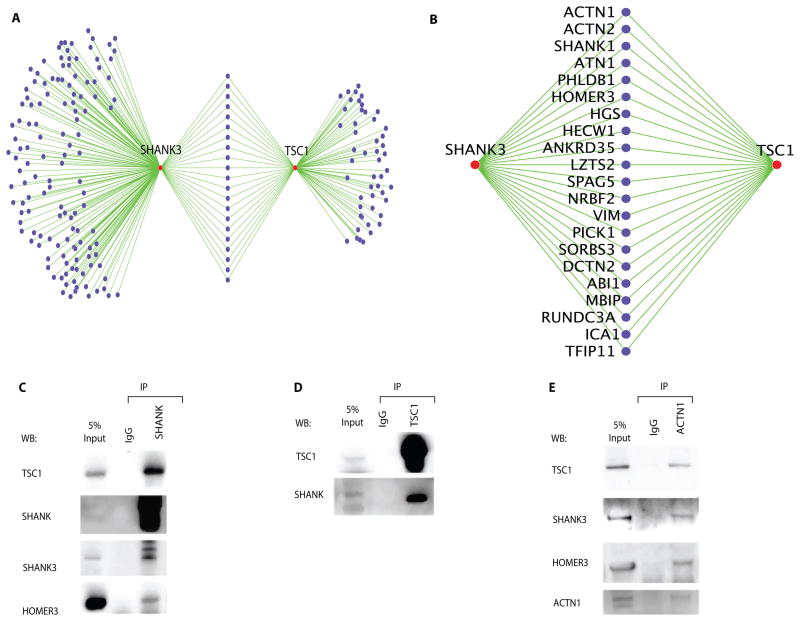
Notably, our ASD interactome revealed previously unsuspected connectivity between two syndromic ASD proteins, SHANK3 and TSC1, which share at least 21 partners (Fig. 2A). SHANK1, the paralog of SHANK3, arose as a potential partner of both TSC1 and SHANK3, which suggested that these syndromic ASD proteins interact in a complex at the postsynaptic compartments—the microenvironment of dendritic spines beneath the specialized membrane structure of the synapse (3, 32) —a suggestion confirmed in vivo by co-immunoprecipitation using mouse brain extracts (Fig. 2C and 2D). In vivo studies further confirmed that ACTN1, a postsynaptic scaffold protein identified as a partner of SHANK3 and TSC1 (Table S2 and Fig. 2A), interacts with TSC1 as well as with the postsynaptic scaffolding proteins SHANK3 and HOMER3 (Fig. 2E). Further, the Y2H screen recapitulated eleven previously reported in vivo interactions (Tables S2 and S3), which we obtained from the Human Protein Reference Database (HPRD) (33) and the Biological General Repository for Interaction Datasets (BioGRID, http://thebiogrid.org/). All except TSC1-AXIN1 were reported in mouse brain tissue. We also identified an additional 21 interactions, previously demonstrated by in vitro co-purifications or Y2H screens (Table S3).
Figure 2. In vivo validation of the network’s connectivity.
(A) The protein interaction data show high connectivity between SHANK3 and TSC1. The shared binding partners (purple nodes) between SHANK3 and TSC1 are annotated with a magnified view (B). ACTN1, SHANK1 and HOMER3 were identified as common binding proteins for the two proteins.
(C–E) In vivo interactions among endogenous TSC1, SHANK, HOMER and ACTN1 in mouse brain extracts. The reciprocal co-immunoprecipitation was performed using anti-pan-SHANK (C) or anti-TSC1 (D) antibodies. Proteins in the immunoprecipitated complex were labeled with the indicated antibodies. Antibodies against SHANK3 and HOMER3 were used to monitor stable protein complex formation in the immunoprecipitated samples with anti-pan-SHANK antibody. Normal rabbit IgG was used for the negative control (IgG). (E) Co-immunoprecipitation with anti-ACTN1 shows that it interacts with TSC1, SHANK3 and HOMER3 in vivo.
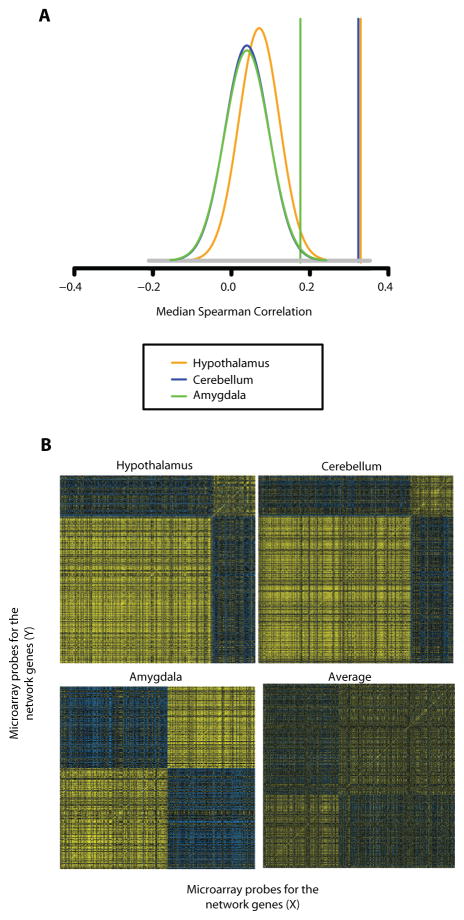
To verify that the syndromic ASD proteins are co-expressed with their binding partners in vivo, we analyzed microarray data from our previous studies of brain tissue from wild-type mice (34, 35). We observed strong correlations among expression profiles for genes in the network, but not for similarly-sized sets of randomly selected probes on the microarrays (p<0.001, Fig. 3A). Further, the majority of the genes that encode the proteins annotated in the network co-clustered into a dominant co-expression group in the hypothalamus (78%), cerebellum (78%), and amygdala (55%) (Fig. 3B). In conjunction with the physical interaction data (Fig. 2B–D), SHANK3, TSC1, ACTN1 and HOMER3 showed highly correlated expression in these brain regions (Table S4). Note that averaged correlation matrices of the genes in the three brain regions did not show such strong correlation, suggesting that subsets of the ASD-associated proteins and their binding partners may enjoy unique relationships in different brain regions, rather than be ubiquitously co-expressed (Fig. 3B). Importantly, 96% of the proteins identified in our primary screen were found to be expressed in brain in the mouse studies, a substantially greater proportion than the expected 59% for randomly sampled genes (p< 1×10−10).
Figure 3. Genes encoding proteins in the autism network are highly co-expressed in mouse brain.
(A) Highly correlated co-expression of the network genes in mouse brain (hypothalamus, cerebellum and amygdala). Vertical lines show the median correlation among 478 mouse-mapped network genes. The plots show the distribution of median correlation among 1,000 Monte Carlo samples of 478 randomly selected genes from the microarray. Insets show the brain tissues subjected to analyses.
(B) Correlation matrices of the genes encoding mouse orthologs of network proteins in three regions. Genes are sorted by PAM clustering. Yellow and blue pixels indicate high and low levels of correlated expression for each pair of genes. For the average correlation, each gene was centered to mean 0 correlation.
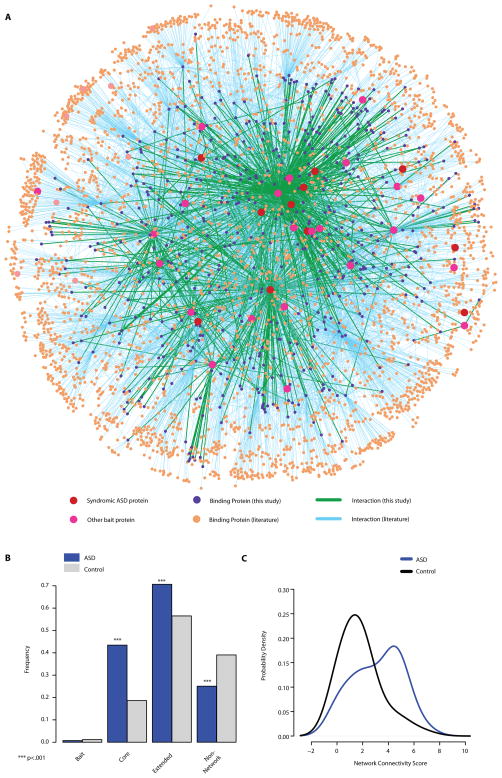
To understand the unique topology (i.e. the pattern of interconnections) of the protein interaction network and systematically assess the connectivity of syndromic ASD proteins, we incorporated literature-curated interaction data for both bait and prey proteins from the HPRD and the BioGRID. This allowed us to produce an extended network consisting of one component with 3,507 proteins connected through 6,881 interactions (Fig. 4A). Of the 35 bait proteins that were used in the Y2H screen, 34 were directly or indirectly connected inside this network; only one protein (SLC6A8) was not connected. Next we calculated the mean path length in the extended network for eight syndromic ASD proteins (Tables S1 and S2) that were in the experimental network (Fig. 1A), and we compared it with the distribution of mean path lengths for 8 randomly sampled proteins selected from the remaining 18 of 26 bait proteins that had at least one binding protein in the primary screen (Table S2). We performed 10,000 random draws of 8 proteins. The eight syndromic ASD proteins showed a significantly shorter mean path length (2.14) than the random samples from the remaining baits (mean of 2.78, p = 0.004, Fig. S2). The close connectivity of the eight syndromic ASD proteins led us to investigate whether different ASD proteins might share common molecular pathways that relate to the pathogenesis of ASD. Indeed, Gene Ontology (GO) analysis of the network (excluding all of the baits) revealed marked enrichment for proteins associated with synapse, postsynaptic density and cytoskeleton under the “Cellular Component” branch of the GO (Fig. S3A), and for small GTPase-mediated signaling and metabotropic glutamate receptor signaling under the “Biological Process” GO branch (Fig. S3B); a biological process describes a series of molecular events or functions. Such coherence between the cellular compartments and biological processes of the ASD baits and their interactome partners underscores the biological value of the network. It also points to key molecular pathways responsible for autistic phenotypes in distinct genetic syndromes and to the biological relevance of the network.
Figure 4. Overlap of the protein interaction network with CNV regions identified in non-syndromic ASD patients.
(A) The extended ASD protein interaction network is shown. Nodes and lines indicate the 11 syndromic ASD proteins (red), the 23 ASD-associated proteins used as bait in Y2H (pink), new binding partners identified in this study (purple), known binding partners (orange), new interactions (green), and previously known interactions (light blue).
(B) Bar plots show the relative frequency (Y) of ASD or control individuals harboring CNV overlap with genes in the interaction network. Bait, the bait proteins used for the Y2H screen; Core, network proteins identified in this study; Extended proteins, network proteins identified by literature searches; Non-network proteins, proteins not present in the network. The values represented by each bar do not sum up to one because an individual may have more than one CNV overlapping the core and extended network.
(C) The curves display density estimates (Y) of the Total Network Connectivity Score (X) for the genes encompassed by all CNVs in each individual from the ASD or control groups. The Total Network Connectivity Score is the log2 of the sum of network connectivity of all genes harbored in CNVs of each individual in each cohort. The ASD and control groups are significantly different with Wilcoxon’s rank sum test (p < 2.2 × 10−16).
It remained to be determined whether a protein interaction network built on syndromic ASD proteins would prove relevant to the pathogenesis of non-syndromic or idiopathic ASDs. To address this question, we collected information from published studies on copy number variations (CNVs) that were observed in normal populations or in non-syndromic ASD patients (36, 37). We then searched for genes that were annotated both in our network and in the intervals of CNVs found in normal individuals or ASD patients. Individuals from the ASD group showed an increased rate of CNVs spanning genes in the Y2H interactome compared to the control group by a factor of 2.4 (incidence 0.43 vs. 0.18, p < 1.13 × 10−23; two-sided Fisher’s Exact Test, “Core”, Fig. 4B) with an odds ratio of 3.3. Conversely, there was a lower rate of individuals in the ASD group whose CNVs failed to encompass genes in the network, i.e., fewer ASD CNVs mapped to loci encoding non-network proteins (0.25 vs. 0.39 for the control group, p = 6.16 × 10−8; “Non-network protein” in Fig. 4B). We also observed a higher rate of overlap between genes encoding proteins in the extended network for ASD than for Control (0.70 vs. 0.565 in frequency, “Extended”, Fig. 4B).
To consider both the connectivity of the network as well as the multi-CNV load in each individual, we computed an additional measure that we defined as the Network Connectivity Score. This score represents the sum of the number of connections in the network of all genes present in CNVs of each individual; the score therefore takes into consideration the contribution of genes by their network relevance. This score was also significantly higher in ASDs vs. control (p < 2.2 × 10−16, Wilcoxon’s rank sum test, Fig. 4C). We mapped the chromosomal locations of the network genes that overlapped with the CNV regions in ASD patients (Fig. 4B and S4). The genes overlapped by CNVs were widely distributed throughout the genome, indicating that our findings were not dominated by hot spots for structural variation in ASDs (Fig. 4B). Interestingly, however, three network genes (MVP, KCTD13 and ALDOA), mapped to the recurrent hot spot of the CNVs in human chromosome 16p11.2, have been reported for ASD and schizophrenia patients in genome-wide hybridization studies (11, 38)(Table S2 and Fig. S4).
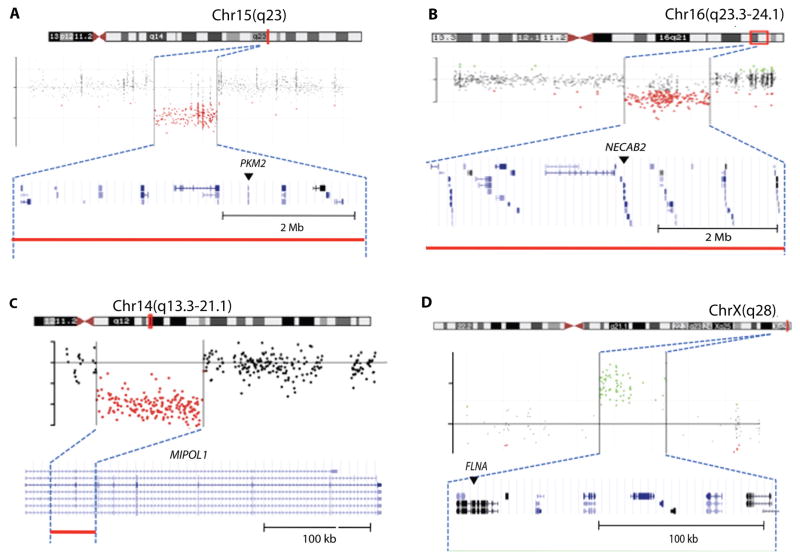
To explore the role of genes in the interaction network in idiopathic ASDs, we performed microarray-based comparative genome hybridization (CGH) for 627 genes in our network using genomic DNA from 288 relatively high-functioning individuals (average IQ 80.94) with a diagnosis of idiopathic ASD (i.e., non-syndromic autism) from the Simons Foundation Simplex Collection (39). These probands do not show any signs of syndromic disorders (systemic malformation, abnormal facies, or severe intellectual disability) on physical examination or brain scanning. We focused on events with large segmental duplications or deletions spanning over 10 kb. This analysis revealed a segmental duplication in chromosome Xq28 involving FLNA and three segmental deletions in chromosomes 15q13.3, 16q23.3-q24.1, and 14q13.3, which involved the PKM2, NECAB2 and MIPOL1 genes, respectively (Fig. 5A–D). CGH analyses of the DNA from the parents of the probands confirmed that the duplication of Xq28 (FLNA), deletions of 15q13.3 (PKM2) and 16q23.3 (NECAB2) were all de novo, whereas the deletion of 14q13.3 (MIPOL1) was maternally inherited (Fig. S5A–D and Table S5). Duplications and point mutations of FLNA cause various degrees of intellectual disability, periventricular heterotopia (a disorder caused by abnormal migration of neurons), and dysmorphic features, but to our knowledge have not been associated with an autistic phenotype in the absence of intellectual deficits (40). By identifying a de novo duplication of FLNA in a patient with an IQ of 109 and autism, we broaden the clinical spectrum of phenotypes associated with such duplications. Furthermore, our discovery that FLNA binds to SHANK3 (mutations in which cause syndromic ASD) using both Y2H assays (Table S2) and co-immunoprecipitation (Fig. S6) in mouse brain extracts validated the physical interaction of these proteins and shows that both syndromic and nonsyndromic ASDs are functionally linked.
Figure 5. Novel CNVs identified in idiopathic ASD that overlap genes encoding proteins in the interactome.
(A) The proband (hz22635) shows 5-Mb segmental deletion in chromosome 15q23, spanning the PKM2 gene. (A–D) The chromosomal location of events (top), plots of log2ratios (middle), and the gene structure for the regions (bottom) are from the UCSC browser. Red (A–C) and green (D) dots in the hybridization plots and the colored lines of each panel indicate the intervals of segmental deletion and duplication. Arrows denote genes in the network.
(B) The proband (hz20685) shows 5.9-Mb segmental deletion in 16q23.3-q24.1 spanning the NECAB2 gene.
(C) The proband (hz20741) shows partial deletion (43 kb) of the MIPOL1 gene at 14q13.3-q21.1.
(D) The proband (hz22625) shows a 200-kb segmental duplication in Xq28 spanning the FLNA gene.
None of the autosomal deletion events were observed in the clinical database of the Molecular Genetics Laboratory at Baylor College of Medicine (BCM), which houses samples from over 15,000 patients with dysmorphology (abnormal form or anatomy) or intellectual and developmental disability screened with a high-density oligonucleotide clinical chromosomal microarray. The BCM clinical array has much greater genome-wide coverage and was able to delineate precise boundaries of the initial CNV findings in our experimental cohort. In the clinical database, only 3 males and 1 carrier mother had the duplication event of Xq28 that involves FLNA but not MECP2, the gene involved in the neurological disorder, Rett syndrome (14, 41). All male patients had developmental disabilities and cognitive deficits, but the female patient was asymptomatic (as expected for an X-linked defect). Thus, all four segmental CNVs confirmed in this study were extremely rare structural variations rather than polymorphic events. We turned to an additional study (42) to examine a set of cognitively normal control individuals for these events using an extremely high-density tiling array. There were no deletion events covering the three genes (PKM2, NECAB2 and MIPOL1) or the FLNA duplication event in these control individuals. Furthermore, there were no deletion events overlapping these 3 deleted genes nor duplication events overlapping with FLNA loci among the CNV data from 1,500 controls (36) that we used for Fig. 4B.
DISCUSSION
We have developed a protein interaction network for ASD-causing and ASD-associated proteins (altogether ASD proteins). We began by identifying protein partners of ASD proteins (1, 2, 8, 13) and determining whether any interact with other ASD proteins. Although the majority of ASD-related proteins identified cause syndromic ASDs, we reasoned that converging pathways might provide insight into non-syndromic or idiopathic ASDs. Indeed, our interactome revealed an unexpected convergence around several proteins. Although the interactome validated several interactions that had been previously identified in the literature, it did not identify others. There are several reasons for this: the screen was not done to saturation; the interactome identifies direct interactions only, unlike co-immunoprecipitation which identifies direct and indirect interactions; protein interactions vary according to tissue; and our system is relatively low-expressing. Thus we are more likely to have false negatives (i.e., missed valid interactions) than false positives. The large number of validated interactions, however, demonstrates that this interactome is a robust framework for future studies exploring relationships among ASD proteins. Further, the significant overlap between CNVs from a previously published ASD cohort and our experimental network supports its utility as a platform for ASD gene discovery. Finally, the network can also shed light on other questions in ASD. For example, we found only one overlapping gene (CBS) on chromosome 21, despite evidence that Down’s syndrome (caused by trisomy of chromosome 21) shows some comorbidity with ASDs. This finding suggests that the ASD phenotype in Down’s syndrome may differ from the phenotypes of other non-syndromic ASDs.
An important aspect of the interactome are the connections it reveals between syndromic ASD-causing proteins and ASD-associated proteins in one network. Higher connectivity between two ASD-causing proteins suggests that they might interact in a protein complex or that they might function in a common molecular pathway. In the present study, we uncovered in vivo interactions between the TSC1 and SHANK proteins. Further exploration of such interactions should shed light on the pathogenic mechanisms underlying the autistic features observed in TSC (8, 43) and PMS (7, 44–47). The TSC1 and TSC2 proteins regulate mTOR, a promoter of protein synthesis in response to growth factors and stress (48); upregulation of protein synthesis due to functional loss of either of the TSC proteins is a likely mechanism for the ASD phenotypes of both disorders (8). The molecular mechanism that specifies the location of abnormal protein synthesis, however, remains to be determined. Our interactome suggests that these two distinct pathways are brought together in one protein complex built upon the postsynaptic protein scaffold, SHANK3.
Links between SHANK3 and other ASD proteins lend support to the idea that common pathways lead to the broader ASD phenotypes (3, 4, 8). In vivo studies of mice carrying mutant alleles of Shank3 and displaying autism-like phenotypes highlight the importance of the SHANK3 protein for maintaining the normal levels of many synaptic proteins that are critical for glutamatergic signaling (44, 45, 47). Recently reported pathogenic mutations in the SHANK2 gene in sporadic ASD cases (9, 49) further support our notion that various ASD-associated proteins, including FLNA, functionally interact with the SHANK proteins. These findings have therapeutic implications. Benefits reported from preclinical trials using rapamycin in Pten or Tsc2 mutant animals (50, 51) raise the possibility that such therapies might be beneficial in broader groups of patients with syndromic and non-syndromic ASDs.
In summary, we developed an ASD interactome that facilitates classification of ASDs according to functional pathways and interacting proteins. The short-term utility of this interactome will be to increase our molecular diagnostic capabilities with a raft of new genes. The mid-term and long-term benefits will come from advancing pathogenesis studies to promote development of rationally designed therapeutics that could be used to treat more than one ASDs.
MATERIALS AND METHODS
Yeast two-hybrid screen
All cDNAs encoding full-length or partial domains of bait proteins (Table S1) were cloned into DB-dest vectors using the Gateway system (Invitrogen) as described (18, 22). The constructs encoding each DB-autism fusion protein were transfected into yeast cells, MaV203 (Mat-alpha), and screened for positive interactions from 1–2 × 106 independent clones in human brain cDNA libraries (Invitrogen).
Co-immunoprecipitation for mouse brain extracts
Whole mouse brains were freshly homogenized in buffer containing 320 mM sucrose, 5 mM HEPES (pH 7.4) and 1 mM EDTA as described (52). One mg of total protein from cytosolic (S2) fractions was incubated at 4°C for 2 hr with either anti-pan-SHANK (5 μl of antisera), anti-TSC1 (1 μl), anti-ACTN1 (5 μl) antibodies, or normal rabbit IgG (5 μg) in the TS buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4) supplemented with 1% hemoglobin. Immune complexes were precipitated with prewashed Protein A agarose beads (Millipore) by incubating at 4°C for another 1 hr, and were washed in the TS buffer 5 times and extracted with SDS-loading buffer.
CGH microarray analysis
We determined the exon coordinates of the genes that were annotated in the protein interaction network, then selected a total of ~42K oligonucleotide probes from Agilent’s open resource library to design a custom microarray for CGH studies. Using the custom 4×44K chips, we performed DNA digestion, labeling and hybridization according as previously described (53, 54). Since the CNV regions for NECAB2, PKM2, and FLNA exceeded the targeted gene loci, we used the BCM MGL clinical array (CMA BAC V8.1, Baylor MGL) to determine boundaries of the events (53). All the intervals of CNV events and other information referred to in this study were defined using the assembly of NCBI36/hg18 at UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
Gene expression analysis
We used Homologene (www.ncbi.nlm.nih.gov/homologene) release 6.1 mapping human genes to their mouse orthologs. Of the 593 in the network we were able to map 478 (81%) to unique mouse genes. We then obtained gene expression data from wildtype mouse hypothalamic, cerebellum and amygdala samples using data from prior studies (34, 35). We computed Spearman’s rank correlation coefficient for all network gene pairs and determined the median. To compare our results with a random set of genes, we sampled 1,000 iterations of an equal number of random genes from the array and computed the same median Spearman correlation measure. The distributions for the median correlation values for random genes are plotted for each brain region. The median values for our mapped network proteins are shown with three vertical lines. In addition, we computed the correlation heat-map between all pairs of genes, and we sorted the genes according to cluster analysis based on pair-wise correlations.
CNV database analysis
We obtained CNV all variant data from dbVAR (www.ncbi.nlm.nih.gov/dbvar/) for ASD (37) and controls (36), which contained CNV data from 419 and 1,552 samples, respectively. We calculated the overlap with the protein interaction network for each CNV in terms the network genes, the network node degree, and network components (Bait, Core, or Extended). Data were processed to show overlapping events per individual. We computed two summaries: overlap indicators for each person with the components of the interaction network (Bait, Core, and Extend) and Network Connectivity Score defined as log2 of the sum of network degree for overlapped genes. Individuals with no network overlap were treated as missing values for the Connectivity Score.
Further details for data analysis are provided in Supplementary Materials.
Supplementary Material
Fig S1. Validation of the interaction data from the Y2H study by co-affinity purification experiments in HEK293T cell
Fig S2. Syndromic ASD proteins are highly connected to each other and have the shortest path length in the protein interaction network
Fig S3. The GO analysis for the extended ASD network
Fig S4. Chromosomal location of the overlapping genes between the protein interaction network and CNV intervals of ASD group
Fig S5. Confirmation of de novo or inherited CNVs
Fig S6. Physical interaction of FLNA with SHANK3 in vivo
Table S1. Summary of yeast-two-hybrid screening for ASD and related disorders
Table S2. The summary of protein interaction data obtained from Y2H screening and Co-AP study
Table S3. Summary of known interaction data in the literature and the overlap with this study
Table S4. Co-expression pattern of the genes in the mouse hypothalamus, cerebellum and amygdala
Table S5. Summary of the Probands with Novel CNVs
Acknowledgments
We thank L. White, L. Liles and M. Hoang at BCM microarray core; L. Lewis at BCM-GSC; and BCM-MGL for the aCGH study; A. McCall, D. Walker, M. Strivens and M. Rao for technical assistance; M. Sheng, W. Dobyns, D. Picketts, R. Gibbons, S. Dindot, A. Beaudet, D. Nelson, S-K. Lee, B. Franco, and I. Bezprozvanny for antisera and expression constructs; M. Sardiello, C. Schaaf, M. Costa-Mattioli and J. Neul for critical comments; C. Schaaf for computing IQ averages of the SSC patients used in this study; V. Brandt for comments on the manuscript; and H.Y.Z. lab members for helpful discussions.
Funding: Supported by the HHMI (H.Y.Z.), the Simons Foundation (H.Y.Z.), and the Ellison Foundation (D.E.H.; awarded to M. Vidal).
Footnotes
Author contributions: Y.S., C.A.S. and H.Y.Z. designed the study, evaluated the data, and wrote the manuscript. Y.S. performed all the experiments with technical support for the Y2H screen by Z.A-M. C.A.S., B.C.D and D.V.D conducted the bioinformatic analysis. D.E.H. provided the ORFeome clone and edited the manuscript.
REFERENCES AND NOTES
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 5.The European Chromosome16 Tuberous Sclerosis Consortium, Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 6.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JB, Ward S, Green AJ, Yates JR, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 7.Phelan MC, Thomas GR, Saul RA, Rogers RC, Taylor HA, Wenger DA, McDermid HE. Cytogenetic, biochemical, and molecular analyses of a 22q13 deletion. Am J Med Genet. 1992;43:872. doi: 10.1002/ajmg.1320430524. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 12.Beaudet AL. Autism: highly heritable but not inherited. Nat Med. 2007;13:534. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- 13.Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135:396. doi: 10.1016/j.cell.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 15.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A, Owley T, Sweeney JA, Brune CW, Cantor RM, Bernier R, Gilbert JR, Cuccaro ML, McMahon WM, Miller J, State MW, Wassink TH, Coon H, Levy SE, Schultz RT, Nurnberger JI, Haines JL, Sutcliffe JS, Cook EH, Minshew NJ, Buxbaum JD, Dawson G, Grant SF, Geschwind DH, Pericak-Vance MA, Schellenberg GD, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009 doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009 doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY, Venkatesan K, Hirozane-Kishikawa T, Dricot A, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Roth FP. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 19.Al-Ramahi I, Perez AM, Lim J, Zhang M, Sorensen R, de Haro M, Branco J, Pulst SM, Zoghbi HY, Botas J. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007;3:e234. doi: 10.1371/journal.pgen.0030234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, Zoghbi HY. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 21.Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 23.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, Venkatesan K, Rual JF, Vandenhaute J, Cusick ME, Pawson T, Hill DE, Tavernier J, Wrana JL, Roth FP, Vidal M. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7:667. doi: 10.1038/nmeth0910-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 31.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 32.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 33.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 39.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Fink JM, Dobyns WB, Guerrini R, Hirsch BA. Identification of a duplication of Xq28 associated with bilateral periventricular nodular heterotopia. Am J Hum Genet. 1997;61:379. doi: 10.1086/514863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, Richman R, Fang P, Glaze DG, Lupski JR, Zoghbi HY. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann Neurol. 2009;66:771. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27:5546. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bangash MA, Park JM, Melnikova T, Wang D, Jeon SK, Lee D, Syeda S, Kim J, Kouser M, Schwartz J, Cui Y, Zhao X, Speed HE, Kee SE, Tu JC, Hu JH, Petralia RS, Linden DJ, Powell CM, Savonenko A, Xiao B, Worley PF. Enhanced Polyubiquitination of Shank3 and NMDA Receptor in a Mouse Model of Autism. Cell. 2011 doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 49.Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, Bonin M, Riess A, Engels H, Sprengel R, Scherer SW, Rappold GA. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 50.Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ljungberg MC, Sunnen CN, Lugo JN, Anderson AE, D’Arcangelo G. Rapamycin suppresses seizures and neuronal hypertrophy in a mouse model of cortical dysplasia. Dis Model Mech. 2009;2:389. doi: 10.1242/dmm.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niu S, Yabut O, D’Arcangelo G. The Reelin signaling pathway promotes dendritic spine development in hippocampal neurons. J Neurosci. 2008;28:10339. doi: 10.1523/JNEUROSCI.1917-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhar SU, del Gaudio D, German JR, Peters SU, Ou Z, Bader PI, Berg JS, Blazo M, Brown CW, Graham BH, Grebe TA, Lalani S, Irons M, Sparagana S, Williams M, Phillips JA, 3rd, Beaudet AL, Stankiewicz P, Patel A, Cheung SW, Sahoo T. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am J Med Genet A. 2010;152A:573. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, Eng P, Li J, Gu Y, Stratton RF, Chinault AC, Shaw CA, Sutton VR, Cheung SW, Nelson DL. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet A. 2007;143A:1358. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Validation of the interaction data from the Y2H study by co-affinity purification experiments in HEK293T cell
Fig S2. Syndromic ASD proteins are highly connected to each other and have the shortest path length in the protein interaction network
Fig S3. The GO analysis for the extended ASD network
Fig S4. Chromosomal location of the overlapping genes between the protein interaction network and CNV intervals of ASD group
Fig S5. Confirmation of de novo or inherited CNVs
Fig S6. Physical interaction of FLNA with SHANK3 in vivo
Table S1. Summary of yeast-two-hybrid screening for ASD and related disorders
Table S2. The summary of protein interaction data obtained from Y2H screening and Co-AP study
Table S3. Summary of known interaction data in the literature and the overlap with this study
Table S4. Co-expression pattern of the genes in the mouse hypothalamus, cerebellum and amygdala
Table S5. Summary of the Probands with Novel CNVs