Abstract
Members of the myc family of cellular oncogenes have been implicated as transcriptional regulators in pathways that govern cellular proliferation and death. In addition, N-myc and c-myc are essential for completion of murine embryonic development. However, the basis for the evolutionary conservation of myc gene family has remained unclear. To elucidate this issue, we have generated mice in which the endogenous c-myc coding sequences have been replaced with N-myc coding sequences. Strikingly, mice homozygous for this replacement mutation can survive into adulthood and reproduce. Moreover, when expressed from the c-myc locus, N-myc is similarly regulated and functionally complementary to c-myc in the context of various cellular growth and differentiation processes. Therefore, the myc gene family must have evolved, to a large extent, to facilitate differential patterns of expression.
Keywords: N-myc, c-myc, murine development, proliferation, knock-in, gene replacement
The myc family of cellular oncogenes is comprised of c-myc, N-myc, and L-myc, three related genes that function in regulation of cellular proliferation, differentiation, and apoptosis (Henriksson and Luscher 1996; Facchini and Penn 1998). A central aspect of Myc oncoprotein function is the capacity to act as a sequence-specific transcription factor governing the regulation of target genes integral to these processes (Dang 1999). Myc family members share gene and protein structural features (Henriksson and Luscher 1996). On the biological level, all three myc family genes can cooperate with a mutant H-RAS gene to transform early passage rat embryo fibroblasts and can generate tumors when overexpressed in transgenic mice (Dang et al. 1999). The use of basic region swap and dominant-negative mutant forms of Myc has shown that Myc family members function through common pathways to transform cells (Mukherjee et al. 1992; Amati et al. 1993; O'Hagan et al. 2000).
Although myc family genes have common structural and transforming features, it is difficult to assess physiological functions based on the limited comparisons that have been done. Several lines of evidence suggest that Myc family members have separable physiological functions. Myc family members are conserved as distinct genes over large phylogenetic distances (Gallant et al. 1996; King et al. 1986, 1993; Schreiber-Agus et al. 1993a,b, 1997). For example, mouse N-myc is more similar to Xenopus N-myc (65% sequence identity), than it is with mouse c-myc (35% sequence identity) (DePinho et al. 1986; Schreiber-Agus et al. 1993). In addition, myc family members have distinct, albeit overlapping, expression patterns with respect to cell types and developmental stages (Zimmerman et al. 1986; Downs et al. 1989). All three myc family transcripts are detectable in preimplantation embryos and show more spatially restricted expression patterns with advancing development (Mugrauer and Ekblom 1991; Stanton et al. 1992; Hatton et al. 1996). At midgestation, c-myc transcripts are broadly distributed; whereas N-myc and L-myc transcripts are abundant in particular organs such as brain and kidney (Schmid et al. 1989). Notably, high level c-myc expression occurs in compartments with high proliferative activity; whereas N-myc and L-myc expression is prominent in postmitotic cells undergoing differentiation (Hirvonen et al. 1990). These expression patterns suggested c-Myc may be particularly important for cellular proliferation, though N-Myc and L-Myc functions may be more relevant for differentiation (Mugrauer et al. 1988; Mugrauer and Ekblom 1991; Morgenbesser and DePinho 1994), a notion supported by transgenic gain-of-function studies (Morgenbesser et al. 1995).
Homozygous null c-Myc and N-Myc mice die at about embryonic days 10 and 12, respectively. The phenotypic consequences c-Myc deficiency included a marked reduction in embryo size and a generalized delay in the early development of many organs, including the heart and neural tube (Davis and Bradley 1993; Davis et al. 1993). N-Myc-deficient embryos also exhibited delayed development and stunted growth, as well as diminished cellularity in organs that normally express abundant N-myc transcripts, most notably the cranial and spinal ganglia, mesonephros, lung, and gut (Charron et al. 1992; Stanton et al. 1992; Sawai et al. 1993). Significantly, the defects associated with N-Myc deficiency occurred despite compensatory c-Myc increases (Stanton et al. 1992), suggesting a unique and essential role for N-Myc. In contrast, L-Myc-deficient mice lacked a discernable phenotype, indicating that this gene is largely dispensable, perhaps because of redundant expression and activities of c-myc and N-myc (Hatton et al. 1996). Redundant functional properties among Myc family members during early embryonic development was proposed to explain survival of individual Myc family gene knockout mice through early embryonic stages (Charron et al. 1992; Stanton et al. 1992; Davis et al. 1993; Sawai et al. 1993). This notion is also consistent with the requirement for Max, the obligate partner of all three Myc proteins, for postimplantation growth and development (Shen-Li et al. 2000).
To address directly the question of functional complementation among Myc family members, we have used gene targeting to generate mice in which the endogenous c-myc coding sequences have been replaced with N-myc coding sequence. Remarkably, our analyses of these c-Myc-deficient mice indicate that N-myc is capable replacing most of the essential c-myc functions required for embryonic development and for proliferation of differentiated cells.
Results
N-myc expression from the c-myc locus rescues embryonic lethality associated with c-myc deficiency
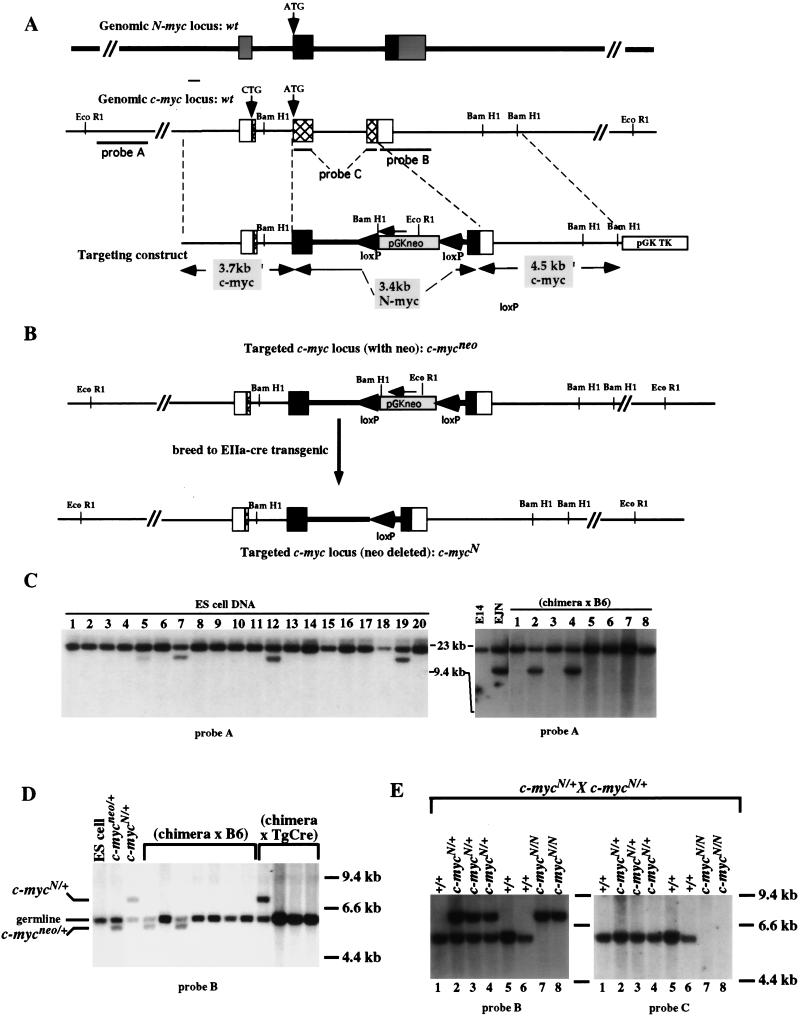
The genomic structures of c-myc and N-myc are similarly organized and are comprised of three exons. Most of the first exon and the 3′ portion of the third exon contain untranslated regions that carry transcriptional or post-transcriptional regulatory sequences (Facchini and Penn 1998). Thus, the targeting construct was designed to replace the entire second exon, the coding portion of the third exon, and the intervening intron of c-myc with the analogous sequences from N-myc (Fig. 1A). It should be noted that the 3′ untranslated region, a region of sequence divergence between mouse N-myc and c-myc genes that has been implicated as a regulator of RNA stability, was retained in the chimeric knock-in gene (Yeilding et al. 1996). In the targeted c-myc allele generated by this construct, the second intron contains the pGKneo gene in the opposite transcriptional orientation (designated c-mycneo) (Fig. 1A,B). Appropriately targeted ES cells were injected into blastocysts and chimeric mice were bred to obtain germ-line transmission of the c-mycneo allele (Fig. 1C). Heterozygous offspring were bred and analyzed for inheritance of c-mycneo alleles. No live-born homozygous c-mycneo/neo mice were found (Table 1). It is likely that the presence of the intronic neor gene interferes with expression of the targeted alleles, leading to the embryonic lethality associated with nullizygous c-myc mice as reported previously (Davis et al. 1993).
Figure 1.
Targeted replacement of c-myc coding sequences with N-myc coding sequences. (A) Schematic diagrams of the intron–exon structures of the genomic N-myc and c-myc loci and the N- into c-myc replacement construct. The majority of the coding sequences of N-myc (black boxes) and c-myc (hatched boxes) are contained within exons 2 and 3. The 5′ and 3′ untranslated regions are represented as shaded boxes for N-myc and open boxes for c-myc. In the targeting construct, called pNCR11, N-myc exons 2 and 3 and the intervening intron are flanked by 5′ and 3′ c-myc homology arms. PGKneo, surrounded by loxP sites, was inserted within the N-myc intron. (B) Schematic diagrams of the targeted c-myc loci before and after Cre-mediated recombination. pNCR11 was transfected into embryonic stem cells and screened for homologous recombination into the c-myc locus. A schematic of the structure of the targeted locus, called c-mycneo, is shown. Mice derived from the targeted ES cells were bred to transgenic mice carrying the EIIa–cre transgene and offspring were screened for transmission of the Cre-mediated recombination, resulting in deletion of the neo gene. A schematic of the structure of the targeted locus, called c-mycN, is shown. (C) Southern blot analysis for germ-line transmission of the c-mycneo targeted locus. Targeting of ES cells was screened by digestion of DNA with EcoRI and hybridization to probe A (left, lanes 5,7,12,19). Appropriate targeting was confirmed by digestion with multiple restriction enzymes and probes (data not shown). Targeted ES cells were injected into blastocysts and chimeric mice were bred either to C57BL/6 or 129Sv/Ev mice. Offspring were tested for germ-line transmission by digesting tail DNA samples with EcoRI and hybridizing to probe A (right, lanes 2,4; data not shown). (D) Southern blot analysis for Cre-mediated deletion of the neo gene to produce c-mycN/N mice. c-mycneo-transmitting chimera were bred to EIIa–cre transgenic mice. Tail-DNA samples were analyzed by digestion with BamHI and Southern blots were hybridized to probe B. The germ line BamHI fragment hybridizing to probe B is ∼6.0 kb; the c-mycneo allele results in a ∼5.5-kb fragment, because of inserted BamHI site in the PGKneo sequence. After Cre-mediated deletion of the PGKneo gene, the elimination of this BamHI site results in a probe B-hybridizing fragment of ∼7.0 kb. (E) Mating of heterozygous c-mycN/+ mice yielded homozygous mice that survived in the absence of a c-myc gene. Tail DNA from 10-day-old offspring of a c-mycN/+ heterozygous mating was digested with BamHI and Southern blots were hybridized to probe B (left). The diagnosed genotypes are designated above each lane and included wild type (c-myc+/+), heterozygous (c-mycN/+), and homozygous (c-mycN/N) (left). The Southern blot was stripped and hybridized to probe C. DNA from c-mycN/N mice failed to hybridize to this probe, which is composed of both coding exons of c-myc DNA, proving that these mice are unable to produce c-myc-encoded transcripts (right).
Table 1.
Inheritance of c-mycneo and c-mycN alleles
|
|
Age
|
Total no.
|
Wild type
|
Heterozygous
|
Homozygous
|
|---|---|---|---|---|---|
| c-mycneo | ≥10 days | 74 | 27 | 47 | 0 |
| c-mycN | ≥10 days | 274 | 92 | 152 | 30 |
| day 1 | 135 | 35 | 74 | 26a | |
| 14 dpc | 39 | 9b | 19 | 8b |
One of these 26 newborns was found dead, but had been born alive as evidenced by the presence of milk in the stomach.
One of 9 wild type and 2 of 8 homozygous embryos were in the process of being reabsorbed.
Transmitting c-mycneo chimera were bred to EIIa–cre transgenic mice to delete the neor gene in germ cells, via expression of Cre-recombinase at early embryonic stages (Fig. 1B) (Lakso et al. 1996). Offspring that transmitted the Cre-mediated neor gene deletion (allele designated c-mycN) (Fig. 1D) were subsequently bred to wild-type mice and screened for loss of the EIIa–cre transgene to ensure germ-line transmission of the c-mycN allele (data not shown). Strikingly, c-mycN/+ intercrosses produced live homozygous offspring (Fig. 1E, left). DNA and RNA isolated from these c-mycN/N offspring failed to hybridize to a probe specific for c-myc coding exons 2 and 3, confirming that c-mycN/N mice survived in the complete absence of c-Myc protein expression (Fig. 1E, right, and Fig. 2).
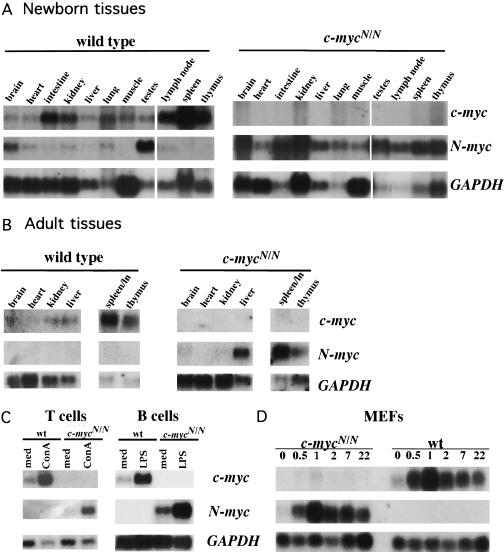
Figure 2.
Expression of c-mycN allele. (A,B) Expression of N-myc and c-myc transcripts in wild-type and c-mycN/N mice. Replicate northern blots of total RNA, extracted from tissues of adult (B) or newborn (A) wild-type and homozygous knock-in mice, were hybridized to probes specific for the c-myc or N-myc coding exons 2 and 3. Blots were subsequently hybridized to GAPDH to control for RNA loading. (C) Mitogen-stimulated up-regulation of N-myc and c-myc genes in lymphocytes. B or T lymphocytes, enriched from spleen or lymph nodes of wild-type or c-mycN/N mice, were incubated in vitro with or without stimulation with LPS or ConA, respectively. RNA was extracted, electrophoresed, and replicate northern blots were hybridized as above. (D) Serum-stimulated up-regulation of N-myc and c-myc genes in MEFs. Subconfluent MEFS were cultured in low (0.5%) serum for 48 hr. Media with 10% serum was added at time 0 and cells were harvested for RNA extraction at various times thereafter and electrophoresed. Replicate Northern blots were hybridized as above.
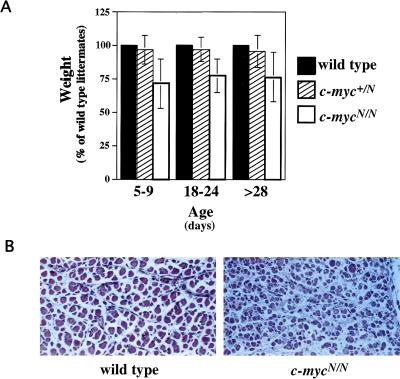
The c-mycN/N mice appeared normal and healthy. Both male and female mutants were fertile when bred with wild-type mice (data not shown). The only difference in appearance between c-mycN/N and their wild-type or heterozygous control littermates was that c-mycN/N newborn and adult mice were slightly smaller in size and average weight as compared to controls (Fig. 3A). Of note, the number of c-mycN/N mice was significantly less than that predicted by Mendelian inheritance, suggesting that the rescue is not complete (Table 1). When mice were analyzed immediately after birth, however, Mendelian inheritance was observed and indicated that a fraction of homozygotes died within the first two days of life (Table 1). Histological analyses of embryos and newborn mice showed that development of most organs—including those that normally express relatively high levels of c-myc (e.g., intestine, kidney, liver)—appeared normal (data not shown). However, detailed histological inspection of newborn mice revealed periodic dystrophy of skeletal muscles in half of the mice analyzed (Fig. 3B). Observation of cyanosis in two c-mycN/N neonates raises the possibility that muscular compromise adversely affects breathing, perhaps explaining the decreased survival.
Figure 3.
Potential differences between c-mycN/N and wild-type mice. (A) The average weight of c-mycN/N mice. Mice from individual litters were weighed at various times after birth. For each litter, the weights of heterozygous or homozygous mutant mice were calculated as a percentage of the average weight of wild-type littermates. The mean (±s.d.) percentage of heterozygous and homozygous mutant mice was calculated. The mean weight of c-mycN/N mice was significantly different from that of wild-type or heterozygous mice (P < 0.001). (B) Histological analysis of c-mycN/N mice. Sections of skeletal muscle from wild-type (left) or homozygous c-mycN/N (right) day 1 newborns are shown. The appearance of muscular dystrophy periodic and observed in 1/2 of the homozygotes.
The ability of the c-mycN allele to rescue the bulk of the c-myc-dependent developmental program would predict that the c-mycN allele should result in N-myc expression in tissues and developmental stages in which c-myc, and not N-myc, expression normally predominates (Zimmerman et al. 1986). In accord with this prediction, Northern blot analysis of RNA from newborn and adult c-mycN/N mice revealed N-myc expression in tissues of c-mycN/N mice (notably, intestine, kidney, muscle, and lung), where c-myc is normally expressed but little or no N-myc expression is normally found (Fig. 2A,B). These findings indicate that the c-myc pattern of developmental and tissue-specific RNA expression from the N-myc-replaced c-myc locus is largely intact (Fig. 2A,B).
Normal development and classical mitogen responses of c-mycN/N lymphocytes
The c-myc and N-myc genes are differentially expressed during lymphocyte development, with c-myc expressed throughout development, whereas N-myc expression is restricted to precursor stages (Zimmerman et al. 1986). Therefore, to assay for potential effects of the c-mycN/N genotype in detail within a specific cellular lineage, we analyzed lymphocyte development in the c-mycN/N mice. Gross size and cellularity of lymphoid organs in c-mycN/N mice were normal (data not shown). In addition, the percentages of T and B lymphocyte precursor populations in the thymus and bone marrow were normal, as were those of mature lymphocytes in the peripheral lymph nodes and spleen (Fig. 4A; data not shown). These data demonstrate that N-Myc can replace all presumptive c-Myc functions required for lymphocyte development.
Figure 4.
Development and proliferation of c-mycN/N lymphocytes. (A) Flow cytometric analysis of lymphocytes from wild-type and homozygous c-mycN/N mice. Single cell populations isolated from thymus (top left), spleen (right), and bone marrow (bottom left) from wild-type or homozygous c-mycN/N mice were stained with a panel of antibodies that identify various subpopulations of T- or B-lineage lymphocytes, as indicated. Live cells were gated based on forward and side-scatter profile. The percentage of gated cells in particular quadrants or boxes are shown. Similar percentages of cell subpopulations were observed for homozygous c-mycN/N and wild-type mice. Absolute numbers of cells in the various fractions were also calculated. No significant differences were found (data not shown). (B,C,D,E) In vitro proliferation of wild-type and homozygous c-mycN/N lymphocytes. T (B,C) and B lymphocytes (D,E) were enriched from lymph node and spleen, respectively, of homozygous c-mycN/N or wild-type littermates and cultured either with medium or various doses of ConA ± IL-2 (B), anti-CD3 ± IL-2 (C), LPS (D), or anti-Ig ± IL-4 (E). The incorporation of 3H-thymidine was measured and indicated as counts per minute.
Mitogenic responses of normal lymphocytes are tightly correlated with c-myc expression (Heikkila et al. 1987; Kelly and Siebenlist 1988; Neckers et al. 1992). To test mitogenic responsiveness, purified T cells were stimulated in vitro by either ConA treatment or via receptor cross-linking with anti-CD3 in the presence or absence of interleukin-2 (IL-2). Purified B cells were stimulated either by lipopolysacharride (LPS) treatment or by receptor cross-linking with anti-Ig in the presence or absence of IL-4. As expected, c-myc, but not N-myc, transcripts were induced in mitogen-stimulated wild-type cells within 3 hr, and no c-myc coding region transcripts were detected in c-mycN/N lymphocytes (Fig. 2C). In contrast, N-myc coding region transcripts were highly induced in mitogen-stimulated B and T cells from c-mycN/N as opposed to wild-type mice (Fig. 2C). Correspondingly, c-mycN/N B and T cells proliferated in response to either mitogen treatment or receptor cross-linking, and these responses were augmented by appropriate lymphokines (Fig. 4B–E). However, responses of c-mycN/N T cells to higher doses of mitogen or antigen receptor stimulation, and responses of c-mycN/N B cells to all doses, appeared slightly dampened compared to wild-type (Fig. 4B–E). Overall, however, these data demonstrate that N-myc can largely compensate for c-myc in lymphocyte development and activation.
Growth potential of embryonic fibroblasts from c-mycN/N mice
To further test the ability of N-myc to compensate for c-myc, we examined the mitogenic responses of mutant (c-mycN/N) and wild-type mouse embryonic fibroblasts (MEFs) following serum stimulation. MEFs derived from 13.5-day-old embryos were serum-starved for 48 hr and RNA was extracted from cells prior to serum stimulation and at multiple time points thereafter. In wild-type MEFs, c-myc transcripts exhibited a characteristic response, high-level induction within 1 hr followed by a plateau at a lower level, which remained higher than initial background levels for up to 24 hr (Fig. 2D). Although N-myc transcripts were undetectable in normal embryonic fibroblasts, serum stimulation of c-mycN/N MEFs induced N-myc transcripts with a profile that closely mimicked that of c-myc in wild-type MEFs (Fig. 2D).
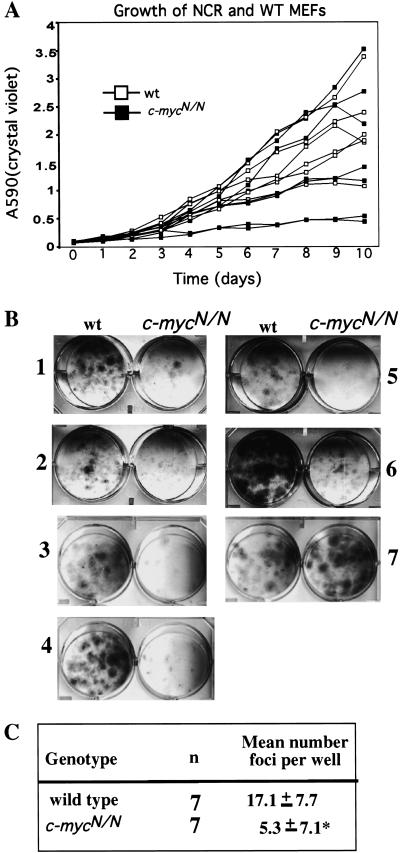
The integrity of the c-myc gene is required for the normal proliferative response of fibroblasts (Mateyak et al. 1997; Bush et al. 1998; Moreno de Alboran et al. unpubl.). To assess relative proliferative potential, we monitored growth rates for multiple independently derived MEF lines from c-mycN/N and wild-type littermates. There was typical variation in growth rates among individual wild-type MEF cultures, and several c-mycN/N cultures grew at least as well as those from wild-type MEFs (Fig. 5A). To further compare growth and survival profiles more stringently, we assayed ability of c-mycN/N and wild-type MEFS to form colonies after seeding at low density. The c-mycN/N MEFs displayed significant colony forming ability by this assay (which again has substantial variation even among wild-type MEFs); however, the colony-forming potential of c-mycN/N MEFs did appear, on average, reduced in comparison to that of wild type (Fig. 5B,C). Finally, we employed flow cytometry to determine whether N-Myc could compensate for c-Myc in maintenance of the cell size of G0/G1 MEFs. No significant cell size differences were detected between wild-type and c-mycN/N MEFs (data not shown). Together, these results demonstrate that N-Myc can largely replace c-Myc functions in regulating cell growth and proliferation of cultured fibroblasts.
Figure 5.
Growth potential of wild type and homozygous c-mycN/N mouse embryonic fibroblasts. (A) P3 MEFS were plated at 5 × 104 cells per well in replicate wells and cultured for the indicated number of days. Triplicate wells were fixed and stained with crystal violet. Adsorption at OD540 was measured for each time point for individual MEF lines and directly correlates with cell numbers (Kamijo et al. 1997). Each symbol represents the mean of triplicate readings of triplicate wells. The standard deviation was <1%. (B,C) P3 MEFS were plated at 3500 cells per well in triplicate, cultured for 12 days, and fixed and stained with crystal violet. A representative plate is shown. The number of foci per well was determined, the mean (±s.d.) was calculated for each cell line. The mean number of foci per well from c-mycN/N MEFs was significantly less than that of wild-type MEFs (*P < 0.01).
Discussion
N-myc can support most c-myc functions necessary for normal murine growth and development
The most dramatic result from our study is that N-myc is able to rescue the essential role of c-Myc in embryonic development and survival. Strikingly, c-mycN/N mice, which completely lack any c-myc coding sequences (or protein), survive into adulthood and are capable of reproduction. With the single exception of the skeletal muscle, no gross defects were consistently found in c-mycN/N mice and detailed examinations showed that lymphocyte development closely resembled that of normal mice. It was also notable that N-myc was largely able to replace c-myc in lymphocyte and fibroblasts with respect to functions related to growth and proliferation. Correspondingly, the expression pattern of N-myc transcripts from the c-mycN allele closely mimicked that of the normal c-myc locus, both with respect to tissue and spatial regulation during embryonic development, as well as with respect to induction in lymphocytes and fibroblasts by a variety of treatments.
Overall, our findings clearly demonstrate that the c-myc transcriptional control elements and 3′ untranslated sequences are sufficient to drive expression of N-myc coding exons in a fashion similar or identical to that of c-myc. Moreover, our results indicate that the differing physiological roles of c-myc and N-myc relate primarily to differences in transcriptional regulation of the genes, rather than distinct biochemical properties of the N-Myc and c-Myc proteins. This interpretation provides an explanation for the survival of c-myc- or N-myc-deficient embryos until midgestation days E9–E11, as this is the time at which myc family gene expression patterns become most divergent (Charron et al. 1992; Stanton et al. 1992; Davis et al. 1993; Sawai et al. 1993). Very recent gene replacement studies also have demonstrated bidirectional complementation of the hox gene paralogs, hoxa3 and hoxd3, leading to the suggestion that the distinct biological functions of these genes results from quantitative differences in expression (Greer et al. 2000).
Apparent differences between c-mycN/N and normal mice and cells
The overall survival of postnatal c-mycN/N mice is modestly reduced compared with wild-type mice. Extensive histologic examination of c-mycN/N embryos and newborn animals revealed periodic dystrophy of skeletal muscles in a subset of the newborn c-mycN/N mice. In addition, we did observe relatively mild, but significant, differences in the average weight of c-mycN/N animals as compared with controls. Moreover, we observed that the c-mycN allele might not fully compensate for the loss of c-myc expression with respect to certain cellular growth responses in lymphocytes or fibroblasts. The skeletal muscle, animal weight, and cell growth response phenotypes were observed despite the fact that c-mycN transcripts were expressed in a c-Myc-like pattern in all organs tested and in cultured lymphocytes and MEFs in response to various growth stimuli. The apparent differences observed between c-mycN/N and normal cells and mice may reflect subtle differences in function of the c-Myc and N-Myc proteins, such as quantitative differences in the activity of their transactivation domains (Barrett et al. 1992). It will be interesting to combine the c-mycN allele with a c-mycnull allele to potentially reveal a more significant phenotype. Alternatively, there may be subtle differences in the steady-state RNA or protein levels generated from the manipulated locus. In either case, the variability in survival of c-mycN/N mice, along with the range of growth and immortalization responses of c-mycN/N MEFs, may implicate the existence of genetic factors that affect the ability of N-myc to compensate for c-myc loss.
Functional redundancy of myc family genes
Our studies clearly demonstrate a major degree of functional redundancy between the N-Myc and c-Myc proteins with respect to most processes in which c-Myc has been implicated. Functional redundancy has also been observed for other gene families, including those encoding Hox, Engrailed, and myogenic regulatory proteins (Hanks et al. 1995; Wang et al. 1996; Greer et al. 2000). The expression patterns of Hox gene paralogs are highly overlapping; whereas differences in expression among members of the engrailed and myogenic families are temporal and largely restricted to one tissue. In contrast, the c-myc and N-myc genes have broader and more divergent expression patterns during particular times in development and inactivation of either gene resulted in adverse developmental consequences that correlated with their specific expression patterns. The high degree of functional redundancy among the c-Myc and N-Myc proteins is particularly notable given their preservation as distinct genes throughout vertebrate evolution. The N-myc genes in mouse and Xenopus are more closely related than are mouse N-myc to mouse c-myc (DePinho et al. 1986; Schreiber-Agus et al. 1993). However, there are short stretches of higher homology (Myc homology boxes) that encode the transactivation, DNA binding, and protein dimerization domains. Our data prove that these are critical functional domains of Myc proteins for the growth and development of diverse cell types and organ systems. In this regard, our findings strongly suggest that the evolutionary maintenance of N-myc and c-myc as distinct genes has been driven largely on the basis of the distinct cis-regulatory elements that impart their specific expression patterns.
Materials and methods
Generation of c-mycneo/neo and c-mycN/N mice
The PGKneor gene flanked by loxP sites was subcloned from pLNTKneor (Gorman et al. 1996) and cloned in opposite transcriptional orientation into the BglII site within the N-myc intron of pEVNH3X (Dildrop et al. 1989). pEVNH3X had previously been constructed to contain the genomic fragment of N-myc exon 2 flanked by an ATG and the c-myc exon 2 splice acceptor site, the coding portion of N-myc exon 3, and the intervening N-myc intron on an XbaI fragment (Dildrop et al. 1989). This XbaI fragment was subcloned into the XbaI site of the pNEB193 vactor, into which the PGKTK gene had been cloned previously. A 4.5-kb XhoI–BglII genomic c-myc fragment that contains the 3′ untranslated portion of the c-myc third exon was cloned into the PmeI polycloning site (PCS), fusing it immediately downstream of the coding portion of the N-myc gene. The resulting product is a chimeric third exon in which the coding sequence is N-myc and the noncoding is c-myc. The 5′ c-myc flanking region is a 3.7-kb XbaI genomic fragment that contains the c-myc first exon cloned into the PacI site of the PCS. In the final spliced mRNA product from this chimeric gene, the entire coding region is N-myc sequence and the 5′ and 3′ flanking sequences are derived from c-myc.
The targeting vector was transfected into E14.1 ES cells (kindly provided by R. Murray) by electroporation, selected in 0.4 mg/ml G418 and 1 mm gancyclovir (kind gift from Syntex Corp.), as described previously (Charron et al. 1992). Homologous recombinants (c-mycneo) were identified by Southern blot analysis of EcoRI-digested DNA, probed with a 1.7-kb XbaI fragment (probe A, Fig. 1A). Homologous recombinants were found at a frequency of 10%–20%. Targeted ES cells were screened with multiple restriction enzymes and probes to ensure that the 5′ and 3′ ends were integrated appropriately. Chimeric male mice were initially bred to C57BL/6 mice and germ-line transmitters were identified by coat color and confirmed by BamHI digest of tail DNA. To delete the PGKneor gene, offspring were bred to EIIa–cre transgenic mice (Lakso et al. 1996). Tail DNA was screened by digestion with BamHI and hybridization to a 3′ flank c-myc probe (1.4-kb XhoI–KpnI fragment, probe B, Fig. 1A). Deletion of the PGKneor gene results in the loss of a BamHI site, leading to a change in the size of the hybridizing fragment from 6 kb to 7 kb. Germ-line transmission of the neo-deleted allele (called c-mycN) was confirmed by breeding to wild-type mice and screening for the absence of the Cre transgene by PCR, as described previously (Gorman et al. 1996)
Southern and Northern blotting
Genomic DNA was prepared as described previously (Laird et al. 1991). RNA was prepared from tissues by the Urea/LiCl2 method (Auffray and Rougeon 1980) or from cell pellets using homogenization in Trizol reagent (GIBCO BRL), as described by the manufacturer. Southern and Northern blot analyses were carried out as described elsewhere. DNA samples were electrophoresed through a 1% agarose gel. Total RNA (∼5–10 μg) was electrophoresed through a formaldehyde/1% agarose gel. DNA or RNA were transferred to Zetaprobe membrane (Bio-Rad) and hybridized to probes labeled by random hexamer priming (Boehringer-Mannheim) with [α-32P]dCTP (NEN). The probes used for Southern blot analyses are shown in Figure 1. Probe A is a 5′ flank genomic c-myc 1.7-kb XbaI fragment; probe B is a 1.4-kb genomic fragment (XhoI–KpnI) that contains the 3′ untranslated of c-myc and hybridizes with the 3′ flank; probe C is a 1.5-kb (PstI) c-myc cDNA fragment that hybridizes to exons 2 and 3. Northern blots were hybridized to either probe C (c-myc cDNA), an N-myc exon 2/3 cDNA fragment, or a 1.25-kb (PstI) GAPDH cDNA fragment.
Histological analyses
Tissues were fixed in Bouin's solution, paraffin embedded, and serially sectioned (5 μm/each section). The sections were then dewaxed in xylene, rehydrated in alcohol of various concentrations, and stained with hematoxylin and eosin following a standard protocol.
FACS analysis
Single cell suspensions from thymus, spleen, and bone marrow were prepared as described (Malynn et al. 1995). Cells in PBS plus 5% FCS were stained for surface expression of the following markers: FITC-conjugated anti-CD8a (53-6.7), PE-conjugated anti-CD4 (RM 4-4), either FITC- or CyC-conjugated anti-B220/CD45R (Ra3-6B2), or FITC-conjugated anti-CD43 (Pharmingen). FACS analysis of the cells was acquired and analyzed on a FACSCaliber flow cytometer (Becton Dickinson) using CellQuest software (Becton Dickinson). Data are presented as dot plots after gating for live cells, using forward versus side scatter plots.
Lymphocyte proliferation assays
Single-cell suspensions of spleen (for B cells) or lymph node (for T cells) cells were prepared as described previously (Malynn et al. 1995). B or T cells were purified using magnetic beads (DYNAL) and purity (∼85%–95%) was determined by FACS analysis. Subsequently, cells were cultured in 96-well plates at a concentration of 5 × 105 cells/ml in RPMI/10% FCS medium (supplemented with 50 mm β-mercaptoethanol). At the initiation of culture, Concanavalin A (Sigma) or anti-CD3 (Pharmingen) to T cells or lipopolysaccharide (Sigma) or Fab goat anti-IgM to B cells were added at the indicated concentrations. Cultures were incubated for a total of 60–72 hr with a 3H-thymidine pulse for the last 8 hr. Cells were harvested and CPM incorporated was determined. The mean (±s.d.) of triplicate cultures is shown. One of three representative experiments is shown.
MEF growth assays
MEFs were isolated from individual 13.5-day-old embryos. For growth curves early passage MEFs were seeded at 5 × 104 cells per 60-mm dish. At the indicated times triplicate plates of cells were washed, fixed in 10% buffered formalin, and stained with crystal violet for 30 min at room temperature. Stained cells were washed in PBS, and stain was extracted using 10% acetic acid. Absorbance was read at 590 nm. For low density seeding assays early passage MEFs were plated at 3500 cells/well in 6-well plates and cultured in DME containing 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine for 15 days. Plates were washed, fixed in 10% buffered formalin, and stained with crystal violet. The mean (±s.d.) number of colonies per well of triplicate cultures was determined.
Acknowledgments
We thank Drs. Averil Ma, Mary Donohoe, and Jean Charron for helpful advice and discussion. We also thank Lawrence Brown, Jonathan Levine, Harrison Lin, William Tran, and Margaret Neuberger for technical assistance and Sherrie Witt and Li Zhang of the Rodent Histopathology Laboratory of the Dana-Farber Harvard Cancer Center for histology sections. This work was supported by the Arthritis Foundation, the Leukemia Society of America, and NIH grants CA61009 (to B.A.M.), HD28317 and EY09300 (to R.A.D.), and AI35714 (to F.W.A.). R.C.O. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. R.A.D. is an American Cancer Society Research Professor. F.W.A. is an Investigator and I.M.A. is an Associate of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL alt@rascal.med.harvard.edu; FAX (617) 355-3432.
References
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Barrett J, Birrer MJ, Kato GJ, Dosaka-Akita H, Dang CV. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992;12:3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, Cole M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes & Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes & Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- Davis A, Bradley A. Mutation of N-myc in mice: What does the phenotype tell us? Bioessays. 1993;15:273–275. doi: 10.1002/bies.950150408. [DOI] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes & Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- DePinho RA, Legouy E, Feldman LB, Kohl NE, Yancopoulos GD, Alt FW. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci. 1986;83:1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildrop R, Ma A, Zimmerman K, Hsu E, Tesfaye A, DePinho R, Alt FW. IgH enhancer-mediated deregulation of N-myc gene expression in transgenic mice: Generation of lymphoid neoplasias that lack c-myc expression. EMBO J. 1989;8:1121–1128. doi: 10.1002/j.1460-2075.1989.tb03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes & Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, Horner J, DePinho RA. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- Henriksson M, Luscher B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Hirvonen H, Makela TP, Sandberg M, Kalimo H, Vuorio E, Alitalo K. Expression of the myc proto-oncogenes in developing human fetal brain. Oncogene. 1990;5:1787–1797. [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kelly K, Siebenlist U. Mitogenic activation of normal T cells leads to increased initiation of transcription in the c-myc locus. J Biol Chem. 1988;263:4828–4831. [PubMed] [Google Scholar]
- King MW, Roberts JM, Eisenman RN. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol Cell Biol. 1986;6:4499–4508. doi: 10.1128/mcb.6.12.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MW, Blackwood EM, Eisenman RN. Expression of two distinct homologues of Xenopus Max during early development. Cell Growth Differ. 1993;4:85–92. [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Demengeot J, Stewart V, Charron J, Alt FW. Generation of normal lymphocytes derived from N-myc-deficient embryonic stem cells. Int Immunol. 1995;7:1637–1647. doi: 10.1093/intimm/7.10.1637. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Morgenbesser SD, DePinho RA. Use of transgenic mice to study myc family gene function in normal mammalian development and in cancer. Semin Cancer Biol. 1994;5:21–36. [PubMed] [Google Scholar]
- Morgenbesser SD, Schreiber-Agus N, Bidder M, Mahon KA, Overbeek PA, Horner J, DePinho RA. Contrasting roles for c-Myc and L-Myc in the regulation of cellular growth and differentiation in vivo. EMBO J. 1995;14:743–756. doi: 10.1002/j.1460-2075.1995.tb07053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G, Ekblom P. Contrasting expression patterns of three members of the myc family of protooncogenes in the developing and adult mouse kidney. J Cell Biol. 1991;112:13–25. doi: 10.1083/jcb.112.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G, Alt FW, Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol. 1988;107:1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B, Morgenbesser SD, DePinho RA. Myc family oncoproteins function through a common pathway to transform normal cells in culture: Cross-interference by Max and trans-acting dominant mutants. Genes & Dev. 1992;6:1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Neckers LM, Rosolen A, Whitesell L. Antisense inhibition of gene expression: A tool for studying the role of NMYC in the growth and differentiation of neuroectoderm-derived cells. J Immunother. 1992;12:162–166. doi: 10.1097/00002371-199210000-00003. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, Meltzer P, DePinho RA. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- Schmid P, Schulz WA, Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989;243:226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Horner J, Torres R, Chiu FC, DePinho RA. Zebra fish myc family and max genes: Differential expression and oncogenic activity throughout vertebrate evolution. Mol Cell Biol. 1993a;13:2765–2775. doi: 10.1128/mcb.13.5.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N, Torres R, Horner J, Lau A, Jamrich M, DePinho RA. Comparative analysis of the expression and oncogenic activities of Xenopus c-, N-, and L-myc homologs. Mol Cell Biol. 1993b;13:2456–2468. doi: 10.1128/mcb.13.4.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N, Stein D, Chen K, Goltz JS, Stevens L, DePinho RA. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc Natl Acad Sci. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Li H, O'Hagan RC, Hou H, Jr, Horner JW, 2nd, Lee HW, DePinho RA. Essential role for max in early embryonic growth and development. Genes & Dev. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes & Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schnegelsberg PN, Dausman J, Jaenisch R. Functional redundancy of the muscle-specific transcription factors Myf5 and myogenin. Nature. 1996;379:823–825. doi: 10.1038/379823a0. [DOI] [PubMed] [Google Scholar]
- Yeilding NM, Rehman MT, Lee WMF. Identification of sequences in c-myc mRNA that regulate its steady state levels. Mol Cell Biol. 1996;16:3511–3522. doi: 10.1128/mcb.16.7.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]