Abstract
In recent studies, the nuclear domain 10 (ND10) components PML and hDaxx were identified as cellular restriction factors that inhibit the initiation of human cytomegalovirus (HCMV) replication. The antiviral function of ND10, however, is antagonized by the IE1 protein, which induces ND10 disruption. Here we show that IE1 not only de-SUMOylates PML immediately upon infection but also directly targets Sp100. IE1 expression alone was sufficient to downregulate endogenous Sp100 independently of the presence of PML. Moreover, cotransfection experiments revealed that IE1 negatively interferes with the SUMOylation of all Sp100 isoforms. The modulation of Sp100 at immediate-early (IE) times of infection, indeed, seemed to have an in vivo relevance for HCMV replication, since knockdown of Sp100 resulted in more cells initiating the viral gene expression program. In addition, we observed that Sp100 was degraded in a proteasome-dependent manner at late times postinfection, suggesting that Sp100 may play an additional antiviral role during the late phase. Infection experiments conducted with Sp100 knockdown human foreskin fibroblasts (HFFs) confirmed this hypothesis: depletion of Sp100 resulted in augmented release of progeny virus particles compared to that from control cells. Consistent with this observation, we noted increased amounts of viral late gene products in the absence of Sp100. Importantly, this elevated late gene expression was not dependent on enhanced viral IE gene expression. Taken together, our data provide evidence that Sp100 is the first ND10-related factor identified that not only possesses the potential to restrict the initial stage of infection but also inhibits HCMV replication during the late phase.
INTRODUCTION
Human cytomegalovirus (HCMV) is an important human pathogen that causes severe disease in situations where the immune system is immature, compromised, or suppressed. It is the leading cause of virus-associated birth defects and still represents a serious problem for immunocompromised individuals, such as allograft recipients, tumor patients, and AIDS patients (29). During lytic HCMV replication, viral gene expression occurs in a strictly coordinated fashion as the HCMV open reading frames are transcribed in a temporally regulated cascade consisting of three sequential phases termed immediate-early (IE), early (E), and late (L) (51).
IE gene expression, which is controlled by a complex DNA element known as the major immediate-early enhancer/promoter (MIEP), has recently been shown to be targeted by an intrinsic immune defense of the host mediated by cellular proteins that localize to a subnuclear structure designated nuclear domain 10 (ND10; alternatively termed PML nuclear bodies [PML-NBs] or PML oncogenic domains [PODs]). ND10 structures are defined by the presence of the three major components PML, hDaxx, and Sp100, which colocalize in distinct foci within the interchromosomal space of the nucleus (31). The PML protein constitutes the core component, serving as a kind of scaffold protein that is necessary for the integrity of ND10 (19, 56). Directly upon entry into the nucleus, HCMV genomes can be found in close spatial proximity to ND10 (18, 20), since incoming viral DNA is rapidly recognized by this novel defense mechanism of the cell. In contrast to the prior concept that viral genomes are transported to static ND10 accumulations, our observation that infection of PML-kd cells (cells in which PML is knocked down), which are ND10 deficient, induces de novo formation of ND10-like hDaxx and Sp100 accumulations and indicates that HCMV triggers active recruitment of ND10 components to sites of viral nucleoprotein complexes (43). This concept is also supported by previous studies on the relocation of ND10 components during herpes simplex virus type 1 infection (10).
Recent data from our group and others demonstrate that PML and hDaxx function as cellular restriction factors that are capable of repressing viral replication, since depletion of either PML or hDaxx results in a substantial increase in permissivity for HCMV infection (6, 35, 38, 43, 44, 54). Furthermore, it has been shown that these ND10 components exert their antiviral activity by inducing a silencing of viral IE gene expression, since more cells initiate the lytic replication cycle of HCMV in the absence of either of these proteins (44). Initial attempts to gain insight into the molecular basis for the antiviral function of ND10 suggest the involvement of epigenetic mechanisms in order to prevent transcription of the viral genome. The hDaxx protein, for instance, has been shown to silence HCMV IE gene expression by inducing a transcriptionally inactive chromatin state around the MIEP via the recruitment of the chromatin-remodeling protein ATRX (alpha thalassemia/mental retardation syndrome X-linked) or chromatin-modifying enzymes, such as histone deacetylases (HDACs), to the viral DNA (28, 54). Consequently, treatment of cells with the HDAC inhibitor trichostatin A or short hairpin RNA (shRNA)-mediated depletion of endogenous ATRX leads to increased IE gene expression after HCMV infection (28, 38, 54).
During coevolution, however, HCMV has evolved mechanisms to efficiently control this ND10-based host response. As a first countermeasure, the tegument component pp71, which is delivered to the nucleus directly upon infection, localizes to ND10 in order to displace ATRX from this subnuclear structure (28). Thereafter, pp71 also antagonizes hDaxx-mediated gene silencing by inducing the proteolytic degradation of hDaxx through an uncommon mechanism that is thought to be proteasome dependent but ubiquitin independent (17, 38, 44). The relief of ATRX- as well as hDaxx-mediated repression, then, allows the initiation of IE gene expression, which results in the synthesis of the viral effector protein IE1. In the next step, IE1 accumulates at ND10 to efficiently overcome PML-mediated repression by inducing the disruption of this subnuclear structure, thereby ensuring the initiation of lytic replication (3, 23, 26, 52). As a mechanism for this process, it has been proposed that IE1 abrogates the SUMOylation of PML (2, 26), since both covalent and noncovalent interactions of PML with SUMO constitute the basis for PML nuclear body formation (19, 39, 57, 58). However, the exact mode of action still remains elusive, since in vitro studies suggest that IE1 does not have intrinsic SUMO protease activity (22).
Here we report that not only PML but also Sp100 is subject to IE1 countermeasures instantly upon infection, since IE1 is capable of directly modulating the SUMO modification status of Sp100. In agreement with this finding, we could demonstrate a repressive effect of Sp100 on viral IE gene expression. Thus, Sp100 contributes to the ND10-based antiviral response of the cell (1). However, our core finding is that the restrictive function of Sp100 is not limited to IE times of infection. We also found enhanced levels of viral true late gene products, which were detectable independently of elevated IE protein levels and led to increased release of viral particles into the cell culture supernatant, in the absence of Sp100. In summary, our data provide evidence that Sp100 not only is able to restrict the initial stage of infection but also inhibits HCMV replication during the late phase.
MATERIALS AND METHODS
Oligonucleotides and plasmid constructs.
The oligonucleotide primers used for this study were purchased from Biomers GmbH (Ulm, Germany) and are listed in Table 1. All Sp100 isoforms were amplified via PCR using pGS5-Sp100A, pGS5-Sp100B, pGS5-Sp100C, and pGS5-Sp100HMG (a kind gift from Gerd Maul, The Wistar Institute, Philadelphia, PA) as templates. The PCR products were cleaved with EcoRV and NotI, followed by ligation into a pcDNA3.1-derived vector that facilitates expression in fusion with a FLAG epitope (pHM971).
Table 1.
Oligonucleotides used for the construction of plasmids encoding Sp100 isoforms in fusion with a FLAG epitope
| Primer | Sequence |
|---|---|
| 5′Sp100-EcoRV | CATAGATATCCATGGCAGGTGGGGGCGGCGACCTG |
| 3′Sp100A-NotI | CATAGCGGCCGCGATCTGAAGTTTTTATTTCTG |
| 3′Sp100B-NotI | CATAGCGGCCGCTTTTTTATTATAAAATCAAATTTTATGATG |
| 3′Sp100C-NotI | CATAGCGGCCGCGACGGTTTTAAAATGGCAATGATA |
| 3′Sp100HMG-NotI | CATAGCGGCCGCTTTGATGGAGGAATAAAGTGACTT |
Cells and viruses.
HEK293T cells were cultivated in Dulbecco's minimal essential medium (DMEM) containing 10% fetal calf serum. Primary human foreskin fibroblasts (HFFs) were prepared from human foreskin tissue as described previously (21) and were maintained in Eagle's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 5% fetal calf serum. HFFs with a small interfering RNA-mediated knockdown of PML (siPML2 cells) and the respective control cells (designated vector or siC) were cultured in Dulbecco's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum and 5 μg/ml puromycin (43). Infection experiments were performed with either the HCMV laboratory strain AD169, a pp71-deficient virus (AD169/del-pp71) (44), or the IE1 deletion mutant CR208 (14). Viral stocks of wild-type (wt) AD169 and the pp71 knockout virus (AD169/del-pp71) were titrated via IE1p72 fluorescence (27). For this purpose, HFFs were infected with various dilutions of virus stocks. After 24 h of incubation, cells were fixed and stained with monoclonal antibody p63-27, directed against IE1p72 (4). Subsequently, the number of IE1-positive cells was determined and was used to calculate viral titers, expressed as IE protein-forming units (IEU). Titers of CR208 (virus stock kindly provided by Michael Nevels, Institute for Medical Microbiology and Hygiene, University of Regensburg, Regensburg, Germany) were determined as described previously (33).
Retrovirus transduction and selection of stably transduced cells.
For the generation of HFFs and siPML2 cells stably expressing the IE1 protein (HFF/IE1 and siPML2/IE1, respectively), replication-deficient lentiviruses were prepared using the pLenti6.4/R4R2/V5-DEST MultiSite Gateway vector kit from Invitrogen (Karlsruhe, Germany). For this purpose, 293T cells were cotransfected with a pLenti6.4/R4R2/V5-DEST MultiSite Gateway vector coding for IE1 under the control of the cellular EF1α promoter together with packaging plasmids pLP1 (coding for HIV-1 Gag and Pol), pLP2 (coding for HIV-1 Rev), and pVSV-G (expressing the vesicular stomatitis virus [VSV] envelope protein) using the Lipofectamine 2000 reagent (Invitrogen, Karlsruhe, Germany). Viral supernatants were harvested 48 h after transfection, clarified by centrifugation, filtered, and stored in aliquots at −80°C. Normal HFFs or PML-kd HFFs (siPML2 cells) were incubated for 24 h with retrovirus supernatants in the presence of 7.5 μg/ml of Polybrene (Sigma-Aldrich, Deisenhofen, Germany). Then blasticidin (2 μg/ml) was added to the cell culture medium in order to select a stably transduced cell population. HEK293T cells with a stable shRNA-mediated knockdown of PML (siPML2 [AGATGCAGCTGTATCCAAG]) and HFFs with a stable shRNA-mediated knockdown of Sp100 (siSp100 [GGAAGCACTGTTCAGCGATGT]) were generated via retroviral gene transfer as described previously (1, 43). Briefly, replication-deficient murine leukemia virus-based retroviruses were prepared by cotransfection of 293FT cells (Invitrogen, Karlsruhe, Germany) with pSIREN-RetroQ plasmids together with packaging plasmids pHIT60 (kindly provided by K. Überla, Bochum, Germany) and pVSV-G (Invitrogen, Karlsruhe, Germany) using the Lipofectamine 2000 reagent (Invitrogen, Karlsruhe, Germany). Forty-eight hours after transfection, the viral supernatants were used for infection of HFFs and 293T cells, respectively. Stably transduced cells were selected by the addition of 5 μg/ml puromycin (PAA, Cölbe, Germany) to the medium.
Antibodies.
Monoclonal antibody (MAb) p63-27 and MAb 28-4, which recognize IE1 and the major capsid protein (MCP), respectively, have been described elsewhere (4, 50). The polyclonal antiserum raised against exon 5 of IE2 (referred to as anti-pHM178) was generated by immunizing rabbits with the prokaryotically expressed protein. MAb-UL44 BS 510, for detection of the viral polymerase processivity factor pUL44, was kindly provided by Bodo Plachter (University of Mainz, Mainz, Germany). MAb-UL69 69-66 was used for the detection of pUL69 (53). Endogenous PML was detected by using either the rabbit polyclonal antibody H-238 (Santa Cruz Biotechnology, Santa Cruz, CA) or the mouse monoclonal antibody 5E10 (kindly provided by Roel van Driel, University of Amsterdam, Amsterdam, The Netherlands). Sp100 was detected by applying one of the following antibodies: (i) polyclonal antibody AB1380 from Chemicon (Schwalbach, Germany), (ii) rabbit polyclonal antiserum GH3 (a kind gift from Hans Will, Heinrich Pette Institute for Experimental Virology and Immunology, University of Hamburg, Hamburg, Germany), or (iii) MaxPab mouse polyclonal antibody B01 (Abnova, Heidelberg, Germany). A rabbit monoclonal antibody from Epitomics (Burlingame, CA) was used for the detection of hDaxx. For the detection of FLAG-tagged versions of Sp100, the mouse monoclonal anti-FLAG-M2 antibody was utilized (Sigma-Aldrich, Deisenhofen, Germany). Monoclonal antibody AC-15, which recognizes beta-actin, was purchased from Sigma-Aldrich (Deisenhofen, Germany). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies for Western blot analysis were obtained from Dianova (Hamburg, Germany), while Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibodies for indirect immunofluorescence experiments were purchased from Molecular Probes (Karlsruhe, Germany).
Indirect immunofluorescence and Western blot analysis.
For indirect immunofluorescence analysis, 3 × 105 HFFs were grown on coverslips for HCMV infection. At the indicated time points postinfection, the cells were washed three times with phosphate-buffered saline (PBS), followed by fixation with 4% paraformaldehyde for 10 min at room temperature. Then cells were permeabilized with PBS-0.2% Triton X-100 on ice for 20 min, followed by incubation with the respective primary or secondary antibodies for 30 min at 37°C. Finally, the cells were mounted by using Vectashield mounting medium plus 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). The samples were examined by using a Leica TCS SP5 confocal microscope, with 488-nm and 543-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported as tagged-image file format (TIFF) files and were then processed using Photoshop.
For Western blotting, extracts from infected cells were prepared in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated on sodium dodecyl sulfate-containing 8% polyacrylamide gels, and transferred to nitrocellulose membranes. Chemiluminescence was detected according to the manufacturer's protocol (ECL Western blot detection kit; Amersham Pharmacia Biotech).
Fluorescence-activated cell sorter (FACS) analysis.
Vector- and siSp100-transduced cells infected with HCMV AD169 were harvested by trypsinization 48 h postinfection (hpi), pelleted, resuspended in ice-cold PBS, and washed twice. Then the cells were fixed by the addition of 2% paraformaldehyde and were incubated for 10 min at room temperature, followed by two PBS washing steps. For cell membrane permeabilization, a saponin-containing buffer (1% saponin in PBS) was added. After incubation for 15 min on ice, cells were again pelleted and were stained for 30 min on ice using the mouse anti-IE1 monoclonal antibody MAb p63-27. After three washing steps with a saponin-containing buffer, an allophycocyanin (APC)-coupled goat anti-mouse secondary antibody from Dianova (Hamburg, Germany) was added, and the mixture was incubated for 20 min on ice in the dark. Finally, 2 × 104 cells per sample were analyzed with a FACSCalibur flow cytometer (BD Biosciences, Heidelberg, Germany), and the results were evaluated with FCS Express V3 (De Novo Software, Los Angeles, CA).
RESULTS
Evidence for a direct effect of IE1 on Sp100 immediately upon infection.
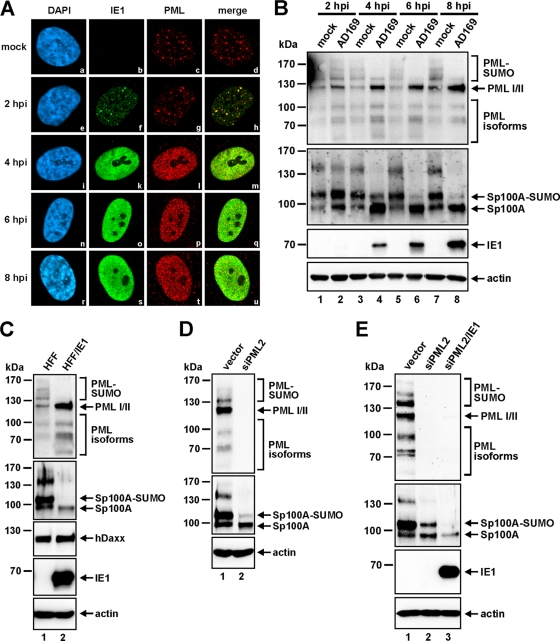
As an early event in HCMV-infected cells, ND10 are known to be dispersed by the virus in an IE1-dependent manner. It is thought that the ability of IE1 to inhibit the accumulation of the SUMO-conjugated forms of PML is relevant for this effect (26, 30), since only SUMOylated PML is capable of assembling ND10 structures (19, 25, 57, 58). These findings are based on previous studies that documented the effect of IE1 on individually overexpressed PML isoforms (26). Here we set out to investigate the effect of HCMV infection on the endogenous PML protein pattern. By performing a time course analysis, we could confirm that the IE1-mediated reorganization of ND10 correlated directly with the loss of the poly-SUMOylated variants of endogenous PML (Fig. 1A and B). For this purpose, HFFs were infected with AD169 at a multiplicity of infection (MOI) of 2, and the cells were harvested at different time points after infection (2, 4, 6, and 8 hpi) for immunofluorescence (Fig. 1A) and Western blot (Fig. 1B) analyses. Starting at 2 hpi, individual IE1-positive cells, in which IE1 accumulated in tiny foci that colocalized with PML aggregates, could be visualized by indirect immunofluorescence analysis (Fig. 1Ae to h), while ND10 structures were still intact, as in mock-infected cells (Fig. 1Aa to d). At this time point, when ND10 was still present, no difference in the SUMOylation pattern of PML could be observed between infected- and uninfected-cell lysates (Fig. 1B, top panel, lanes 1 and 2). However, as early as 4 hpi, the continuous production of IE1 led to disruption of ND10 in the whole-cell population, as evidenced by the microdispersed localization pattern of PML and the diffuse distribution of IE1 (Fig. 1Ai to u). In parallel with the disintegration of ND10, we could detect a loss of the SUMOylated variants of PML (Fig. 1B, top panel, lanes 3 to 8). In accordance with previous findings (26), PML turned out to be only partially de-SUMOylated: only the poly- SUMOylated PML species disappeared, while the mono- SUMOylated form of PML remained unaffected. In addition, this experiment revealed an interesting, novel finding: not only PML but also Sp100 protein levels were affected by IE1-based ND10 disruption (Fig. 1B, second panel). ND10 dispersal was paralleled by a loss of the low-mobility isoforms of Sp100 and a substantial reduction in the level of SUMO-modified Sp100A, which most likely accounted for the concomitant increase in the level of unmodified Sp100A (Fig. 1B, second panel, lanes 3 to 8). Furthermore, the generation of HFFs that stably expressed the IE1 protein by retroviral transduction (HFF/IE1) indicated that IE1 alone was sufficient to induce these effects on the abundances of the PML (Fig. 1C, top panel) and Sp100 (Fig. 1C, second panel) proteins, while hDaxx, in contrast, remained unaffected (Fig. 1C, third panel). Interestingly, similar changes in the Sp100 protein pattern could also be observed in cells where PML was depleted by shRNAs (Fig. 1D). This finding has also been reported by Everett and colleagues, and it prompted the authors to speculate that the metabolism of Sp100 seems to be intimately connected to that of PML (13). Therefore, we had to clarify whether Sp100 is directly targeted by IE1 or is only indirectly affected by the ability of IE1 to deplete the SUMO-conjugated variants of PML. To address this issue, we retrovirally transduced PML-kd HFFs (siPML2 cells) with the IE1 expression construct (Fig. 1E). While depletion of PML again induced the characteristic changes in the Sp100 protein pattern (Fig. 1E, second panel, siPML2), the stable expression of IE1 in these cells, nonetheless, led to a further downregulation of Sp100 abundance (Fig. 1E, second panel, siPML2/IE1). In the presence of IE1, the overall level of unmodified Sp100A was clearly diminished, and the SUMOylated form of Sp100A vanished completely, compared with levels in IE1-negative siPML2 cells (Fig. 1E, second panel). Thus, our data indicate that not only PML but also Sp100 is a direct target of IE1 during the initial postinfection stage.
Fig. 1.
The effects of IE1 on Sp100 occur in parallel with ND10 disruption but are not dependent on the presence of PML. (A and B) HFFs were infected with wt AD169 (MOI, 2) for indirect immunofluorescence analysis (A) as well as for Western blotting (B) using identical viral inocula. At the indicated times following infection (2 to 8 hpi), samples were harvested in parallel for dissection of IE1-mediated induction of ND10 reorganization by immunofluorescence costaining of PML and IE1 (A) as well as by detection of the abundances of the PML and Sp100 proteins by immunoblotting (B). In the immunofluorescence experiments (A), cell nuclei were identified by detection of DAPI signals. (C) Analysis of the effect of IE1 expression alone on ND10 protein levels. Lysates of retrovirally transduced HFFs stably expressing the IE1 protein (HFF/IE1) were compared to lysates of nontransduced control HFFs with regard to PML, Sp100, and hDaxx protein patterns. (D) Immunoblotting of PML and Sp100 in empty vector- or siPML2-transduced cells. (E) Detection of PML and Sp100 in lysates of control cells (vector), PML-kd HFFs (siPML2), and PML-kd HFFs stably expressing IE1 (siPML2/IE1). For all Western blot experiments, beta-actin was used as a loading control and IE1 as an infection (B) or transduction (C and E) control.
IE1 negatively interferes with the SUMOylation of all individual Sp100 isoforms.
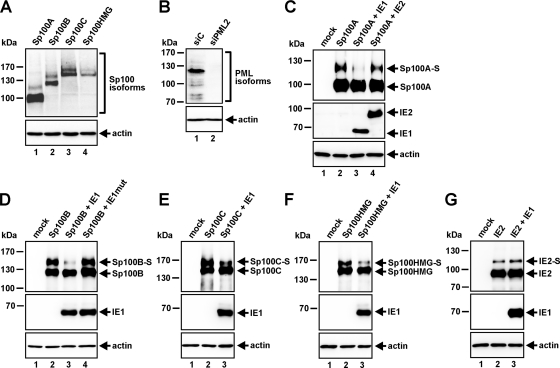
The identity of the high-molecular-weight species of endogenous Sp100 is not totally clear yet. They could represent less abundantly expressed Sp100 isoforms, such as Sp100B, Sp100C, and Sp100HMG, or even further modified variants of Sp100A. The latter assumption is supported by a recent publication showing that shRNA-mediated knockdown of all Sp100 isoforms (Sp100B, -C, and -HMG) except Sp100A had no impact on the Sp100 protein expression profile normally observed in cells (32). Thus, in an effort to evaluate the effects of IE1 on the individual Sp100 isoforms, we generated constructs encoding the four different Sp100 variants and tested their expression in 293T cells (Fig. 2 A). In every case, a low-mobility species representing the SUMO-modified variant of the respective Sp100 isoform could be detected (Fig. 2A). Then we transfected the Sp100 variants either alone or in combination with IE1 (Fig. 2C to F). To exclude any effects of PML on Sp100, PML-kd 293T cells, in which the loss of endogenous PML was verified by Western blotting, were used for this purpose (Fig. 2B). As is evident from Fig. 2C to F, coexpression of IE1 resulted in clear reductions in the levels of the SUMOylated species of all Sp100 isoforms (designated Sp100A-S, Sp100B-S, Sp100C-S, and Sp100HMG-S). In contrast, an IE1 mutant that has been shown not to interact with PML (30), and thus not to localize at ND10 and not to disrupt this subnuclear structure, was not capable of influencing Sp100 SUMOylation (Fig. 2D). Abrogation of SUMOylation also could not be observed upon coexpression of Sp100 with the viral IE protein IE2 (Fig. 2C, lane 4), which is also known to associate with ND10 and, consequently, is specific for IE1. Moreover, the fact that IE1 had no impact on the SUMOylation status of IE2 (Fig. 2G) argues against a general de-SUMOylating activity of IE1 and supports the notion that Sp100 is a specific target of the viral regulatory protein. Taken together, our findings suggest that IE1 has the ability to de-SUMOylate not only the ND10 factor PML but also Sp100.
Fig. 2.
IE1 expression leads to clear reductions in the SUMOylation of all Sp100 isoforms. (A) Western blot analysis for the detection of the various Sp100 isoforms in extracts from HEK293T cells transfected with expression constructs coding for the individual Sp100 variants. (B) Verification of the PML-kd in HEK293T cells transduced with the siPML2 shRNA compared to siC-transduced 293T cells by use of the mouse monoclonal anti-PML antibody 5E10. (C to F) Extracts from PML-kd/293T cells transfected with Sp100 isoform Sp100A (C), Sp100B (D), Sp100C (E), or Sp100HMG (F), alone or in combination with IE1 (C to F), IE2 (C), or IE1mut (D), were analyzed by Western blotting. Lysates from nontransfected PML-kd/293T cells (mock) served as a negative control. (G) Western blotting of PML-kd/293T cell lysates expressing IE2 alone or in combination with IE1. Lysates from untransfected PML-kd/293T cells served as a negative control. For all Western blot experiments, detection of beta-actin expression was included as an internal loading control.
Sp100 restricts the initiation of viral IE gene expression in an MOI-dependent manner.
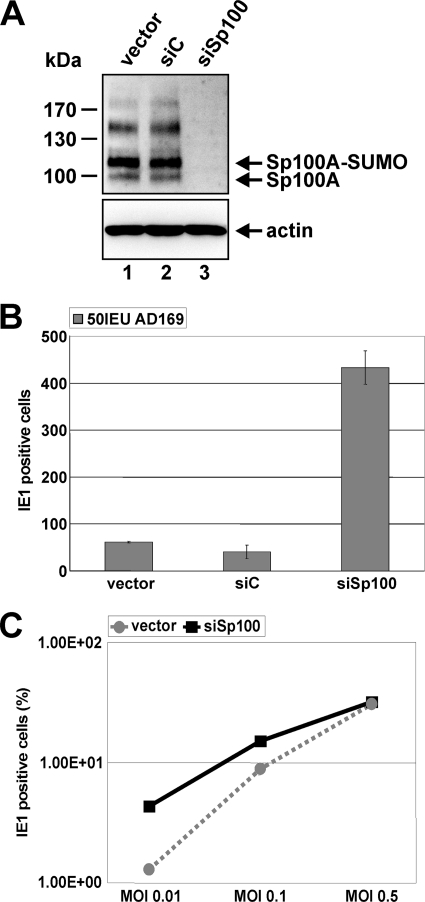
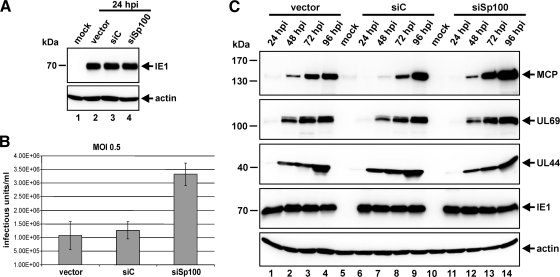
In order to explore whether the targeting of Sp100 by IE1 directly upon infection correlates with an antiviral role of Sp100 for HCMV replication, we applied an shRNA-directed approach to construct HFFs devoid of Sp100 (1) (Fig. 3A). Stable expression of an shRNA targeting all isoforms resulted in complete downregulation of Sp100 in siSp100 HFFs (Fig. 3A, top, lane 3) compared to Sp100 expression in the corresponding control HFFs transduced with an empty vector (designated “vector”) (Fig. 3A, top, lane 1) or with a nonfunctional shRNA fragment (siC) (Fig. 3A, top, lane 2). Next, Sp100-kd and control cells were infected with 50 IEU/well of AD169 (Fig. 3B). Twenty-four hours later, cells were immunostained for IE1, and the number of IE1-positive cells was determined (Fig. 3B). As was found for PML and hDaxx in previous studies (43, 44), depletion of Sp100 facilitated the initiation of HCMV gene expression, leading to an approximately 8-fold higher number of infected cells (Fig. 3B). Importantly, however, the effect of more cells initiating the lytic replication cycle in the absence of Sp100 was strictly dependent on the virus dose and could be observed only under low-MOI conditions (Fig. 3C, MOI 0.01 or MOI 0.1). Figure 3C shows the percentage of IE1-expressing siSp100 cells in comparison to that of vector-transduced cells infected with increasing amounts of AD169 as determined by FACS analysis. At an MOI of 0.5, the difference between Sp100-kd and control HFFs in the ability of HCMV to enter the viral gene expression program was no longer detectable, suggesting that the antiviral activity of Sp100 was already efficiently overcome by the virus under these infection conditions (Fig. 3C). Hence, Sp100, like PML and hDaxx, acts as a repressor of viral IE gene expression and, together with PML and hDaxx, contributes to the antiviral response instituted by ND10.
Fig. 3.
Sp100 inhibits viral IE gene expression in an MOI-dependent manner. (A) (Top) Detection of endogenous Sp100 by Western blotting using cell lysates derived from primary human fibroblasts stably transduced with the shRNA expression vectors indicated. (Bottom) Beta-actin was used as a loading control. (B) Enhancement of HCMV IE gene expression in the absence of Sp100. HFFs, retrovirally transduced as indicated, were infected with 50 IEU/well of HCMV AD169. Cells were fixed at 24 hpi, and the number of IE1-expressing fibroblasts was determined via indirect immunofluorescence analysis. (C) FACS analysis for the detection of IE1-positive vector- and siSp100-transduced cells harvested 48 h after inoculation with increasing amounts of wt AD169 (MOIs, 0.01, 0.1, and 0.5). Results are given as the percentages of all cells analyzed that expressed IE1.
HCMV differentially affects the abundances of the major ND10 factors during the course of infection.
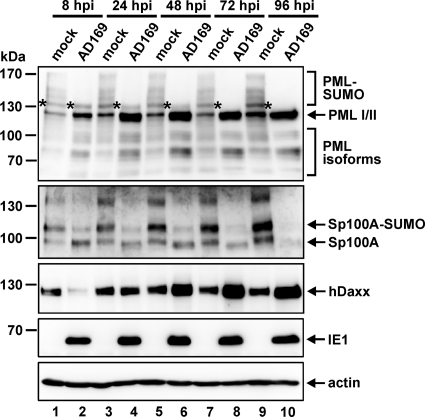
In contrast to infection with herpes simplex virus 1 (HSV-1), HCMV infection does not have the potential to completely eliminate ND10 constituents such as PML or Sp100 in order to efficiently abolish their repressive activity (46). Since hDaxx protein levels also recover from pp71-mediated depletion as infection progresses (38, 44), we questioned whether these host factors might still play a role during later stages of replication. To address this issue, we set out to elucidate the fates of PML, hDaxx, and Sp100 during the entire course of the HCMV infection cycle by performing Western blot experiments (Fig. 4). At first, this approach revealed that HCMV has diverse effects on the PML protein level compared to that in mock-infected cells (Fig. 4, top panel). For instance, we observed HCMV-induced depletion of the presumed SUMO-modified species of PML. Interestingly, however, while in the beginning only a partial de-SUMOylation of PML could be observed, since only the PML variants containing multiple SUMO adducts were no longer detectable (Fig. 4, top panel, lanes 2, 4, and 6), at later stages of infection the mono-SUMOylated form of PML (indicated by an asterisk) also disappeared (Fig. 4, top panel, lanes 8 and 10). Furthermore, upregulation of PML became apparent, which, however, occurred in a selective manner; only the protein levels of the major isoforms (I and/or II) were enhanced in virus-infected cells over those in mock-infected controls (Fig. 4, top panel). In the case of Sp100 (Fig. 4, second panel), HCMV infection resulted in downregulation of the lower-mobility isoforms during the first hours after infection, as shown above (Fig. 1B, second panel). Intriguingly, the continuous downregulation of Sp100 finally culminated in a complete loss of all Sp100 variants at late stages of the viral replication cycle (Fig. 4, second panel, lane 10). As shown in the third panel of Fig. 4, hDaxx protein levels, in contrast, dropped initially due to the action of the tegument protein pp71 (Fig. 4, third panel, lane 2) but were then upregulated as the infection progressed (Fig. 4, third panel, lane 4). As a consequence, hDaxx abundance was finally increased over that in mock-infected cells (Fig. 4, third panel, lanes 6, 8, and 10). This is in direct contrast to the downregulation of Sp100 at the late stage. Thus, HCMV diversely influences the abundances of the three major ND10 factors PML, Sp100, and hDaxx during the course of infection.
Fig. 4.
Examination of the relative abundances of the major ND10 factors PML, Sp100, and hDaxx during the entire course of the HCMV replication cycle. HFFs either were mock infected or were infected with wt AD169 at an MOI of 3. Lysates were harvested at the indicated times postinfection (8 to 96 hpi) and were analyzed by Western blotting for PML, Sp100, and hDaxx protein levels. IE1 and beta-actin were included as infection and internal loading controls, respectively. The asterisks in the top panel indicate the position of a mono-SUMOylated PML isoform.
IE1 plays a major role in the HCMV-induced modulation of ND10 protein levels during infection.
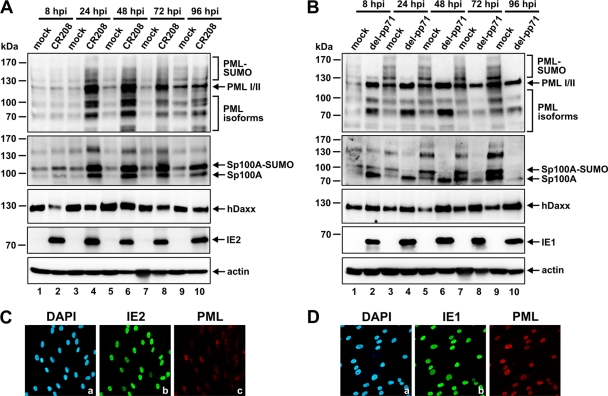
Next, we set out to clarify which viral effector proteins are involved in modulating the abundances of the three major ND10 factors following HCMV infection. First, we analyzed the role of IE1 in this context, since IE1 targets PML and Sp100 immediately upon infection. In order to investigate whether IE1 has additional impacts on ND10 protein expression levels, we infected HFFs with the IE1 mutant virus CR208 (Fig. 5A). As expected, in the absence of IE1, the loss of the SUMO-modified species of PML, as observed during infection with the wt virus (Fig. 4, top panel), was no longer detectable (Fig. 5A, top panel). Moreover, since the SUMOylation of PML remained unaffected throughout the course of the CR208 infection cycle, it seems that IE1 is also responsible for the loss of the mono-SUMOylated variant of PML during the late stage of HCMV replication (Fig. 4, top panel, lanes 8 and 10, asterisks). In addition, CR208 infection resulted in a more general upregulation of PML, in that all of the individual isoforms were affected, while in wt HCMV-infected cells, only the major PML variants were preferentially upregulated (compare the top panel of Fig. 4 to that of Fig. 5A). Moreover, CR208 also failed to downregulate Sp100 during the course of infection (Fig. 5A, second panel). Instead, as with PML, an overall upregulation of each of the individual Sp100 isoforms could be observed (Fig. 5A, second panel). Finally, the enhancement of hDaxx protein levels at late stages of wt HCMV infection was also abolished in the absence of IE1 (Fig. 5A, third panel). This led us to conclude that in fact, the alterations observed in the levels of the PML, Sp100, and hDaxx proteins during the course of HCMV infection all occur in an IE1-dependent manner.
Fig. 5.
(A and B) Examination of the relative abundances of the major ND10 factors PML, Sp100, and hDaxx during the entire course of the HCMV replication cycle after infection with the IE1 mutant virus CR208 (A) or with a pp71 deletion virus (del-pp71) (B). HFFs either were mock infected or were infected with CR208 or a pp71 deletion virus at an MOI of 3. Lysates were harvested at the indicated times postinfection (8 to 96 hpi) and were analyzed by Western blotting for PML, Sp100, and hDaxx protein levels. IE1/IE2 and beta-actin were included as infection and internal loading controls, respectively. (C and D) In order to control the infection efficacies of the mutant viruses, HFFs were infected with CR208 (C) or the pp71 deletion virus (D) at an MOI of 3, followed by fixation at 24 hpi and indirect immunofluorescence analysis of IE1, IE2, or PML as indicated. Cell nuclei were detected by DAPI staining.
pp71 is responsible only for the degradation of hDaxx directly upon HCMV infection.
Given that the viral regulatory protein pp71 likewise functions as an important antagonist of the antiviral response of ND10 immediately upon virus entry, by promoting hDaxx degradation, we wanted to determine whether pp71 has any further effects on ND10 protein levels during the course of HCMV replication (Fig. 5B). However, infection experiments with a pp71 deletion virus revealed no major difference from wt AD169 with regard to the modulation of PML protein levels in infected cells (compare the top panels of Fig. 4 and 5B). As shown in the top panel of Fig. 5B, AD169/del-pp71 still efficiently inhibited the accumulation of the SUMOylated forms of PML and promoted the upregulation of specific PML variants. Moreover, it became apparent that pp71 also was not involved in the HCMV-induced downregulation of Sp100, which still occurred in the absence of pp71 (Fig. 5B, second panel). Consequently, pp71 is responsible only for the HCMV-induced depletion of hDaxx, which occurs directly upon infection (Fig. 4 and 5A, third panel, lane 2) but is abrogated when pp71 is missing (Fig. 5B, third panel, lane 2).
Proteasomal activity is required for the downregulation of Sp100 at late times of infection.
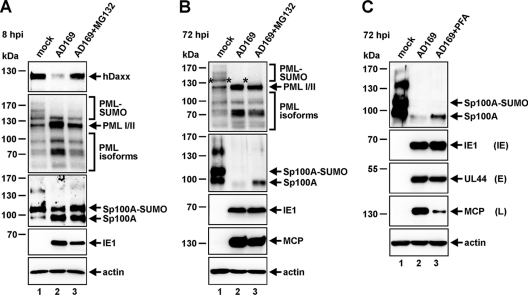
In order to further clarify the mechanism by which HCMV modulates ND10 protein levels during the course of the replication cycle, we next elucidated the role of the proteasome in ND10 protein abundance (Fig. 6A and B). As expected, during the early stage of infection (8 hpi) (Fig. 6A), the pp71-mediated degradation of hDaxx, which is known to occur via the proteasome (38), could be reversed by addition of the proteasome inhibitor MG132 (Fig. 6A, top panel), thus serving as a positive control for the inhibitory effect of MG132. At the same time, however, the SUMOylated variants of PML could not be restored by treatment of cells with MG132 (Fig. 6A, second panel). This correlates with previous findings showing that proteasomal activity is not required for IE1 to induce the de-SUMOylation of PML and the concomitant disruption of ND10 (26, 55). Furthermore, the IE1-triggered modulation of Sp100 protein abundance, observed initially upon infection (Fig. 6A, third panel, lane 2), also turned out not to be affected by the addition of MG132 (Fig. 6A, third panel, lane 3). Similarly, as shown in Fig. 6B, inhibition of the proteasome at the late stage of infection (72 hpi) did not result in reconstitution of the mono-SUMOylated form of PML (indicated by an asterisk) (Fig. 6B, top panel, lane 3), which starts to disappear during later time points of the replication cycle (Fig. 6B, top panel, lane 2). However, in contrast, addition of MG132 led to at least a partial rescue of Sp100, which had already completely vanished at 72 hpi (Fig. 6B, second panel, lane 2), since considerable amounts of unmodified Sp100A became detectable again (Fig. 6B, second panel, lane 3). Of note, since proteasomal activity is known to be required at multiple steps of the HCMV life cycle, we ensured identical IE1 expression (8 hpi) (Fig. 6A) and identical MCP expression (72 hpi) (Fig. 6B) in MG132-treated versus untreated cells. Thereby, detrimental effects of the proteasome inhibitor on HCMV replication could be excluded. Finally, incubation of cells with the viral DNA synthesis inhibitor phosphonoformic acid (PFA) abolished the HCMV-induced downregulation of Sp100 (Fig. 6C, top panel, compare lanes 2 and 3). Since PFA treatment is known to inhibit true late gene expression (Fig. 6C, fourth panel, lane 3), while IE and E protein levels remain unaffected (Fig. 6C, second and third panels), these data suggest that a true late gene product is necessary for the degradation of any Sp100 remaining in the presence of IE1. Taken together, these data allow the assumption that Sp100 is downregulated in a proteasome-dependent manner, requiring the activity of a late HCMV protein during the final stage of infection.
Fig. 6.
Contribution of the proteasome to HCMV-induced changes in ND10 protein abundance. (A and B) HFFs were infected with wt AD169 at an MOI of 3 and were treated the proteasome inhibitor MG132 at 5 μM either simultaneously with virus inoculation (A) or at 48 hpi (B). Thereafter, the levels of the hDaxx, PML, Sp100, and MCP proteins relative to those in mock-infected samples were analyzed at immediate-early (IE) times of infection (8 hpi) (A) or during the late (L) stage of the viral replication cycle (72 hpi) (B) in the presence or absence of MG132. (C) The degradation of Sp100 at the late stage of infection can be abrogated by addition of the viral DNA synthesis inhibitor phosphonoformic acid (PFA). HFFs were either mock infected or infected with wt AD169 at an MOI of 3. In parallel with virus inoculation, 250 μM PFA was added as indicated to the infected-cell sample. Cell lysates were harvested 72 hpi and were subjected to SDS-PAGE and immunoblotting using antibodies directed against Sp100, the viral immediate-early protein IE1, the viral early (E) protein UL44, and the viral late protein MCP. For all experiments, IE1 served as an infection control and beta-actin as a loading control.
Antiviral activity of Sp100 during the late phase of HCMV infection.
The observation that HCMV infection causes a complete loss of Sp100 at late times of the replication cycle prompted us to speculate that Sp100 could play an additional antiviral role during later stages of infection, which, again, has to be defeated by the virus. To test this hypothesis, we first analyzed the impact of Sp100 on the release of infectious progeny virus into the cell culture supernatant (Fig. 7B). In order to compare the amounts of virus particles released in the presence and absence of Sp100, we infected siSp100 HFFs in parallel with control cells with AD169 (MOI, 0.5) and harvested the supernatant 96 hpi (Fig. 7B). Importantly, since we could demonstrate an inhibitory influence of Sp100 on viral IE gene expression (Fig. 3B), we had to exclude the situation in which more cells initiate the lytic replication cycle when Sp100 is absent, which per se would lead to enhanced particle release. To circumvent this problem, we took advantage of the MOI-dependent antiviral effect of Sp100 at IE times of infection (Fig. 3C). By choosing a high viral load (MOI, 0.5), we guaranteed that HCMV would initiate IE gene expression equally efficiently in vector-, siC-, and siSp100-transduced cells, since similar IE1 protein levels could be detected in all three cell types at 24 hpi by Western blot experiments (Fig. 7A). Under these infection conditions, approximately three times more virus particles were released by siSp100 HFFs than by control cells (Fig. 7B). This confirmed our hypothesis of an antiviral function of Sp100 during later stages of the HCMV replicative cycle. Furthermore, dissection of the expression kinetics of HCMV revealed enhanced levels of true late gene products in Sp100-depleted cells; these late gene products are known to include structural components of the virus, such as MCP (Fig. 7C, top panel). Accordingly, this finding could account for the increased virion release observed following infection of siSp100 cells. Finally, it should be noted that the repressive effect of Sp100 actually seems to target the final stage of the viral replication cycle, since Sp100 specifically affected the expression of true late genes. At the same time, no difference in the expression profiles of pUL69 (expressed with E/L kinetics) (Fig. 7C, second panel) and pUL44 (expressed with E kinetics) (Fig. 7C, third panel) could be detected between vector-, siC-, and siSp100-transduced cells. Monitoring of IE1 abundance, again, served as a control to ensure identical numbers of infected Sp100-kd HFFs and control cells (Fig. 7C, fourth panel). In summary, our findings clearly argue in favor of an additional antiviral role of Sp100 during the late phase of HCMV replication.
Fig. 7.
Sp100 exerts antiviral activity during the late stage of the HCMV replication cycle. (A and B) Sp100-kd and control HFFs, as indicated, were infected in parallel with HCMV AD169 at an MOI of 0.5 for Western blotting (A) and for the quantification of virus particle release (B) using identical viral inocula. (A) At 24 hpi, cell lysates were harvested for detection of the IE1 protein. (B) To determine the numbers of progeny particles released upon infection of Sp100-kd and control cells, cell culture supernatants were harvested 96 hpi and were used for the reinfection of HFFs. The infected cells were fixed 24 hpi to determine the number of IE1-expressing cells by indirect immunofluorescence analysis. (C) Investigation of HCMV expression kinetics following infection of vector-, siC-, and siSp100-transduced cells with AD169 at an MOI of 0.5. Cell lysates were harvested at the indicated times postinfection (24 to 96 hpi) and were subjected to Western blotting for the detection of the viral proteins MCP (L), UL69 (E/L), UL44 (E), and IE1 (IE). Beta-actin was included as an internal loading control in all immunoblot experiments.
DISCUSSION
Research performed during recent years has convincingly demonstrated that proteins of the subnuclear structure ND10 participate in an intrinsic defense mechanism of the cell with the potential to restrict the lytic replication of herpesviruses in general (for reviews, see references 9, 45, 46, and 47). Herpesviruses, in turn, have developed differential evasion strategies to cope with the antiviral functions of ND10. While HSV-1 has evolved mechanisms to completely eliminate PML in order to inactivate ND10 (5, 12), HCMV is capable only of interfering with the SUMOylation of PML; unmodified PML remains unaffected (26). Interestingly, a recent publication demonstrated that the activity of specific PML isoforms in restricting HSV-1 replication was dependent on SUMO modification, suggesting that SUMOylated PML represses herpesviral gene expression (8). Here we provide evidence that the IE1-based dispersal of ND10 that occurs at IE times of infection not only correlates with the loss of the poly-SUMOylated variants of PML but also influences Sp100 abundance (Fig. 1A and B). Endogenous Sp100 is characterized by a number of bands, the most prominent of which represent unmodified Sp100 isoform A and a derivative of isoform A conjugated to a single SUMO-1 moiety (41), while the identities of the lower-mobility bands are not totally clear yet. In parallel with ND10 disruption, besides SUMO-modified Sp100A, it was especially these low-mobility variants of Sp100 that were observed to be downregulated (Fig. 1A and B). In fact, IE1 expression was necessary (Fig. 5A) and sufficient (Fig. 1C) for this effect. However, in the beginning, it was not possible to conclude that Sp100 is directly targeted by IE1, because depletion of PML is known to cause similar changes in Sp100 protein abundance (11, 13). Thus, loss of the SUMO-modified forms of PML induced by IE1 could be sufficient to alter Sp100 protein levels. To exclude any PML-dependent effects, we analyzed the influence of IE1 on Sp100 in PML knockdown cells. Western blot analyses confirmed that these cells were devoid of endogenous PML. Since stable expression of IE1 in the PML-depleted background resulted in an additional overall decrease in the level of Sp100 (Fig. 1E), it is reasonable to conclude that the modulation of Sp100 by IE1 can be separated from its effects on PML.
Given the still uncertain nature of the high-molecular-weight species of endogenous Sp100, we set out to investigate the influence of IE1 on each of the individual Sp100 isoforms. Interestingly, this experiment revealed that IE1 exerts a broad de-SUMOylating activity that affects all of the existing Sp100 variants and that, again, is not dependent on the presence of PML (Fig. 2C to F). Thus, IE1 has the potential to annihilate not only the SUMOylation of PML but also that of Sp100. However, the exact mode of action of IE1 still remains enigmatic. Studies on the mechanism underlying the IE1-mediated de-SUMOylation of PML have demonstrated that IE1 does not harbor an intrinsic SUMO protease activity (22) and that proteasomal activity is not required for this process (26). In contrast, Lee and colleagues could show that a physical interaction between IE1 and PML is absolutely necessary (26). Intriguingly, no direct protein interaction has been described for IE1 and Sp100 to date. Hence, one can only speculate how IE1 abrogates the SUMOylation of Sp100. However, in light of the fact that IE1 was not able to influence the SUMOylation of IE2 (Fig. 2G), it is rather unlikely that IE1 possesses a general de-SUMOylating activity, which might operate, for instance, by targeting a component of the SUMO conjugation pathway. Furthermore, while IE1 is known to interfere with PML SUMOylation in order to compromise ND10 integrity, the functional consequences of Sp100 de-SUMOylation remain elusive, too. Initial studies suggested that SUMO modification determines the subcellular distribution of Sp100, since RanBP2 has been found to stimulate Sp100 SUMOylation during its nuclear entry (34, 42). However, a recent report by Cuchet and coworkers cast doubt on this theory, since they could also detect unmodified Sp100 in the nucleus (8).
Nonetheless, the direct targeting of Sp100 by IE1 immediately upon virus entry seems to have significance for HCMV replication. Prior to this study, PML and the hDaxx-ATRX chromatin-remodeling complex were regarded as the major players in the cellular antiviral resistance against HCMV infection. Here we report that the core component Sp100 likewise contributes to the intrinsic immunity of ND10 by inhibiting viral IE gene expression. shRNA-mediated depletion of Sp100 increased the replication efficiency of HCMV, because more cells initiated the lytic replication cycle in the absence of Sp100 (1) (Fig. 3B). Importantly, however, the antiviral activity of Sp100, exerted at IE times of infection, turned out to be dependent on the virus dose, since it could be observed only under low-MOI conditions (Fig. 3C). This could be explained by efficient viral antagonism of Sp100. Consistent with this assumption, we observed that the impaired growth phenotype of the IE1 mutant CR208 was complemented by Sp100 depletion (data not shown).
Analysis of the abundances of PML, Sp100, and hDaxx after the dispersal of ND10 revealed that they are differentially regulated by HCMV as infection progresses (Fig. 4). Especially during the late stage of infection, we observed a loss of the mono-SUMOylated form of PML as well as of unmodified Sp100A, both of which initially remained unaffected by IE1-based antagonism directly upon infection. This gave rise to the idea that during later stages of infection, ND10 components could still possess inhibitory functions that the virus must overcome in order to efficiently complete the replication cycle. However, this seems to account for PML and Sp100 only, since hDaxx protein levels, in contrast, recovered from the pp71-mediated depletion at IE times of infection, so that hDaxx abundance was finally even greater than that in mock-infected cells. The reaccumulation of hDaxx during the course of infection could be a means to prevent the induction of hDaxx-mediated apoptosis, since silencing of hDaxx expression has been shown to sensitize cells to apoptosis (7).
In accordance with a putative antiviral role of Sp100 at late times of infection, we could show that Sp100 is degraded by HCMV in a proteasome-dependent manner during the final stage of the viral life cycle (Fig. 6B). Interestingly, it became apparent that, except for the initial pp71-promoted proteasomal degradation of hDaxx upon virus inoculation and the loss of Sp100 at late times of infection, all HCMV-induced changes in the abundances of PML, Sp100, and hDaxx observed during the course of infection occur in a proteasome-independent manner (Fig. 6A and B). In contrast, it turned out that most ND10 protein alterations are strictly dependent on the expression of IE1 (Fig. 5A). In the absence of IE1, we could observe a general upregulation of all PML and Sp100 isoforms, while hDaxx protein levels, in contrast, remained constant or were even reduced during the final stage of infection. Since the IE1 protein is also known to counteract the interferon (IFN) pathway in order to inhibit the activation of IFN-stimulated genes (ISG) (16, 24, 33), and since PML and Sp100 are prominent members of this family of genes (15, 40), it is very likely that the general upregulation of PML and Sp100 can be ascribed to a failure of the IE1 mutant virus to efficiently antagonize the IFN response. This assumption is further supported by our observation that a similar upregulation of all the individual PML and Sp100 isoforms, but not of hDaxx, can be found upon stimulation of fibroblasts with IFN (data not shown). Therefore, one potential scenario might be that the overall increase in the abundances of PML and Sp100 as a consequence of IFN stimulation masks the downregulation of both host factors at late time points of the replication cycle, which might still occur in the absence of IE1. Thus, since we determined that a late viral function is required for the proteasome-dependent degradation of Sp100 (Fig. 6C), this late viral protein may exert its effect independently of IE1. Alternatively, however, the qualitative changes in PML and Sp100 (i.e., de-SUMOylation) induced by IE1 may constitute a prerequisite for the late viral protein to exert its degradative function. Hence, although IE1 might only indirectly influence ND10 protein levels at the late stage of infection, our data clearly show that IE1 plays a pivotal role in the HCMV-induced modulation of ND10 protein abundance during the entire course of the HCMV replication cycle.
Finally, evidence is accumulating in the literature that the repressive potential of ND10 components is not restricted to the initial stage of virus infections. The antiviral strategy of ND10 in adenovirus type 5 (Ad5) infection, for instance, is not based solely on transcriptional repression of Ad5 early gene expression, which is only modestly affected, to a degree that is not sufficient to compromise early protein function (48). Instead, ND10 suppresses Ad5 replication mainly by negatively influencing viral DNA synthesis (48, 49). Moreover, Ad5 has evolved a unique strategy to sustain the neutralization of ND10 bodies after they have initially been disrupted by E4ORF3: PML and Sp100 are sequestered into nuclear inclusions in order to ensure optimal viral proliferation throughout the replicative cycle (37). With regard to herpesviral infections, only recently have Reichelt and colleagues provided the first evidence that PML has the ability to target the late stage of varicella-zoster virus (VZV) replication, since they could demonstrate the entrapment of nucleocapsids in cages formed by PML (36). In addition, a series of data exists that connect ND10 with all stages of the papillomavirus life cycle, although the biological relevance of these findings is still controversial (45). Here we show that the repressive capacity of Sp100 in HCMV infection also is not limited to the inhibition of viral IE gene transcription. We found that Sp100, in addition, negatively interfered with viral true late gene expression, as evidenced by the fact that larger amounts of virus structural components, such as MCP, could be detected in infected Sp100-kd cells than in infected cells in which Sp100 was present (Fig. 7C). Importantly, this effect could be observed independently of elevated IE protein levels and could account for the concomitant increase in the release of progeny virus particles from cells lacking Sp100. Consequently, proteasomal degradation of Sp100 at late times of HCMV replication could provide a means to inactivate Sp100. Taking our findings together, we conclude that Sp100 is the first ND10 component identified that exerts an antiviral function during different stages of the HCMV replicative cycle. Thus, our data further support the notion that ND10-related restriction factors have the ability to target various steps of virus replication even after the integrity of ND10 bodies has been compromised by the virus. Therefore, it is tempting to speculate that intact ND10 structures are not a prerequisite for the contribution of these factors to the antiviral response of the cell.
ACKNOWLEDGMENTS
We thank Roel van Driel (Amsterdam, The Netherlands) and Peter Hemmerich (Jena, Germany) for the generous gifts of antibodies.
This work was supported by SFB796.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Adler M., Tavalai N., Muller R., Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the ND10 component Sp100. J. Gen. Virol. 92:1532–1538 [DOI] [PubMed] [Google Scholar]
- 2. Ahn J. H., Brignole E. J., III, Hayward G. S. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn J. H., Hayward G. S. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andreoni M., Faircloth M., Vugler L., Britt W. J. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157–167 [DOI] [PubMed] [Google Scholar]
- 5. Boutell C., Orr A., Everett R. D. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantrell S. R., Bresnahan W. A. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L. Y., Chen J. D. 2003. Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol. Cell. Biol. 23:7108–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuchet D., et al. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J. Cell Sci. 124:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Everett R. D., Chelbi-Alix M. K. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 10. Everett R. D., Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Everett R. D., Parada C., Gripon P., Sirma H., Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Everett R. D., Parsy M. L., Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Everett R. D., et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greaves R. F., Mocarski E. S. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grötzinger T., Jensen K., Will H. 1996. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-γ activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J. Biol. Chem. 271:25253–25260 [DOI] [PubMed] [Google Scholar]
- 16. Huh Y. H., et al. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82:10444–10454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hwang J., Kalejta R. F. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334–338 [DOI] [PubMed] [Google Scholar]
- 18. Ishov A. M., Maul G. G. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishov A. M., et al. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishov A. M., Stenberg R. M., Maul G. G. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jahn G., et al. 1984. Predominant immediate-early transcripts of human cytomegalovirus AD 169. J. Virol. 49:363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang H., et al. 2006. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J. Gen. Virol. 87:2181–2190 [DOI] [PubMed] [Google Scholar]
- 23. Korioth F., Maul G. G., Plachter B., Stamminger T., Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158 [DOI] [PubMed] [Google Scholar]
- 24. Krauss S., Kaps J., Czech N., Paulus C., Nevels M. 2009. Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J. Virol. 83:12854–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lallemand-Breitenbach V., et al. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee H. R., et al. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lorz K., et al. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lukashchuk V., McFarlane S., Everett R. D., Preston C. M. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mocarski E. S., Shenk T., Pass R. F. 2007. Cytomegaloviruses, p. 2701–2772In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 30. Müller S., Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Negorev D., Maul G. G. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242 [DOI] [PubMed] [Google Scholar]
- 32. Negorev D. G., et al. 2010. Sp100 as a potent tumor suppressor: accelerated senescence and rapid malignant transformation of human fibroblasts through modulation of an embryonic stem cell program. Cancer Res. 70:9991–10001 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Paulus C., Krauss S., Nevels M. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U. S. A. 103:3840–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109–120 [DOI] [PubMed] [Google Scholar]
- 35. Preston C. M., Nicholl M. J. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113–1121 [DOI] [PubMed] [Google Scholar]
- 36. Reichelt M., et al. 2011. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 7:e1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosa-Calatrava M., Puvion-Dutilleul F., Lutz P., Dreyer D., de Thé H., Chatton B., Kedinger C. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 4:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saffert R. T., Kalejta R. F. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stadler M., et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565–2573 [PubMed] [Google Scholar]
- 41. Sternsdorf T., Jensen K., Will H. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tatham M. H., et al. 2005. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 12:67–74 [DOI] [PubMed] [Google Scholar]
- 43. Tavalai N., Papior P., Rechter S., Leis M., Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tavalai N., Papior P., Rechter S., Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tavalai N., Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 46. Tavalai N., Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tavalai N., Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 157:128–133 [DOI] [PubMed] [Google Scholar]
- 48. Ullman A. J., Hearing P. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82:7325–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ullman A. J., Reich N. C., Hearing P. 2007. Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J. Virol. 81:4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waldo F. B., Britt W. J., Tomana M., Julian B. A., Mestecky J. 1989. Non-specific mesangial staining with antibodies against cytomegalovirus in immunoglobulin-A nephropathy. Lancet i:129–131 [DOI] [PubMed] [Google Scholar]
- 51. Wathen M. W., Stinski M. F. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J. Virol. 41:462–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilkinson G. W., Kelly C., Sinclair J. H., Rickards C. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79(Pt 5):1233–1245 [DOI] [PubMed] [Google Scholar]
- 53. Winkler M., aus Dem Siepen T., Stamminger T. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Woodhall D. L., Groves I. J., Reeves M. B., Wilkinson G., Sinclair J. H. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660 [DOI] [PubMed] [Google Scholar]
- 55. Xu Y., et al. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhong S., et al. 1999. PML is essential for proper formation of the nuclear body. Blood 94:489A [Google Scholar]
- 57. Zhong S., et al. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2753 [PubMed] [Google Scholar]
- 58. Zhong S., Salomoni P., Pandolfi P. P. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85–E90 [DOI] [PubMed] [Google Scholar]