Abstract
Fission yeast (Schizosaccharomyces pombe) centromeres are composed of large (40–100 kb) inverted repeats that display heterochromatic features, thus providing a good model for higher eukaryotic centromeres. The association of three proteins that mediate region-specific silencing across centromere 1 has been mapped by quantitative chromatin immunoprecipitation. Swi6 and Chp1 are confined to the flanking outer repeats and Swi6 can spread across at least 3 kb of extraneous chromatin in cen1. In contrast, Mis6 coats the inner repeats and central core. tRNA genes demarcate this transition zone. These analyses clearly define two distinct domains within this complex centromere which interact with different proteins.
Keywords: Centromere, spreading, chromatin immunoprecipitation, chromodomain, silencing, Schizosaccharomyces pombe
Accurate chromosome segregation is dependent on the activity of the centromere, a highly specialized region of chromatin that assembles kinetochore proteins to mediate spindle attachment. Mammalian and Drosophila centromeres are large, cytologically distinct structures, which are composed of repetitive sequences, and show features of heterochromatin (Brown et al. 1994; Karpen 1994; Murphy and Karpen 1995; for review, see Weiler and Wakimoto 1995; Lee et al. 1997; Sun et al. 1997). Little is known about exact sequence requirements for centromere function in these large regional centromeres. In sharp contrast, the centromeres of the budding yeast are very small and well defined, both in terms of their DNA sequence requirements and the localization of centromeric proteins (Espelin et al. 1997; Meluh and Koshland 1997; Meluh et al. 1998 and references therein). However, fission yeast centromeres are more evocative of higher eukaryotic centromeres than those of budding yeast. They are composed of large inverted repeats with a central core of more unique sequence, surrounded by inner (imr/B) and outer (otr/K+L) repeats (Clarke et al. 1986; Nakaseko et al. 1986, 1987; Fishel et al. 1988). Marker genes inserted within the centromere are transcriptionally repressed (Allshire et al. 1994, 1995). This silent centromeric chromatin is underacetylated, and perturbation of this state by transient treatment with a histone deacetylase inhibitor leads to the epigenetic inheritance of defective centromere function (Ekwall et al. 1997). Histone deacetylase inhibition also causes delocalization of Swi6, a chromodomain protein that normally localizes to silent chromatin at mating type and telomeres, in addition to centromeres (Ekwall et al. 1995, 1997). Proper localization of Swi6 is dependent on Clr4 and Rik1, and mutation of any of these genes leads to defective centromeric silencing and impaired centromere function (Allshire et al. 1995; Ekwall et al. 1995, 1996).
Clr4 is the fission yeast homolog of Su(var)3-9, a protein with both a chromo and SET domain, involved in transcriptional regulation in Drosophila (Tschiersch et al. 1994; Ivanova et al. 1998). The human homolog (SUV39H1) localizes at centromeres in metaphase-arrested cells, and coimmunoprecipitates with M31, an HP1 family member and homolog of fission yeast Swi6 (Aagaard et al. 1999).
Recently, two essential centromere-specific proteins (Mis6 and Mis12) have been described (Saitoh et al. 1997; Goshima et al. 1999). These proteins are required for determining correct metaphase spindle length and orienting sister chromatids for efficient segregation. Here, a quantitative chromatin immunoprecipitation (ChIP) assay is used to dissect the protein–DNA architecture of centromere 1 (cen1). The relative distribution of three proteins, Swi6, Mis6, and Chp1 across this centromere is presented. In addition, we demonstrate that both Swi6 and Mis6 are capable of spreading over marker genes inserted within the centromere, and show that Swi6 can coat and mediate silencing of large noncentromeric DNA inserts. Thus, proteins important for full centromere function can spread, and this lends support to current models for the plasticity of sites of centromere formation.
Results
Mis6 and Chp1 silence distinct centromeric domains
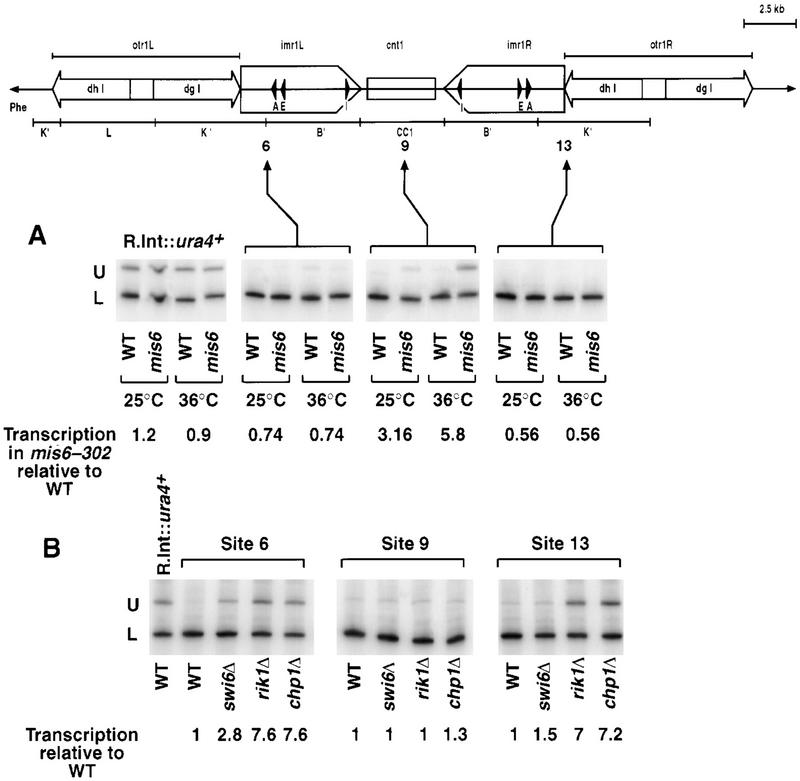
The effect on centromeric silencing of a mutation in the gene encoding Mis6 (Takahashi et al. 1994) was examined. Quantitative RT–PCR (Fig. 1) was performed on cDNA generated from strains with ura4+ located at three sites within cen1 (U) and an expressed ura4–DS/E minigene located at the normal ura4 locus (L). A fully expressed random integrant of ura4+, R.Int::ura4+ served as a control. The PCR assay uses one primer pair to amplify different products from full-length ura4+ (U; 694 bp) and the ura4–DS/E minigene (L; 426 bp). At the restrictive temperature, 36°C, mis6-302 significantly derepressed the ura4+ marker gene when positioned within the central core (site 9, Fig.1A) but not when inserted in the flanking outer and inner repeats (sites 6 and 13). Even at 25°C, mis6-302 alleviated central core silencing, which correlates with elevated rates of chromosome loss (Takahashi et al. 1994). mis6-302 is the first mutation identified to strongly alleviate silencing specifically within the central core of the centromere.
Figure 1.
Chp1 and Mis6 silence distinct centromeric domains. (A) mis6-302 specifically alleviates central core silencing, but not silencing of the outer repeats. (B) Chp1, like Clr4, Rik1 and Swi6 is required for silencing of the outer repeats, but not the central core. For both A and B, competitive radioactive PCR was performed on cDNA generated by RT–PCR from strains with ura4+ (U) inserted within the central core (site 9), the inner and outer repeats of centromere 1 (sites 6 and 13) or in euchromatin (R.Int::ura4+) and a fully expressed ura4–DS/E minigene (L) at the ura4 locus. Separated PCR products were quantified. Levels of ura4 (U) were normalized to ura4–DS/E (L) in the mutant strains and expressed relative to values obtained for the wild-type background for each insertion site. In A, mis6-302 and wild-type strains were grown at 25°C, or shifted to the nonpermissive temperature for 4 hr prior to RNA extraction.
Another chromodomain protein, Chp1, has been identified by sequence homology (Doe et al. 1998; B. Borgstrom and R. Allshire, unpubl.). Like clr4Δ, rik1Δ, and swi6Δ, strains lacking chp1+ show little alleviation of central core silencing (site 9, Fig.1B), but strong derepression of ura4+ within the outer and inner repeats of cen1 (sites 6 and 13, Fig. 1B). This and previous analyses suggests that Mis6 may act and interact exclusively through the central core, whereas Chp1, Clr4, Rik1, and Swi6 might be confined to the flanking repeats.
Mis6 and Swi6 associate with distinct regions of cen1
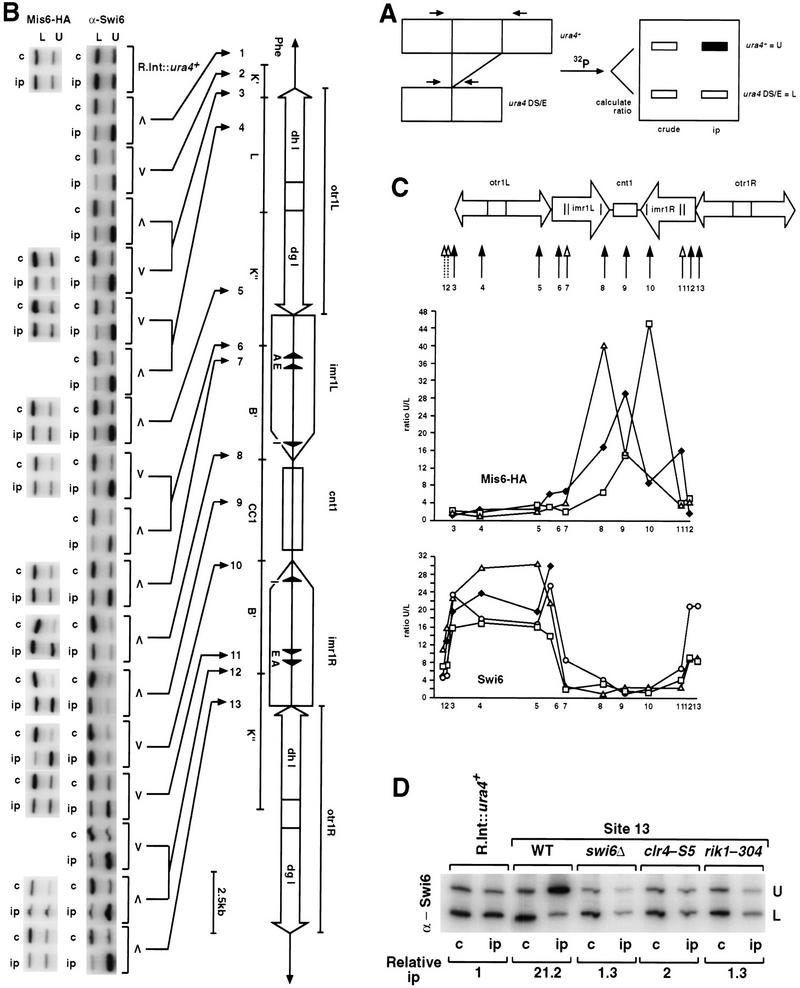
To test whether the region-specific silencing effects reflect differences in the distribution of proteins, ChIP was used. ChIP has been previously used to examine proteins associated with centromeres and telomeres in budding yeast and fission yeast (Hecht et al. 1996; Ekwall et al. 1997; Meluh and Koshland 1997; Saitoh et al. 1997; Strahl-Bolsinger et al. 1997; Meluh et al. 1998; Goshima et al. 1999). However, determining the relative distribution of Chp1, Mis6, and Swi6 proteins across an entire fission yeast centromere is hampered by the fact that all three centromeres share flanking DNA repeats (Clarke et al. 1986; Nakaseko et al. 1986, 1987; Fishel et al. 1988). Thus, little information on protein localization over the outer repeats can be generated from ChIP experiments, as probes that detect specific centromere outer repeats cannot be designed. To overcome this difficulty, strains with the ura4+ marker located at 13 different positions within, or adjacent to cen1 were utilized (Allshire et al. 1995). Each ura4+ insertion site provides a specific tag for different cen1 regions and all strains also carry the ura4–DS/E minigene at the normal ura4 locus. Quantitative competitive PCR is performed on immunoprecipitated chromatin to assess enrichment of each centromeric ura4+ relative to the ura4 locus (Fig. 2A). A strain with the ura4+ gene inserted at a fully expressed site (R.Int::ura4+) serves as a control. To test this assay, we examined the distribution of Mis6 tagged with 3xHA epitopes across cen1 as Mis6–3xHA is known to immunoprecipitate central core and imr, but not otr, chromatin (Saitoh et al. 1997). Mis6–3xHA efficiently immunoprecipitated ura4+ at the central core (site 9) and at imr (sites 8 and 10), but little association of Mis6–3xHA was detected at other insertion sites (Fig. 2B,C). Quantification of three independent experiments demonstrates that central core and inner repeat ura4+ insertions are enriched in Mis6–3xHA ChIPs. These data correlate well with the pattern of mis6+ dependent silencing across cen1 (Fig.1A).
Figure 2.
Mis6 and Swi6 associate with distinct domains of cen1. (A) Strains with ura4+ at different positions within cen1 (U) and the ura4–DS/E minigene at the ura4 locus (L), were formaldehyde fixed. Chromatin was prepared and sheared to 500–1000 bp prior to immunoprecipitation with anti-Swi6 or anti-HA antibodies (Mis6–3xHA strains). Recovered immunoprecipitated DNAs (ip) were compared with input crude DNA (c) by competitive PCR of ura4+. (B) Quantitative chromatin immunoprecipitation shows that Swi6 associates with the outer repeats, and Mis6 with the inner repeats and central core of cen1. Chromatin immunoprecipitation from strains with ura4 inserted at different positions within cen1 (represented by cartoon at right) is shown. For some insertion sites, two strains bearing different orientations of ura4+ were tested (open arrowheads; see Allshire et al. 1995). Results for both Swi6 and HA immunoprecipitation of Mis6–3xHA strains is shown. (C) Quantitation of these results is plotted. Levels of enrichment of ura4 (U) at each insertion site were quantified relative to ura4–DS/E (L) and normalized to values obtained for strains bearing a random integrant of ura4 (R.Int::ura4+). Results for several experiments investigating Swi6 and Mis6–3xHA immunoprecipitation are plotted. (D) Swi6 immunoprecipitation of centromeric chromatin is Clr4, Rik1, and Swi6 dependent. Swi6 chromatin immunoprecipitation of ura4+ (U) at centromeric site 13 is abolished in yeast bearing mutations in clr4 and rik1.
Next, the distribution of Swi6 across cen1 was examined (Fig. 2B,C). Swi6 antibodies (Ekwall et al. 1995) were used to immunoprecipitate chromatin extracts prepared from cen1–ura4+ tagged strains. Analysis of the immunoprecipitated chromatin showed strong association of Swi6 with ura4+ tags in the outer repeats (sites 3–6, 12–13). Little or no association of Swi6 was observed with the inner repeat (sites 7, 8, 10, 11) or central core domains (site 9). The symmetry of both cen1 and the Swi6 distribution profile underscores the reproducibility of this procedure.
Swi6 immunolocalization requires functional Clr4 and Rik1 proteins (Ekwall et al. 1996). Figure 2D shows that Swi6 immunoprecipitation of ura4+ at site 13 is also abolished in cells lacking functional Clr4 and Rik1. This correlates with the loss of silencing seen at this site in clr4Δ and rik1Δ mutants (Fig. 1B; Allshire et al. 1995), and demonstrates the specificity of anti-Swi6 ChIP. These data reveal two distinct domains of silencing within cen1, with Swi6 and Mis6 distributed in a reciprocal fashion across the centromere. No change in Mis6 association across cen1 was seen in clr4Δ, rik1Δ, or swi6Δ mutants (data not shown), suggesting that Clr4, Rik1, or Swi6 do not act to restrict Mis6 to the central domain.
Defining the borders of Swi6 and Mis6 cen1 association
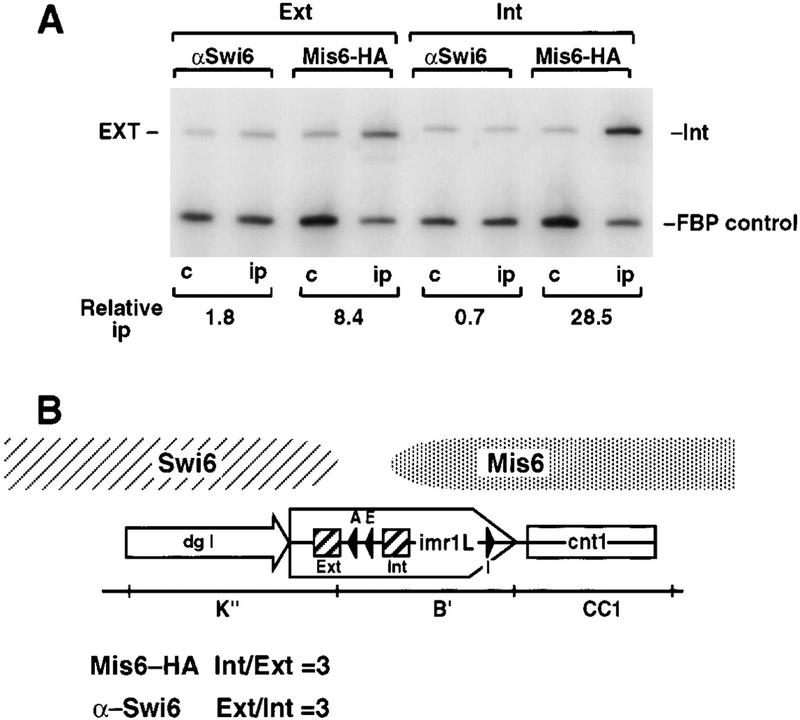
Our analyses indicate that Swi6 does not associate with regions internal to sites 7 (LHS) and 11 (RHS) located between the tRNAAla and tRNAGlu genes. In contrast, Mis6 appears to be confined to these inner repeats and central core regions. Using the available cen1–ura4+ tags, we cannot clearly define the limits of Mis6 association with cen1. To map the boundaries of protein association, primers were designed to specifically amplify sequence adjacent to both the left and right copies of the tRNAAla and tRNAGlu genes and to quantify enrichment of these sequences in Mis6 and Swi6 chromatin immunoprecipitates (Fig. 3). One example of these immunoprecipitations is shown (Fig. 3A), with the average results for several experiments presented in Figure 3B. Mis6 associates threefold more with sequence just interior to the tRNAs (Int) than sequence exterior to the tRNAs (Ext). In contrast, Swi6 immunoprecipitates gave a threefold enrichment of product Ext over product Int. These figures are an underestimate because the chromatin is only sheared to 500–1000 bp and the two probes lie just 700 bp apart. Because both Swi6 and Mis6 are capable of spreading (see below), the observation that these proteins show differential centromere association within a 700-bp region suggests that the region encompassing the tRNAAla and tRNAGlu genes defines a transition zone between distinct centromere domains.
Figure 3.
tRNA genes demarcate a transition zone between Mis6 and Swi6 cen1 association. (A) PCR primers were designed to amplify sequences internal (Int) or external (Ext), indicated by the hatched boxes in imr1L, surrounding the tRNAAla and tRNAGlu genes (filled arrowheads, B) from immunoprecipitated samples. Swi6 and Mis6–3xHA immunoprecipitates were assessed for the levels of Ext and Int PCR products relative to the fbp1+ control. (B) Diagram of cen1 showing the mapping of the transition zone between centromeric Mis6–3xHA and Swi6 to a region containing tRNA genes. Mis6–3xHA immunoprecipitates gave an average threefold enrichment of Int product over Ext, and Swi6, a threefold enrichment of Ext over Int.
Chp1 associates with flanking centromere repeats
Because deletion of chp1 derepresses centromeric silencing (Fig. 1B), attempts were made to use ChIP to examine the distribution of functional-tagged Chp1 (Chp1–6xMyc, B. Borgstrøm and R. Allshire, unpubl.) across cen1. However, no enrichment of any cen1–ura4+ sites was detected in Chp1–6xmyc immunoprecipitates (data not shown). The primers used for PCR to detect ura4+ in these ChIP experiments were ∼570 and 500 bp from the ends of the ura4+ tags and the released chromatin–DNA was sonicated to 500–1000 bp prior to immunoprecipitation. Thus, detection of cen1–ura4+ in immunoprecipitates requires that proteins of interest contact the main body of the ura4+ marker. Swi6 and Mis6 are clearly able to encroach into centromeric ura4+ genes, but our failure to detect association of Chp1–6xMyc might be because Chp1 cannot spread from neighboring centromeric chromatin. We therefore tested for direct interaction of Chp1 with centromeric sequences at two positions within cen1 that can be specifically amplified by PCR (Fig. 4A). Chp1–6xMyc, like Swi6 immunoprecipitates, are enriched for centromeric imr/otr repeat chromatin, but not the central core region, when compared with immunoprecipitation of the euchromatic control fbp1+ gene locus. This is in contrast to Mis6, which associates only with the central core, and not the centromeric repeat region in this assay.
Figure 4.
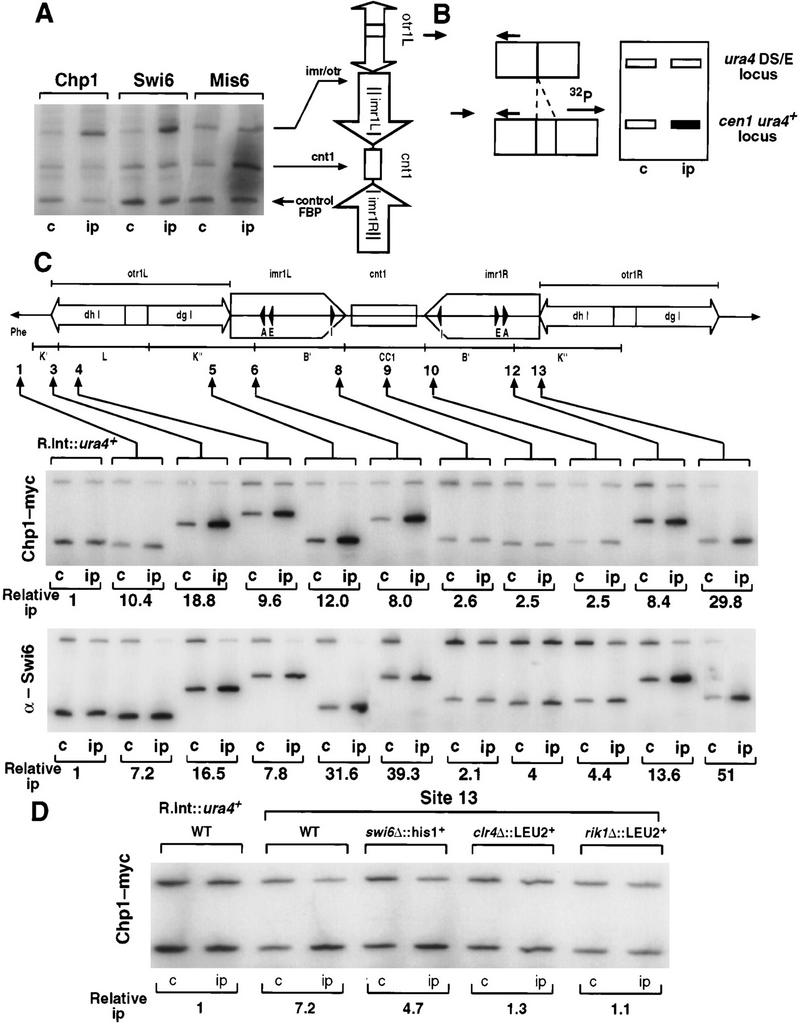
Chp1 displays a similar cen1 association pattern as Swi6. (A) Multiplex PCR was performed to detect association of Chp1–6xMyc with centromeric chromatin. Primers were designed to two sites (imr/otr junction and a region of cnt1), which give amplification specifically from cen1 sequences, and to the euchromatic fbp1+ gene locus to act as a control for nonspecific association. Chp1–6xMyc and Swi6 immunoprecipitates both showed enrichment of the imr/otr product relative to fbp1 and showed no enrichment for the central core sequence (cnt1). In contrast, Mis6–3xHA immunoprecipitates showed enrichment for cnt1 and not for imr/otr. (B) Chp1–6xMyc interactions across cen1 were mapped by specific PCR from immunoprecipitates of strains with different cen1–ura4 insertions using various primers from cen1 and one primer anchored in the ura4+ gene. Enrichment of centromeric ura4+ by immunoprecipitation is reflected by increased intensity of the smaller PCR products, which vary in size from different strains, depending on the location of the centromeric primers, relative to the large PCR product of constant size that reflects association with the euchromatic ura4–DS/E locus. (C) Using this assay, Chp1–6xMyc and Swi6 associate with the flanking repeats but not the central core of cen1. Relative ip values are an average of 2 (Swi6), and 3 (Chp1–myc) experiments. (D) Chp1–6xMyc immunoprecipitation at site 13 is dependent on Clr4 and Rik1 but not Swi6.
Chp1 can clearly associate with centromeric chromatin, but cannot encroach on ura4+ genes inserted within the centromere. Therefore, an alternative PCR assay was devised to map interactions of Chp1–6xMyc across cen1 (Fig. 4B). This assay examines Chp1–6xMyc association with centromeric sequences flanking each cen1–ura4+ tag. A semicompetitive PCR assay was set up with one primer anchored at the end of ura4+ (which also recognizes ura4–DS/E) and a primer homologous to centromeric sequences neighboring each centromeric ura4+ insertion site. The centromeric primer is different for each insertion site, and thus generates PCR products of various sizes. The ura4+ primer provides specificity for each cen1 insertion site. Inclusion of an additional primer from sequence adjacent to the ura4–DS/E locus in all reactions yields the larger (550 bp) control product (see Fig. 4B). This assay was tested with anti-Swi6 immunoprecipitates and the results (Fig. 4C) demonstrate specific association of Swi6 with the outer repeats and fully support those presented in Figure 2B. Our data suggests that there is no significant difference in the ability of Swi6 to associate with centromeric sequences compared with exogenous DNA inserted within the centromere (cf. Fig. 2B,C with Fig. 4C). In contrast, when Mis6–3xHA immunoprecipitates were tested, we saw up to eightfold more enrichment for binding the flanking centromeric sequences than for binding centromeric ura4+ (data not shown).This indicates that there may be a gradient of Mis6 over these ura4+ inserts. This same PCR assay was then applied to Chp1–6xMyc immunoprecipitates and analyses of all centromeric sites revealed a similar distribution of Chp1–6xMyc and Swi6 across cen1 (Fig. 4C). Chp1 interacts strongly with flanking repeat regions but not with the central domain of cen1. Immunolocalization of Chp1–6xMyc, like Swi6, depends on functional Clr4 and Rik1, but is independent of Swi6 (B. Borgstrøm and R. Allshire, unpubl.). The ability of Chp1–6xMyc to immunoprecipitate cen1-flanking repeat chromatin is also lost in clr4Δ and rik1Δ strains but maintained in swi6Δ, validating this modified ChIP–PCR assay (Fig. 4D).
Thus, two chromodomain proteins, Swi6 and Chp1, associate with the outer repeats of cen1, and require Clr4 and Rik1 for localization. However, they differ in their behavior, because Swi6, but not Chp1, can spread across exogenous ura4+ chromatin assembled within the centromere. The absence of apparent Chp1 spreading could reflect the sensitivity of the method and less total Chp1 in the cell, rather than an absolute difference in the behavior of the proteins.
Swi6 can spread over large insertions of exogenous DNA
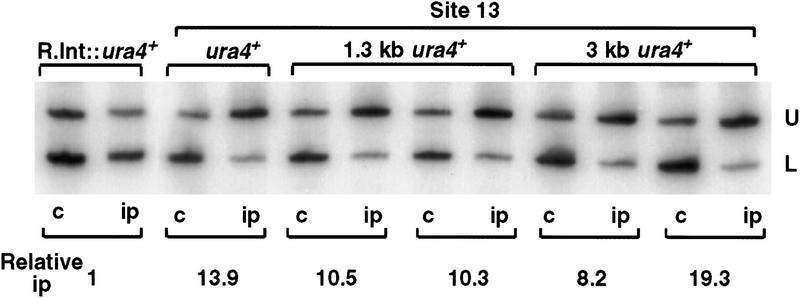
To further test this ability of Swi6 to spread, two strains were constructed. In one strain (1.3 kb), 1.3 and 1.7 kb of noncentromeric sequence were used to flank ura4+ at site 13 and in the other (3 kb), this flanking sequence was increased to 3 and 5 kb. The anti-Swi6 ChIP assay showed (Fig. 5) that equivalent levels of ura4+ at site 13 are associated with Swi6 irrespective of the length of the surrounding noncentromeric DNA. Thus, Swi6 can spread over at least 3 kb of additional noncentromeric sequence flanking the ura4+ gene inserted within cen1, and this spreading correlates with efficient silencing of the ura4+ gene (data not shown).
Figure 5.
Swi6 can spread over noncentromeric DNA. Strains with 1.3 kb or 3 kb surrounding ura4+ and inserted at site 13 showed equivalent levels of Swi6 chromatin immunoprecipitation of ura4+ as strains with only ura4+ at that site.
Discussion
Quantitative mapping by ChIP of three proteins across a large and complex fission yeast centromere has been performed. These analyses reveal the presence of discrete domains of protein interaction within cen1, with Mis6 restricted to the central core and inner repeats, and Swi6 and Chp1 associated with the outer repeats. Swi6 and Mis6 are both capable of spreading over silent ura4+ chromatin placed within cen1, and Swi6 can efficiently coat and silence >3 kb of exogenous DNA inserted within cen1.
Mutational analysis of cen1 on minichromosomes has revealed the importance of both a central core and some flanking repeat sequence for the formation of a mitotically stable centromere. This minimal centromere corresponds to sequences distal to site 5 and encompasses sequence close to site 12 (Baum et al. 1994). Further truncation of this minimal centromere, causing removal of site 5 and some core proximal sequence, leads to loss of mitotic function (Hahnenberger et al. 1991). Our protein mapping data suggests, therefore, that both inner repeats and central core sequences (capable of being bound by Mis6), and sequences capable of high levels of Swi6/Chp1 association, are required for efficient mitotic segregation of minimal centromere constructs.
The transition between the outer repeat and central core/inner repeat protein complexes is demarcated by a region encompassing the inner repeat tRNA genes. tRNA genes are present within the inner repeat sequences of all three centromeres (Takahashi et al. 1991), and may contribute to the definition of distinct protein domains in natural centromeres. However, hybrid minimal centromeres can be formed that lack these tRNA genes, but these minichromosomes require additional spacer DNA to maintain spatial separation of the domains for function (Baum et al. 1994). Recently, a 1-kb region including a tRNA gene was identified as having insulator function at the silent HMR locus in Saccharomyces cerevisiae (Donze et al. 1999). An interesting possibility is that the tRNAAla and tRNAGlu genes play a role in demarcating the two distinct protein interaction domains in fission yeast cen1.
There have been several reports of proteins that are able to spread from specific nucleation points to either activate or suppress gene expression of neighboring chromatin. Silencing at S. cerevisiae telomeres is mediated by a complex including Sir3p, which is recruited to the telomere by binding of Rap1p to telomeric repeats. The Sir3p complex normally spreads from the terminal 300-bp nucleation sites, and associates with and promotes silencing of 3 kb of telomere adjacent sequence. Upon overexpression, Sir3p has been detected 16-kb away. This spreading is thought to be mediated by interaction of Sir3p with the underacetylated tails of histones H3 and H4 (Hecht et al. 1995, 1996; for review, see Grunstein 1998). It has been reported recently that the dosage compensation complex MSL, which binds the X chromosome in Drosophila, shows high affinity binding to ∼35 sites on the chromosome. These sites are thought to be chromatin entry and nucleation sites, from which the MSL complex spreads to coat the X chromosome, and activate gene expression (Kelley et al. 1999). Interestingly, two of the MSL components (MSL3 and MOF) contain chromodomains (Gorman et al. 1995; Hilfiker et al. 1997). In addition, Polycomb, a Drosophila chromodomain protein can spread from its nucleation sequence, the Polycomb response element, to alter gene expression at a distance (for review, see Moehrle and Paro 1994). Drawing on these models, we propose that at fission yeast centromeres, Clr4 and Rik1 serve as nucleation factors for the assembly of a protein complex containing both Chp1 and Swi6 on the outer repeats of the centromere. Swi6 can then spread from these nucleation points, possibly via underacetylated centromeric chromatin (Ekwall et al. 1997), simultaneously creating a chromatin structure that blocks transcription of embedded genes, and which mediates the assembly of a fully functional kinetochore.
Current evidence suggests that the formation of centromeres is subject to epigenetic regulation with a certain degree of plasticity in the sites selected for centromere assembly. For example, active new centromeres (neocentromeres) can be formed in the absence of any recognizable centromeric DNA sequences in both humans and Drosophila. These noncentromeric sequences may be activated by spreading of centromere character in cis from nearby functional centromeres (Murphy and Karpen 1998; Williams et al. 1998). This centromeric character may take the form of specialized chromatin, or be induced by the spreading of specific centromere proteins, which promote the formation of an active kinetochore. The clear spreading of fission yeast centromere proteins reported here suggests that the plasticity in centromere formation may be mediated in part by spreading of chromatin associated proteins.
Materials and methods
Strain construction
Strains were obtained by crossing centromeric ura4+ insertion strains (Allshire et al. 1995) with mis6-302 (Takahashi et al. 1994), chp1Δhis3+ (B. Borgstrøm and R. Allshire, unpubl.), Mis6–3xHA (Saitoh et al. 1997) and Chp1–6xMyc (B. Borgstrøm and R. Allshire, unpubl.) and verified by the presence of markers and PCR analysis. To generate strains with additional sequence flanking ura4+ at centromeric insertion site 13, homologous recombination was used to replace a centromeric otr1R::ade6+ marker at site 13 (Allshire 1996). Replacement was performed with a BamHI–XhoI fragment of ade6 with ura4+ inserted within the HindIII site to generate the strain with 1.3 kb flanking ura4. To insert 3 kb flanking ura4+, the BamHI–XhoI fragment of ade6+ was inserted into pJK210 (Keeney and Boeke 1994), and NdeI linearized plasmid was integrated at otr1R::ade6+.
RNA analysis
Yeast were grown in YES at 32°C to 5 × 106 cells/ml. mis6-302 and controls were grown at 25°C to 5 × 106 cells/ml or shifted to 36°C for 4 hr prior to RNA extraction. cDNA was prepared by oligo dT primed RT–PCR and competitive PCR of ura4+ and ura4–DS/E was performed as described previously (Ekwall et al. 1997). ura4+ levels (U) were normalized to ura4–DS/E (L) and quantified relative to wild-type strains.
Formaldehyde cross-linked ChIP
Fission yeast were grown at 32°C in YES to 5 × 106 cells/ml, shifted to 18°C for 2 hr prior to 30-min fixation with 3% formaldehyde. Soluble chromatin was prepared and immunoprecipitated as described for nonabundant antigens (Ekwall and Partridge 1999) using affinity-purified Swi6 antibodies (Ekwall et al. 1995), α-HA (12CA5) or α-myc (9E10). Recovered DNAs were assayed by PCR including [32P]dCTP, resolved on 4% native polyacrylamide gels, dried, and exposed to a PhosphorImager cassette. Bands were quantified on a Storm PhosphorImaging system (Molecular Dynamics) using Imagequant software. Swi6 and Mis6–3xHA immunoprecipitates (Fig. 2) were assayed by competitive PCR of ura4+ as described previously (Ekwall et al. 1997). For direct analysis of centromeric chromatin in immunoprecipitates (Fig. 4A), primer pairs for core (5′-AACAATAAACACGAATGCCTC-3′, 5′-ATAGTACCATGCGATTGTCTG-3′), imr/otr junction (5′-CACATCATCGTCGTACTACAT-3′, 5′-GATATCATCTATATTTAATGACTACT-3′) and fbp1+ euchromatic control (5′-AATGACAATTCCCCACTAGCC-3′, 5′-ACTTCAGCTAGGATTCACCTGG-3′) were used in multiplex PCR with crude and immunoprecipitated chromatin from R.Int::ura4+ strains. Centromeric chromatin flanking ura4+ insertion sites (Fig. 4B,C,D) was assayed with the ura4 primer (5′-TATACAAACAAATACACTAGG-3′), the ura4–DS/E locus primer (5′-GCCATGTCAGATTTGACACAACTC-3′) and centromeric primers for each insertion site (sequences available on request). PCR reactions were set up with 200 ng of the ura4 primer, 100 ng of ura4–DS/E locus primer and 100 ng of centromeric primer in 20-μl reactions including 1.9 mm MgCl2 and 4 μl of immunoprecipitated material or 4 μl of 1/100 dilution of crude material. Relative immunoprecipitation values were calculated for 2 (Swi6) and 3 (Chp1-myc) experiments.
Immunoprecipitation of sequences around the tRNA genes (Fig. 3) was assessed using Interior (Int, 5′-GTCGAATTGAGATGTAAACG-3′, 5′-GCCGGAATAGTAATTATGGC-3′) and Exterior primers (Ext, 5′-GGGCACAAATATTTATGGAC-3′, 5′-GCAGTTGGTACAACGAATGACCTG-3′) with control fbp1+ primers in duplex radioactive PCR on Swi6 and Mis6–3xHA immunoprecipitates. Enrichment of Int and Ext in immunoprecipitated samples relative to fbp1 was calculated, normalized to ratios obtained for crude samples, and ratios averaged from 8 (Swi6) and 3 immunoprecipitations (Mis6–3xHA).
Acknowledgments
We thank Professor M. Yanagida for the gift of mis6 strains, and the Photography Department for their good-humored help. Thanks to Allshire laboratory members, both past and present, for enthusiastic support, and W. Bickmore, H. Cooke, K. Hardwick, N. Hastie, and C. Gordon for comments on the manuscript. This work was supported by the UK Medical Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL robin.allshire@hgu.mrc.ac.uk; FAX 44-(0)131-343-2620.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. Transcriptional silencing in the fission yeast: A manifestation of higher order chromosome structure and functions; pp. 443–466. [Google Scholar]
- Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat J-P, Cranston G. Mutations derepressing silent domains within fission yeast centromeres disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WR. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- Clarke L, Amstutz H, Fishel B, Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Wang G, Chow C-M, Fricker MD, Singh PB, Mellor EJ. The fission yeast chromo domain encoding gene chp1+ is required for chromosome segregation and shows a genetic interaction with alpha-tubulin. Nucleic Acids Res. 1998;26:4222–4229. doi: 10.1093/nar/26.18.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Partridge JF. Fission yeast chromosome analysis: Fluorescence in situ hybridisation (FISH) and chromatin immunoprecipitation (CHIP) In: Bickmore WA, editor. Chromosome structural analysis, a practical approach. Oxford, UK: Oxford University Press; 1999. pp. 39–57. [Google Scholar]
- Ekwall K, Javerzat J-P, Lorentz A, Schmidt H, Cranston G, Allshire RC. The chromodomain protein Swi6: A key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat J-P, Borgstrom B, Egel R, Cranston G, Allshire RC. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localization of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M, Franke A, Baker BS. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development. 1995;121:463–475. doi: 10.1242/dev.121.2.463. [DOI] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hahnenberger KM, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJS. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- Karpen GH. Position effect variegation and the new biology of heterochromatin. Curr Opin Genet Dev. 1994;4:281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Moehrle A, Paro R. Spreading the silence: Epigenetic transcriptional regulation during Drosophila development. Dev Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Kinoshita N, Yanagida M. A novel sequence common to the centromere regions of Schizosaccharomyces pombe chromosomes. Nucleic Acids Res. 1987;15:4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen GH. Molecular structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol. 1991;218:13–17. doi: 10.1016/0022-2836(91)90867-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]