Abstract
We describe here the Arabidopsis gene DAG1, encoding a zinc finger transcription factor of the Dof family, and show that it is involved in the control of seed germination. By a reverse genetics approach, we isolated an Arabidopsis mutant line with one T-DNA insertion in DAG1. Seeds from homozygous knockout dag1-1 plants do not develop dormancy and germinate also in the absence of light. Segregation analysis indicates that the effect of the mutation is maternal. Accordingly, in situ mRNA hybridizations reveal expression of DAG1 in the vascular tissue of the flower and maturing fruit but not in the seed.
Keywords: Arabidopsis, seed germination; dormancy; abscisic acid; zinc finger; Dof proteins
The seeds of several plant species do not germinate immediately after detachment from the mother plant, because the embryos that they contain are dormant. Dormancy, which is initiated during the final stages of seed development, is a trait of considerable adaptive significance—it prevents seeds from germinating under unfavorable conditions—as well as of great agricultural and biotechnological interest. A considerable body of evidence indicates that seed dormancy is induced by the plant hormone abscisic acid (ABA), which is also capable of inhibiting seed germination; mutants impaired in either ABA biosynthesis (aba) or sensitivity (abi) show reduced dormancy (Koornneef and Karssen 1994; Bewley 1997). Another class of plant growth factors, gibberellins (GA), are necessary for germination, and severe mutant alleles of the ga1 locus, which are required for GA biosynthesis, fail to germinate (Koornneef and Karssen 1994). Dormancy can be relieved by a period of dry storage of the seeds, and in some plant species, including Arabidopsis, germination requires light (Koornneef and Karssen 1994; Bewley 1997). However, the molecular mechanisms controlling seed dormancy and germination are, as yet, very poorly understood. Here, we report the identification of the Arabidopsis gene DAG1 (Dof affecting germination), encoding a Dof zinc finger transcription factor, and show that is involved in the control of seed dormancy.
The Dof proteins are a recently discovered family of plant transcription factors, ubiquitously present in the plant kingdom but absent from yeast and animals. These proteins are characterized by a strikingly conserved 52-amino-acid segment, denominated Dof domain, encompassing a single CX2CX21CX2C zinc finger (Yanagisawa 1995; Zhang et al. 1995; De Paolis et al. 1996; Vicente-Carbajosa et al. 1997; Mena et al. 1998); in contrast, their sequences diverge widely outside the Dof domain. These features suggested that the Dof proteins are involved in the regulation of different functions typical of and of general relevance for plants. The few Dof proteins studied so far have been shown to be involved in gene regulation by light in maize (Yanagisawa and Sheen 1998), in the activation of storage protein genes in maize and barley (Vicente-Carbajosa et al. 1997; Mena et al. 1998), and in the the regulation of stress-related genes in Arabidopsis (Chen et al. 1996). Very recently, we have shown that the tobacco Dof protein NtBBF1 controls expression of the oncogene rolB in plants (Baumann et al. 1999).
We identified the Arabidopsis Dof gene DAG1 by sequence homology with the Dof domain of NtBBF1, and we isolated a dag1 knockout mutant by reverse genetics. The seeds of this mutant are nondormant and capable of germinating in the absence of light. This phenotype and its segregation pattern, as well as the pattern of expression of the gene, point to DAG1 as a gene that plays a novel and unexpected maternal role in controlling seed germination.
Results
Isolation and structure of DAG1
An incomplete Arabidopsis DAG1 cDNA isolated previously by sequence homology with the Dof domain of the tobacco gene NtBBF1 (De Paolis et al. 1996) was utilized to isolate the corresponding genomic clone from an Arabidopsis library. Southern blot analysis indicated that the DAG1 protein is encoded by a single gene (not shown). The DAG1 gene was then mapped on chromosome 3 by screening of a YAC library (Camilleri et al. 1998) and localized between markers m424 and nga6. Analysis of the genomic sequence (EMBL accession no. AJ224122) revealed that DAG1 has two introns and encodes a 295-amino-acid putative protein with the single Dof zinc finger domain between amino acid positions 76 and 104.
Isolation of a knockout mutant of DAG1
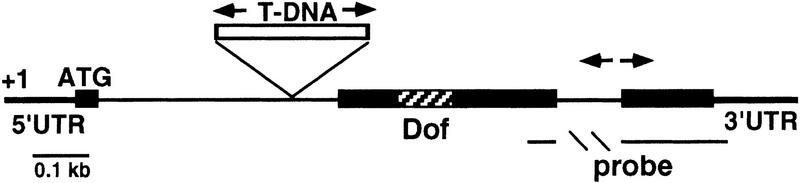
To assess the function of the DAG1 gene, we utilized a reverse genetics approach based on PCR screening of a transferred-DNA (T-DNA) mutagenized population of Arabidopsis plants (Bechtold et al. 1993). Screening of ∼22,000 independent tagged Arabidopsis lines of the Wassilewskija (WS) ecotype, led to the identification of one line with a T-DNA insertion in the DAG1 gene and subsequently to the isolation of the homozygous mutant dag1-1. The mutant was backcrossed with the wild type, and all subsequent characterizations were carried out on the F2 homozygous progeny. Segregation of the kanamycin resistance marker associated with the T-DNA indicated the presence of a single insertion in DAG1, which was confirmed by Southern blot analysis (not shown). Sequencing revealed that the T-DNA is inserted close to the 3′ splice site of the first intron of the DAG1 gene, as schematized in Figure 1. No DAG1 mRNA is detectable by either Northern or RT–PCR analysis in homozygous dag1-1 plants (not shown).
Figure 1.
Structure of the DAG1 gene and position of the T-DNA insertion, which is not drawn to scale. (Solid rectangles) exons; (arrows) primers utilized for the isolation of the mutant line. The DAG1 cDNA fragment was utilized as a probe for Northern analysis and to derive the sense and antisense riboprobes for in situ mRNA hybridizations.
Phenotypic analysis
Development and growth of the dag1-1 knockout plants appear normal, and throughout the rosette (vegetative) stage mutants are indistinguishable from wild-type WS plants. Flowering dag1-1 plants are less branched than WS, but the number of inflorescences per branch is substantially the same and the length of the floral stalk is reduced only modestly (in average, 28.5 ± 0.7 cm for dag1-1 as compared to 32.8 ± 0.7 cm for WS plants). Flowering time and flower morphology of mutant and WS plants do not show appreciable differences.
Remarkably, however, all of the siliques (fruits) from dag1-1 plants have a characteristically twisted appearance, as shown in Figure 2, and are shorter than normal WS siliques (10.8 ± 0.2 mm vs. 13.1 ± 0.3 mm). Kanamycin-resistant heterozygous plants segregated the twisted silique phenotype as a recessive trait with a 3:1 ratio and plants with aberrant siliques proved to be homozygous for the T-DNA insertion, indicating that the mutant phenotype is linked to the insertion.
Figure 2.
Siliques from dag1-1 mutant and WS plants.
dag1-1 seeds are nondormant and do not need light to germinate
The morphological modification of the siliques observed in the dag1-1 mutant prompted us to check whether disruption of the DAG1 gene also affects seed development and/or viability. The twisted siliques were shown to carry a somewhat reduced number of seeds—on average 47.7 ± 4.5 seeds each, as compared to 58.0 ± 5.0 seeds in WS siliques—of normal appearance and containing morphologically normal embryos.
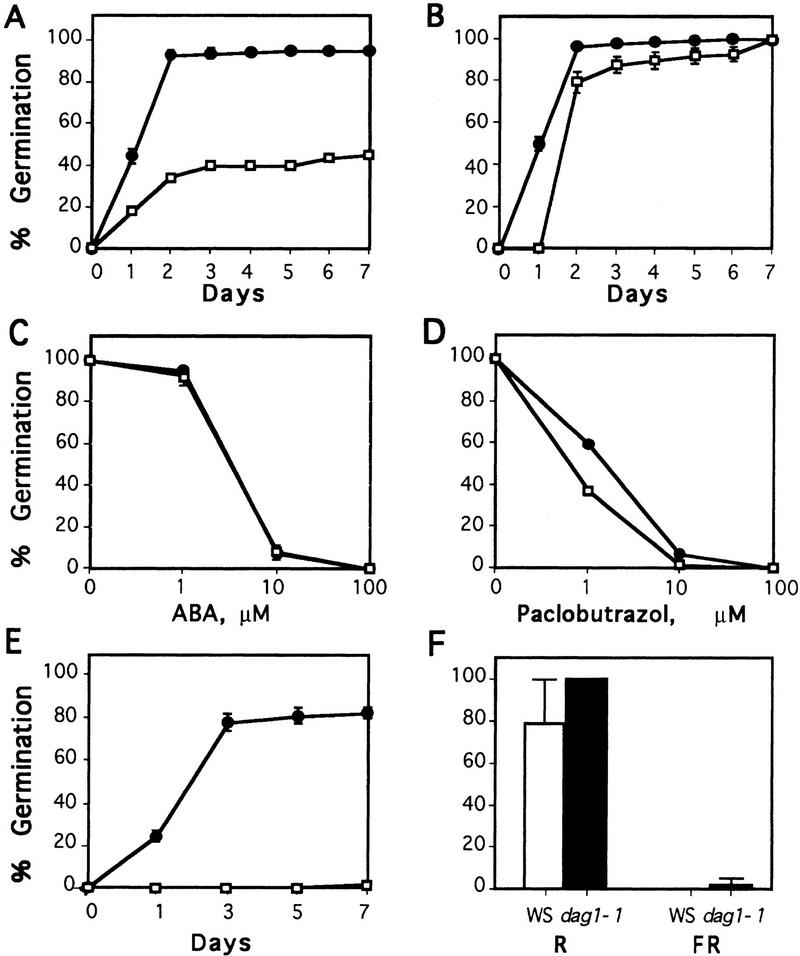
Disruption of the DAG1 gene has, in contrast, strong consequences on the germination of the seeds. As shown in Figure 3A, due to the effect of dormancy the rate of germination of freshly harvested WS seeds is rather low. In contrast, germination of dag1-1 mutant seeds reaches >95%, indicating that they are substantially nondormant. Because storage gradually releases Arabidopsis seeds from dormancy, germination tests were repeated 4 weeks after seed harvest. The capability to germinate normal seeds is, as expected, strongly enhanced by storage, whereas that of dag1-1 mutant seeds, being almost maximal upon harvest, is only fractionally affected (Fig. 3B).
Figure 3.
Germination rates of WS and dag1-1 mutant seeds. (A) Freshly harvested seeds; (B) seeds dry-stored for 4 weeks after harvest; (C) stored seeds, germination in the presence of ABA, scored at day 7; (D) stored seeds, germination in the presence of paclobutrazol, scored at day 7; (E) stored seeds, germination in total darkness [(□) WS seeds; (●) dag1-1 seeds]; (F) stored seeds, germination in total darkness after a red (R) and a far-red (FR) light pulse [(open bars) WS seeds; (solid bars) dag1-1 seeds].
Seed dormancy is induced by ABA, which also inhibits seed germination. Disruption of the DAG1 gene does not appear to affect sensitivity of the seeds to the hormone, as germination of both WS and dag1-1 knockout seeds is inhibited by the same concentration of ABA, as shown in Figure 3C.
Like WS seeds, dag1-1 seeds require GA to germinate: Figure 3D shows that germination of dag1-1 seeds is as sensitive as that of WS seeds to 10 μm paclobutrazol, an inhibitor of GA biosynthesis.
Most strikingly, although stored seeds from WS plants are absolutely incapable of germinating in the absence of light, those from dag1-1 mutant plants germinate in total darkness almost as well as in the presence of light (Fig. 3E). For normal Arabidopsis seeds to germinate, a pulse of red light is necessary to convert the (inactive) Pr form of the light receptor phytochrome to the active Pfr form (which is inactivated by far red light). We assessed that this is indeed the case for the WS ecotype seeds utilized in this work, as their negligible dark germination rate increased to 80% when WS seeds were exposed to a pulse of red light before performing the dark germination tests (Fig. 3F). The high germination rate of dag1-1 seeds in the dark is stimulated further by this initial pulse of red light and reaches 100%. We then tested whether the capability of mutant dag1-1 seeds to germinate in the absence of light is mediated by active phytochrome, by exposing the seeds to a Pfr-inactivating pulse of far red light prior to incubation in the dark. Figure 3F shows that Pfr is required for both the WS and the mutant seeds to germinate in the dark, as no germination of either was observed under these conditions.
The dag1-1 phenotype is due to disruption of DAG1
Conclusive evidence that the phenotype observed in dag1-1 plants is due to disruption of the DAG1 gene was obtained by transforming mutant plants with a genomic clone containing a wild-type copy of DAG1. F3 transformants homozygous for the DAG1 transgene—identified through segregation of hygromycin-resistance trait conferred by the cloning vector—all showed normal-appearing siliques, indicating that DAG1 complements the twisted phenotype. The seeds from these siliques germinated in total darkness at the same low level of freshly harvested wild-type WS seeds, indicating that the exogenous DAG1 complements also the phenotype of dag1-1 seeds.
The effect of the disruption of DAG1 is maternal
To determine whether the phenotype of dag1-1 seeds is due to the mother plant or to the developing embryo, we analyzed the germination of stored seeds from heterozygous DAG1/dag1-1 plants. Two hundred seeds from DAG1/dag1-1 plants were sown in triplicate and scored for germination after 7 days in total darkness. The results are consistent with a maternal effect of this mutation, as none of the seeds from heterozygous DAG1/dag1-1 plants is capable of germinating in the absence of light. Instead, up to one-fourth (dag1-1 homozygous) of the total seeds from heterozygous mothers would be expected to germinate in the dark, if the mutant phenotype were embryo-determined.
DAG1 is expressed in the mother plant but not in the embryo
In view of the maternal effect of the mutation and because seed dormancy initiates during embryo development, we analyzed expression of the DAG1 gene at different stages of development of Arabidopsis flowers, fruits, and embryos. The RT–PCR analysis shown in Figure 4A revealed that the DAG1 gene is expressed in flowers, as indicated by the amplification bands from flowers at stages 12 (lane F12) and 13 (lane F13), that is, before and, respectively, after flower opening (Smyth et al. 1990). After fertilization, DAG1 is expressed in developing siliques (stage 16, lane Si16) but not in fully mature ones (stage 17, lane Si17). As also shown in Figure 4A, in dag1-1 mutant plants no expression of the DAG1 gene is detectable in the corresponding developmental stages and organs. Control amplifications of the cDNA of ATL18 ribosomal protein from the same samples were all positive (not shown).
Figure 4.
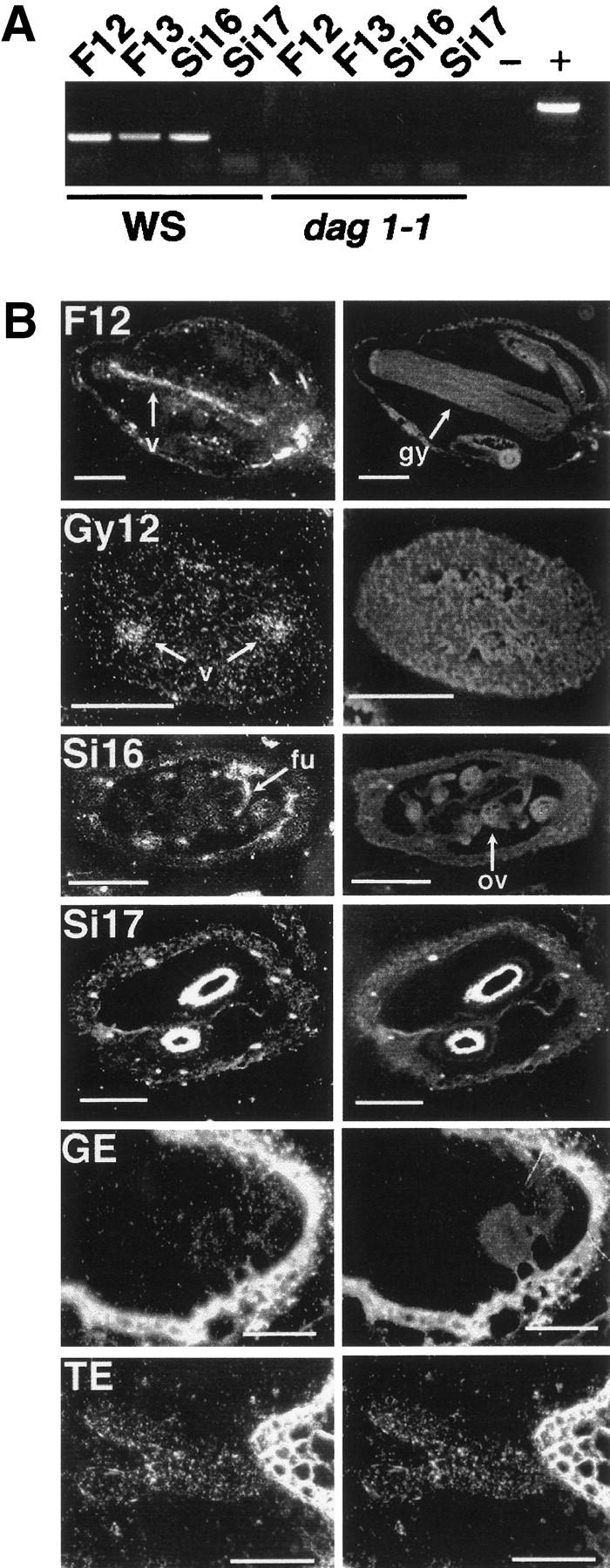
Analysis of DAG1 expression in different organs and developmental stages. (A) RT–PCR with DAG1-specific primers on RNA from WS and dag1-1: (F12, F13) flowers at stages 12 and 13; (S16, S17) siliques at stages 16 and 17. (−) RT–PCR negative control; (+) PCR performed with WS DNA as a template. (B) In situ mRNA hybridizations with tritium-labeled antisense DAG1 riboprobe on sections of WS flowers at stage 12 (F12, longitudinal section), gynoecium of F12 flowers (Gy12, transverse section), siliques at stages 16 (Si16, transverse section) and 17 (Si17, transverse section), late globular-stage embryos (GE, longitudinal section), and torpedo-stage embryos (TE, longitudinal section). (Left) Dark-field images of the hybridizations; (right) corresponding fluorescence micrographs. Arrows point to the vasculature (v), gynoecium (gy), funiculus (fu) and ovule (ov). Bar 250 μm in F12 panels, 200 mm in Gy12, 200 μm in Si16, 400 μm in Si17, 60 μg in GE and TE.
A more detailed view of the pattern of expression of DAG1 was obtained by in situ mRNA hybridizations. As shown in the longitudinal section of a stage 12 flower reported in Figure 4B, panel F12, a clear signal of DAG1 mRNA is visible in the gynoecium, specifically localized in the vascular tissue. Transverse sections of the gynoecium (panel Gy12) confirmed that the DAG1 mRNA signal is localized in the vasculature. After fertilization, expression of DAG1 in stage 16 developing siliques (panel Si16) also is clearly localized in the vascular bundles and extends in the funiculus that connects the placenta to the ovule, but no DAG1 mRNA signal can be detected in the ovule. In agreement with the RT–PCR data, no DAG1 mRNA signal is detectable in any tissue of stage 17 mature siliques (panel Si17). No DAG1 mRNA signal is visible at any stage of development in the embryo, as also shown in Figure 4, where sections of globular (panel GE) and torpedo (panel TE) embryos are reported. No DAG1-specific signal was detectable in any of the above sections with the DAG1 sense riboprobe (not shown). Thus, the fact that the pattern of expression of the DAG1 gene is limited to the mother plant is fully consistent with the maternal effect of the dag1-1 mutation.
Discussion
Relatively few Arabidopsis mutants with reduced or suppressed seed dormancy have been identified so far. In the seed maturation mutants of lec (leafy cotyledon) class lec1 (Meinke 1992) and fus3 (Bäumlein et al. 1994; Keith et al. 1994), dormancy is affected as a pleiotropic effect of mutations in (as yet unidentified) genes controlling embryo development. Nondormant lec1 and fus3 seeds contain morphologically abnormal embryos that develop into seedlings whose anatomical characteristics suggest a homeotic (Meinke 1992) and, respectively, heterochronic (Keith et al. 1994) interpretation of these two mutations. In contrast, embryos of the dag1-1 mutant appear normal and develop into normal seedlings.
Other seed dormancy mutants are affected in either ABA biosynthesis (aba, Koornneef et al. 1982; Léon-Kloosterziel et al. 1996a) or ABA sensitivity (abi, Koornneef et al. 1984)—major evidence supporting the view that ABA is the main plant hormone responsible for the induction of dormancy. Of the abi mutants, only abi3 shows an effect limited to the embryo (Koornneef et al. 1984) and the ABI3 gene encodes a seed-specific transcription factor (Giraudat et al. 1992). ABI1 and ABI2 encode protein phosphatases 2C (Leung et. al. 1994, 1997; Meyer et al. 1994), and their mutation alters several responses of the plant to ABA (Koornneef et al. 1984), as is the case for aba mutants (Koornneef et al. 1982; Léon-Kloosterziel et al. 1996a). In contrast, disruption of DAG1 does not affect sensitivity of the seeds to ABA—germination of dag1-1 seeds is as sensitive to the hormone as that of WS seeds—and does not lead to the pleiotropic effects on water relations in adult plants characterictic of ABA-deficient mutants. In addition, although aba seeds do not require de novo synthesis of GA to germinate, dag1-1 seeds do: Like WS seeds, dag1-1 seeds are incapable of germinating in the presence of a concentration (10 μm) of paclobutrazol that inhibits GA biosynthesis (Léon-Kloosterziel et al. 1996a).
The reduced dormancy mutants rdo1 and rdo2, whose genes have not been isolated yet, are not affected in their response to ABA. In this respect, they are similar to the dag1-1 mutant, but their reduced dormancy trait is embryo-determined (Léon-Kloosterziel et al. 1996b). In contrast, both the segregation pattern of the mutant phenotype in seeds from heterozygous plants and the lack of expression of the gene in the seeds indicate that DAG1 exerts its role from the mother plant.
To our knowledge, DAG1 is the first gene encoding a transcription factor that is specifically involved in the maternal control of seed germination. As yet, the only example of maternally determined loss of seed dormancy is in the mutants of the testa—the seed coat, which is of maternal origin. However, the enhanced germination of most of the testa mutants is due to altered physicochemical and/or mechanical properties of their seed coat—evidence that the seed coat acts in preventing the embryonic radicle from protruding from the seed prematurely (Koornneef 1981, 1990; Léon-Kloosterziel 1997). Although testa mutations result in altered coloration, shape, or composition of the seed coat, dag1-1 seeds appear normal, suggesting that the maternal effect of the disruption of DAG1 does not involve the seed testa.
A most interesting trait of the nondormant dag1-1 mutant seeds is their capability to germinate in total darkness. In contrast, nondormant WS seeds (i.e., after storage-induced release from dormancy) still require (red) light to germinate. Red light is needed to turn the inactive Pr form of the phytochrome into the active Pfr form, which then triggers germination via a pathway, as yet totally unknown. The lack of light requirement of dag1-1 seeds, however, does not imply that they bypass the phytochrome-mediated pathway to initiate germination. When Pfr is inactivated by a pulse of far-red light, dag1-1 seeds fail to germinate in the dark.
On the other hand, DAG1 is expressed in the vascular tissue of the mother plant, suggesting that its mode of action involves transport to the seed of some component necessary for the control of dormancy. This evidence can be rationalized by a model whereupon DAG1 normally would ensure the inactivity (for dormancy initiation) of a (protein) component—transported from the mother plant to the seed—that would be activated (to trigger germination) as a consequence of red light perception. In plants where DAG1 is disrupted, this component would be in the active form independent of red light: dormancy would not be established and seeds would germinate in the absence of light but germination could still be repressed by far-red light.
Further work is needed to verify this model or test alternatives. However, the identification of the first regulatory protein involved in the maternal control of seed germination opens a new avenue of research in this important and still largely obscure aspect of plant adaptation.
Materials and methods
Plant material and seed germination
Seeds from Arabidopsis thaliana Heyhn, ecotype WS, were surface-sterilized as described by Santoni et al. (1994), sown into petri dishes on a solid medium containing Murashige and Skoog salts (Sigma), vitamins (Sigma), 0.8% agar, and 1% sucrose, and kept in the dark at 4°C for 2 days. The dishes were then transferred into the growth chamber (24°C/21°C, 16/8 hr day/night, 300/μEinstein/m2). Seedlings were transferred into individual pots 8 days after germination, and maintained in the growth chamber. Thirty WS and dag1-1 flowering plants were analyzed for morphometric parameters, and 30 siliques from both types of plants were measured and scored for the number of seeds contained.
For germination experiments, surface-sterilized seeds were sown in triplicate in Petri dishes (30–60 per dish) over one layer of Whatmann paper saturated with 2.5 ml of sterile distilled water and kept in the dark at 4°C for 2 days. Germination tests were subsequently carried out under the illumination conditions utilized for plant growth or in total darkness. In the text, stored seeds are seeds stored for 4 weeks after harvest in the dark under dry conditions at room temperature. For ABA and paclobutrazol sensitivity tests, seeds were sown on 0.8% agar in the presence of the ABA and paclobutrazol concentrations indicated in Figure 3, C, and, D, respectively; germination was scored after 7 days of incubation under standard illumination conditions.
For light-pulse experiments, light sources were from Phylips (bulb), and fluence rates were as follows: red light, bulb TL (40 W), fluence, 316 μW/cm2; far-red light, bulb Phylinea (60 W), fluence, 350 μW/cm2. Seeds were exposed to red or far-red light for 5 min and kept in the dark for 7 days before scoring for germination.
Molecular techniques
Molecular techniques were perfomed according to standard protocols. Arabidopsis DNA was extracted according to Bouchez et al. (1993). Total RNAs were extracted from wild-type and mutant plants as described in Vittorioso et al. (1998). The RT–PCR analysis was performed with DAG1-specific primers, according to the manufacturer's instructions (Perkin-Elmer).
Isolation of DAG1
The Dof domain of the previously isolated partial cDNA of DAG1 (De Paolis et al. 1996), was utilized for the screening of an Arabidopsis genomic library (Clontech). Several positive clones were isolated. An 8-kb SalI insert containing the entire DAG1 gene was cloned into pBluescript KS+ vector (Stratagene) for restriction analysis. The 1.5- and 3.0-kb EcoRI fragments containing the promoter (1 kb) and the DAG1 gene, respectively, were subcloned and sequenced. Full-length sequence of the cDNA was obtained by 5′-RACE, according to the manufacturer's instructions (GIBCO-BRL). DNA sequencing was performed with an ABI373A automated DNA sequencer as recommended by the manufacturer (Applied Biosystems). The analysis of DAG1 cDNA and derived protein sequence was performed using GCG and BLAST, or the National Center for Biotechnology Information (NCBI, Bethesda, MD).
Isolation of the dag1-1 knockout plant
A collection of T-DNA insertion mutants (Laboratoire de Génetique et Amélioration des Plantes, INRA, Versailles, France) containing ∼22,000 individual lines, was screened by PCR utilizing pairs of primers corresponding to both the T-DNA right and left borders and the DAG1 3′-specific region. One positive line was identified and isolated. The position of the T-DNA within the DAG1 gene was determined by sequencing the PCR-amplified fragment. The DAG1-tagged line was shown to contain a single T-DNA insert and checked for linkage of the T-DNA insert to the DAG1 gene. More than 100 putative heterozygous plants (kanamycin resistant with normal siliques) segregated 25% mutant progeny (twisted siliques). Plants heterozygous for the mutation were self-fertilized, and the transmission of the phenotype was confirmed in the F3 generation.
Complementation of the dag1-1 mutant
A 2.7-kb genomic fragment corresponding to the DAG1 gene was cloned in the pCAMBIA binary vector (Canberra, Australia). dag1-1 homozygous plants were transformed by in planta infiltration (Bechtold et al. 1993) and transformants selected on 40 μg/ml hygromycin. The F3 progeny from two individual hygromycin-resistant plants was analyzed for complementation of the dag1-1 phenotypes.
In situ mRNA hybridizations
A 400-bp PCR fragment containing the carboxy-terminus of the DAG1-coding sequence up to the 3′ UTR, was cloned in the pCR2.1 vector (Stratagene). For the antisense riboprobe, the construct was linearized and in vitro transcribed with T7 RNA polymerase in the presence of 3H-labeled UTP. The sense probe was derived from a construct containing the same PCR-amplified fragment cloned in the opposite orientation. Conditions for tissue fixation, paraffin embedding, hybridization (on 8-mm sections), and washings were as described by Drews et al. (1991).
Acknowledgments
We thank T. Berleth and C. Hardtke for help with the in situ hybridizations. This work was partially supported by EC BIO4 contracts and by grants from MIPA, MURST, and CNR (to P.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL costantino@axrma.uniroma1.it; FAX 39 06 4440812.
References
- Bäumlein H, Misera S, Luersen H, Kolle K, Horstmann C, Wobus U, Muller AJ. The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryiogenesis. Plant J. 1994;3:379–387. [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G. The DNA binding site of Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell. 1999;11:323–334. doi: 10.1105/tpc.11.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser II. 1993;316:1194–1199. [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. A binary vector based on basta resistance for in planta transformation of Arabidopsis thaliana. CR Acad Sci, Ser II. 1993;316:1188–1193. [Google Scholar]
- Camilleri C, Lafleuriel J, Macadré C, Varoquaux F, Parmentier Y, Picard G, Caboche M, Bouchez D. A YAC contig map of Arabidopsis thaliana chromosome 3. Plant J. 1998;14:633–642. doi: 10.1046/j.1365-313x.1998.00159.x. [DOI] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh K. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- De Paolis A, Sabatini S, De Pascalis L, Costantino P, Capone I. A rolB regulatory factor belongs to a new class of single zinc finger plant proteins. Plant J. 1996;10:215–223. doi: 10.1046/j.1365-313x.1996.10020215.x. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. Fusca3: A heterocronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M. The complex syndrome of ttg mutants. Arabidopsis Inf Serv. 1981;18:45–51. [Google Scholar]
- ————— Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf Serv. 1990;27:1–4. [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin-sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Koorneef M, Karssen CM. Seed dormancy and germination. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Léon-Kloosterziel KM. “Genetic analysis of seed development in Arabidopsis thaliana”. Ph.D. thesis. Wageningen, Germany: Wageningen Agricultural University; 1997. [Google Scholar]
- Léon-Kloosterziel KM, Alvarez GA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwatz SH, Zeevaart JAD, Koornneef M. Isolation and characterization of abscissc acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996a;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, van de Bunt GA, Zeevaart JAD, Koornneef M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996b;110:233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chelfdor F, Giraudat J. Arabidopsis ABA-response gene ABI1: Features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE 2 (ABI2) and ABI1 genes encode redundant protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science. 1992;264:1452–1455. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt R, Carbonero P. An endosperm-specific Dof protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Santoni V, Bellini C, Caboche M. Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta. 1994;192:557–566. [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose S, Parsons RL, Schmidt R. A maize zinc finger protein binds the prolamin box in zein gene promoters and interacts with basic leucine zipper transcriptional activator Opaque2. USA Proc. Natl Acad Sci. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C. Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol Cell Biol. 1998;18:3034–3043. doi: 10.1128/mcb.18.5.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. A novel DNA binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995;23:3403–3410. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10:75–99. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley R, Büttner M, Singh K. Interactions between distinc types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]