Abstract
Self-renewal of rodent embryonic stem (ES) cells is enhanced by partial inhibition of glycogen synthase kinase-3 (Gsk3)1 2. This effect has variously been attributed to stimulation of Wnt signalling via β-catenin1, stabilisation of cMyc3, and global de-inhibition of anabolic processes4. Here we demonstrate that β-catenin is not necessary for ES cell identity or expansion, but its absence eliminates the self-renewal response to Gsk3 inhibition. Responsiveness is fully restored by truncated β-catenin lacking the C-terminal transactivation domain5. However, requirement for Gsk3 inhibition is dictated by expression of Tcf3 and mediated by direct interaction with β-catenin. Tcf3 localises to many pluripotency genes6 in ES cells. Our findings confirm that Tcf3 acts as a transcriptional repressor and reveal that β-catenin directly abrogates Tcf3 function. We conclude that Gsk3 inhibition stabilises the ES cell state primarily by reducing repressive influence on the core pluripotency network.
Canonical Wnt signalling is a key regulator of stem cells in epithelial tissues (reviewed in7). This pathway has also been proposed to play a major role in self-renewal of pluripotent embryonic stem (ES) cells. Wnt ligands promote nuclear accumulation of β-catenin, which associates with DNA-bound Tcf/Lef factors and activates transcription8-10. Glycogen synthase kinase-3 (Gsk3)1, 11 negatively regulates Wnt signalling by phosphorylating β-catenin leading to its ubiquitination and proteolysis12, 13. This is prevented by inhibitors of Gsk3 which thereby act as mimetics of Wnt stimulation. Gsk3 inhibitors such as BIO or CHIRON99021 (CH) support short term expansion of mouse ES cells1, 2 and this has been interpreted as evidence for canonical Wnt function in self-renewal1, 6, 14. Differentiation is only partially suppressed, however, and cultures collapse upon passaging2. Robust and long-term self-renewal additionally requires the cytokine leukaemia inhibitory factor (LIF), which activates the transcription factor Stat315, 16, or inhibition of the mitogen activated protein kinase (Mapk) cascade17.
Deletion of Tcf3 can delay ES cell differentiation18, 19 but, unlike other Tcfs, evidence that β-catenin directly activates Tcf3 target genes is lacking. Significantly, genetic analyses in the embryo20, 21 have not revealed a requirement for Wnt in the naïve epiblast, the counterpart of ES cells. Furthermore, Gsk3 is a negative regulator of proteins involved in metabolism, transcription, translation, cell cycle, anti-apoptosis and signal transduction4. Its inhibition therefore has potentially much broader effects than canonical Wnt signalling. Importantly, Gsk3 inhibition is not necessary for ES cell propagation if LIF and inhibition of the Mapk cascade are combined2 or if LIF is used with serum or Bmp422.
Selectivity is a general concern with the use of kinase inhibitors. CH has limited cross-reactivity with many other kinases23, but similar information is not available for BIO. We tested 7 proprietary Gsk3 inhibitors (Supplementary Information, Fig. S1). These compounds have distinct chemical structures, reducing the likelihood of shared off-target effects. In combination with the Mek inhibitor PD0325901 (PD), all promoted undifferentiated ES cell expansion over several passages in bulk culture and enabled colony formation at clonal density in a dose-dependent manner (Fig. 1a). Some of these compounds are effective at nanomolar concentrations. Interestingly, at slightly higher concentrations colony formation was reduced (see compounds C-G, Fig. 1a). We have also observed this effect for CH (data not shown) and infer that incomplete inhibition of Gsk3 is optimal. To test further whether Gsk3 is the critical target we carried out a genetic perturbation. We previously showed that ES cells lacking both isoforms of Gsk324 can be maintained using a Mek inhibitor alone without CH2. However, adaptation of these cells during repeated gene targeting manipulations cannot be excluded. We therefore transfected ES cells with siRNAs against Gsk3α, Gsk3β or both, and scored formation of undifferentiated ES cell colonies at low density in the presence of PD. Immunoblotting confirmed specific knockdown of Gsk3α and β (Supplementary Information, Fig. S2). In wild-type ES cells double knock down produced a small increase in colony number while single knock downs had no effect (data not shown). We then tested ES cells in which one Gsk3α and both Gsk3β alleles have been inactivated24. Gsk3α siRNAs reproducibly increased undifferentiated colony formation by approximately three-fold in these cells while Gsk3β siRNAs had no effect, testifying to the specificity of the siRNA-response (Fig. 1b). Collectively these results validate the conclusion that reduced activity of Gsk3 enhances ES cell self-renewal.
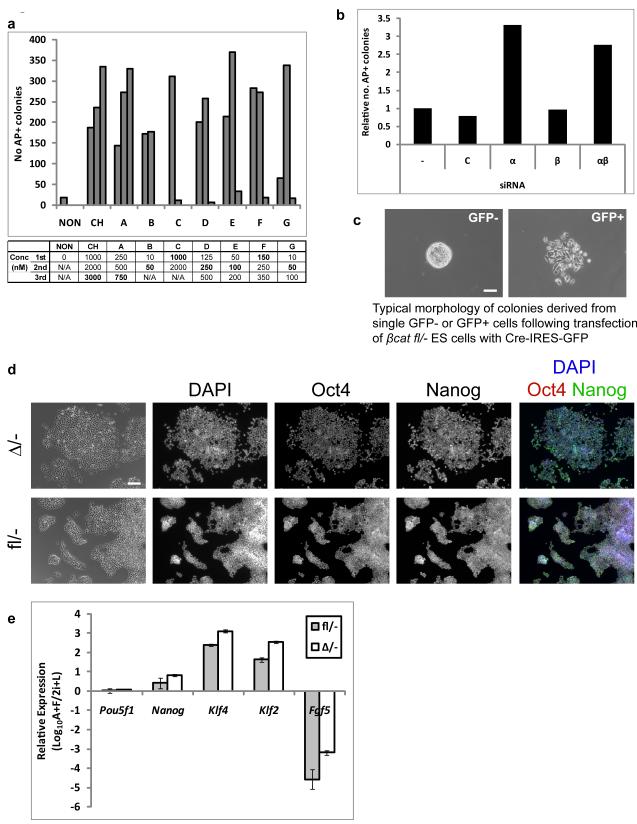
Figure 1. Suppression of Gsk3 mediates enhanced ES cell self-renewal but β-catenin is dispensable for ES cell maintenance.
(a). Histogram showing number of undifferentiated (alkaline phosphatase positive, AP+), colonies formed from 600 E141VC ES cells plated in N2B27 with Mek inhibitor PD0325901 (PD, 1 μM) plus CHIRON99021 (CH); or alternative Gsk3 inhibitors A, B, C, D, E, F, and G (see methods for details). Concentrations (Conc) of the inhibitors (nM) are indicated in the table beneath the graph. 1st, 2nd and 3rd correspond to the bars from left to right. Optimum concentrations are shown in bold print.
(b). Histogram showing relative number of undifferentiated (alkaline phosphatase positive, AP+), colonies formed from 600 3/4KO ES cells plated in N2B27 plus PD. Cells were untreated (−) or transfected with control siRNA (C) or siRNAs against Gsk3α (α), Gsk3β (β), or both (αβ). Mean of two biological replicates.
(c). Phase contrast images showing typical morphology of primary colonies isolated from GFP-negative (left) and –positive (right) fractions of βcatfl/− ES cells transiently transfected with Cre-IRES-GFP. Note the lack of cell-cell contacts in colonies from the GFP-positive fraction. Scale bar, 200μm
(d). Phase contrast and fluorescent images showing immunostaining of βcatfl/− and βcatΔ/− ES cells for Oct4 and Nanog. Scale bar, 100μm.
(e). Histogram showing gene expression in βcatfl/− and βcatΔ/− ES cells cultured in 2i+LIF relative to EpiSC-like cells derived by culture in activin+FGF2 (A+F) for 7 passages. Klf4 and Klf2 are specific for naive ES cells while Fgf5 is up-regulated and Nanog down-regulated in EpiSCs27. Mean ± s.d. of three biological replicates.
To investigate the involvement of β-catenin we used ES cells carrying null and floxed alleles (βcatfl/−). Following transient transfection with CreIresGFP, cells were sorted by flow cytometry and plated in N2B27 with PD and CH (2i) plus LIF (2i+LIF, Supplementary Information, Fig. S3a). The GFP negative (non-transfected) population produced compact colonies typical of ES cells in 2i, while the GFP fraction yielded colonies of dispersed cells (Fig. 1c). Both expanded after picking and clonal lines were maintained thereafter in 2i+LIF. Immunoblotting confirmed that the dispersed cells had undergone deletion (Supplementary Information, Fig. S3b). Interestingly, these β-catenin-null (βcatΔ/−) ES cells re-established cell-cell contacts and normal ES cell colony morphology within 3 passages, even though β-catenin protein remained undetectable (Supplementary Information, Fig. S3b and Fig. 1d). As reported in the accompanying manuscript by Lyashenko et al., this adaptation is most likely due to compensatory up-regulation of plakoglobin21 which can replace β-catenin in adherens junctions.
βcatΔ/− cells expressed Nanog and Oct4 (Fig. 1d,e; Supplementary Information Fig. S3b) and readily formed alkaline phosphatase-positive colonies at clonal density (Fig. 2a). By immunostaining, Nanog appears at a similar level in all cells, suggesting that βcatΔ/− ES cells in 2i+LIF are uniformly undifferentiated (Fig. 1d). We compared marker gene expression in 2i+LIF to post-implantation epiblast stem cell (EpiSC)s25-27 (Supplementary Information, Fig. S3c) and to cultures in activin and FGF2 which induces ES cell differentiation into EpiSCs (Fig. 1e). Null cells in 2i + LIF expressed ground state ES cell markers Klf4, Klf2 and Nr0b1 and lacked the early differentiation marker Fgf5, whereas after culture in activin and FGF2 they showed the reciprocal pattern characteristic of EpiSCs. Previous reports of a partly differentiated phenotype of β-catenin-null cells28, 29 may therefore reflect characterisation of EpiSCs rather than ES cells. Interestingly the null cells failed to up-regulate brachyury in EpiSC culture, consistent with this being a canonical Wnt target30.
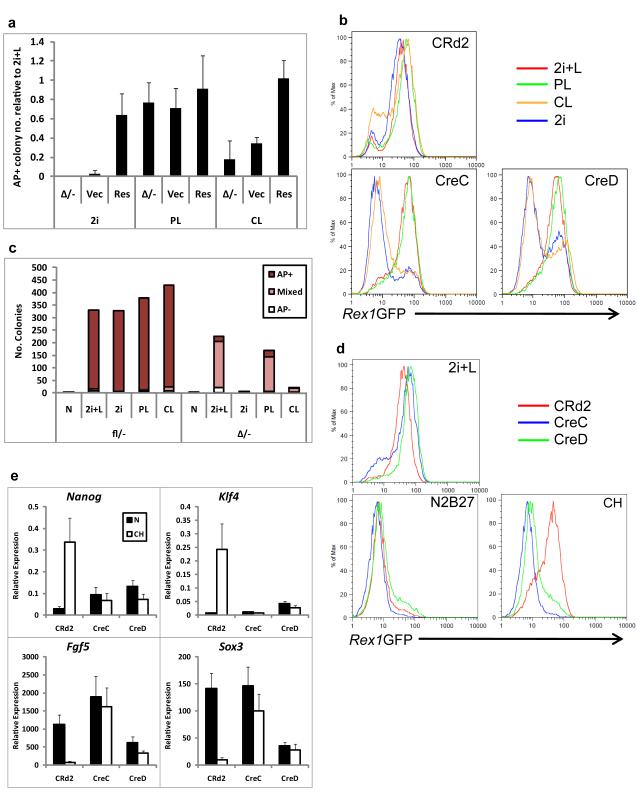
Figure 2. βcatΔ/− ES cells do not resist differentiation upon Gsk3 inhibition.
(a). Histogram showing number of undifferentiated (alkaline phosphatase positive, AP+), colonies formed by parental βcatΔ/− or βcatΔ/− ES cells expressing a β-catenin transgene (Res) or the corresponding vector control (Vec) plated in N2B27 plus 2i, PD+LIF (PL) or CH+LIF (CL). Data are expressed relative to the number of colonies in 2i+LIF. Mean ± s.d. of three biological replicates.
(b). Flow cytometry analysis of Rex1GFP expression in βcatfl/− (CRd2) or βcatΔ/− (CreC and CreD) Rex1GFP reporter ES cells cultured in 2i+LIF, PD+LIF (PL), CH+LIF (CL) or 2i for 96 hours.
(c). Histogram showing number of undifferentiated (alkaline phosphatase positive, AP+), mixed and differentiated (AP−) colonies formed from 600 βcatfl/− or βcatΔ/− ES cells plated in 2i+LIF following 48 hours culture in N2B27 alone or plus 2i+LIF (2i+L), 2i, PD+LIF (PL) or CH+LIF (CL).(d). Flow cytometry analysis of Rex1GFP expression in βcatfl/− (CRd2) or βcatΔ/− (CreC and CreD) Rex1GFP reporter ES cells cultured in 2i+LIF, N2B27 alone or CH for 48 hours.
(e). Histogram showing relative expression of pluripotency markers Nanog and Klf4 and early differentiation markers Fgf5 and Sox3 in βcatfl/− (CRd2) or βcatΔ/− (CreC and CreD) Rex1GFP reporter ES cells cultured in 2i+LIF, or N2B27 alone or plus CH for 48 hours. Mean ± s.d. of three biological replicates.
Wild type ES cells can be cultured from single cells in 2i + LIF or with slightly lower efficiency using any two of these components31. βcatΔ/− cells in contrast formed no colonies in 2i and very few in CH+LIF. Only when both PD and LIF were present was colony formation robust (Supplementary Information, Fig. S3d and Fig. 2a). Stable transfection with a β-catenin transgene (Supplementary Information, Fig. S3e) restored clonal expansion in 2i or CH+LIF (Fig. 2a). These results establish that the effect of Gsk3 inhibition is in large part mediated via β-catenin. Nonetheless, colony yield from βcatΔ/− cells was rather higher in 2i+LIF than in PD+LIF. This result was consistent with three alternative Gsk3 inhibitors (Supplementary Information, Fig. S4). Therefore β-catenin-independent effects of Gsk3-inhibition contribute to maximise ES cell clonogenicity.
We inserted destabilised GFP (GFPd232) into the Rex1 locus of βcatfl/− ES cells and then generated βcatΔ/− derivative clones, CreC and CreD (Supplementary Information, Fig. S5a,b). Rex1 is down-regulated at the onset of ES cell differentiation33. In 2i+LIF or PD+LIF, both the βcatfl/− and βcatΔ/− ES cells were almost uniformly GFP-positive (Fig. 2b), as reported for other markers in these defined conditions31. In 2i or CH+LIF, however, while βcatfl/− cells retained GFP, CreC and CreD cells lost expression. The pluripotency-associated cell surface marker, PECAM34, behaved in the same manner (Supplementary Information, Fig. S6a). We then assayed colony formation after a 48h period in the different conditions. βcatfl/− cells produced undifferentiated colonies in all cases except after culture in N2B27 alone (Fig. 2c). βcatΔ/− cells produced colonies after 48h in 2i+LIF or PD+LIF but not following culture in 2i or CH+LIF.
We cultured Rex1GFPd2 cells for 48 hours in CH, PD or LIF alone. PD or LIF delayed down-regulation of Rex1GFP in βcatfl/− and βcatΔ/− cells (Supplementary Information, Fig. S5c) whereas CH maintained GFP in floxed but not in null cells (Fig. 2d). Similar results were obtained in the presence of serum (Supplementary Information, Fig. S5d). In line with Rex1GFP, down-regulation of Nanog was delayed by PD or LIF but not by CH in βcatΔ/− cells (Supplementary Information, Fig. S6b). qRT-PCR analysis confirmed that CH maintained expression of naïve pluripotency markers and suppressed up-regulation of early differentiation markers in βcatfl/− ES cells (Fig. 2e). In contrast, βcatΔ/− ES cells, lost naive markers and up-regulated ectodermal markers Fgf5 and Sox3. Activated caspase-3 was not increased (Supplementary Information, Fig. S6c) indicating that apoptosis is not induced. These results indicate that Gsk3 inhibition resists exit from naïve pluripotency and that this response is dependent on β-catenin.
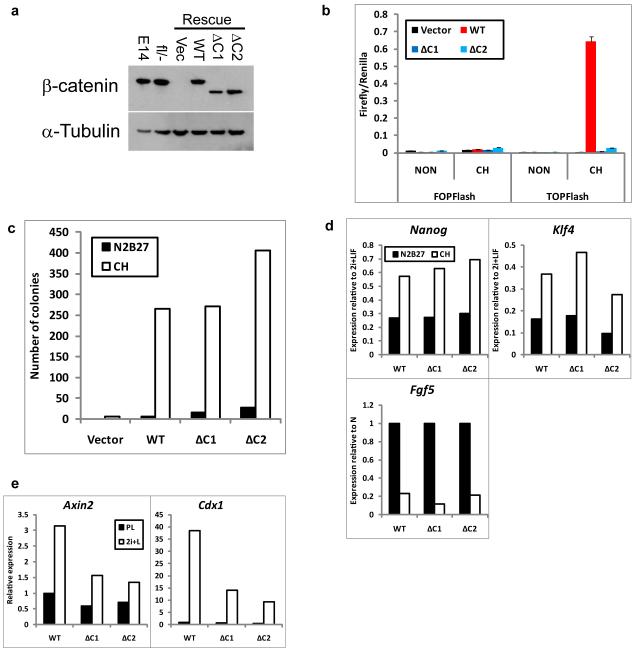
β-catenin interacts with Tcf/Lef transcription factors and can activate their targets via its C-terminal transcriptional activation domain10. To test the requirement for β-catenin-mediated transcriptional activation we established clonal βcatΔ/− “rescue” lines stably expressing full-length β-catenin (RescueWT) or truncated β-cateninΔC (RescueΔC), which lacks the C-terminal transactivation domain5 (Fig. 3a). Both full-length and β-cateninΔC localised primarily to the cell membrane (Supplementary Information, Fig. S7). RescueWT but not RescueΔC transfectants activated TOPFlash in response to CH (Fig. 3b). We tested differentiation sensitivity by culture for 48h with CH only followed by replating. Both RescueWT and RescueΔC cells produced undifferentiated colonies indicating restored response to CH (Fig. 3c). Down-regulation of the pluripotency markers Nanog and Klf4 and up-regulation of the early differentiation marker Fgf5 were similarly suppressed by CH in RescueWT and RescueΔC cells (Fig. 3d). These data demonstrate that the canonical transcriptional activation mechanism is not required for β-catenin to increase ES cell resistance to differentiation.
Figure 3. β-catenin inhibits differentiation independent of its transcriptional activation domain.
(a). Western blot showing β-catenin and α-Tubulin (loading control) expression in wild-type (E14), βcatfl/− or “Rescue” βcatΔ/− ES cells expressing randomly integrated wild-type (WT) or C-terminal-deleted β-catenin (ΔC) transgenes or the corresponding vector control (Vec). ΔC1 and ΔC2 are independent clones. Uncropped images of blots are shown in Supplementary Fig. S9.
(b). Histogram showing TOPFlash and FOPFlash reporter activity in βcatΔ/− cells expressing randomly integrated wild-type (WT) or C-terminal-deleted (ΔC) β-catenin transgenes in PD+LIF with or without CH. ΔC1 and ΔC2 are independent clones. Mean ± s.d. of three biological replicates.
(c). Histogram showing number of undifferentiated colonies formed from 600 βcatΔ/− cells expressing randomly integrated wild-type (WT) or C-terminal-deleted (ΔC) β-catenin transgenes plated in 2i+LIF after 48 hours in N2B27 alone or plus CH.
(d) Histograms showing relative expression of Nanog, Klf4, and Fgf5 in βcatΔ/− cells expressing randomly integrated wild-type (WT) or C-terminal-deleted β-catenin (ΔC) transgenes cultured in 2i+LIF or in N2B27 alone or plus CH for 24 hours. Expression is shown relative to levels in 2i+LIF. ΔC1 and ΔC2 are independent clones.
(e). Histogram showing relative expression of Axin2 and Cdx1 in βcatΔ/− cells expressing randomly integrated wild-type (WT) or C-terminal-deleted (ΔC) β-catenin transgenes cultured in PD+LIF or 2i+LIF for 48 hours.
Data in (c)-(e) are mean of two biological replicates.
β-cateninΔC cannot function as a transcriptional activator, but it retains domains for interaction with Tcf/Lef factors. Tcf3 is the predominant Tcf/Lef in ES cells and has been reported to behave primarily as a repressor18. β-catenin might relieve Tcf3-mediated repression18. This can explain why Axin2 and Cdx1, which are induced by CH in RescueWT cells are also more modestly induced in RescueΔC cells (Fig. 3e). Intriguingly Tcf3 deletion results in delayed ES cell differentiation18, 19. We deployed siRNA in Rex1GFPd2 reporter cells to investigate whether the Tcf3 loss-of-function phenotype18 is dependent on β-catenin. Rex1GFP down-regulation following withdrawal from 2i+LIF was reduced by Tcf3 siRNA in both βcatfl/− and βcatΔ/− ES cells (Fig. 4a). Tcf3 knock down (Supplementary Information, Fig. S8a) also maintained Nanog and Klf4 expression in both βcatfl/− and βcatΔ/− ES cells (Fig. 4b). Expression of canonical Wnt pathway targets Axin2 and Cdx1 was increased by Tcf3 siRNA, independent of β-catenin (Fig. 4b), confirming that increased transcription of some Tcf targets does not require direct activation by β-catenin.
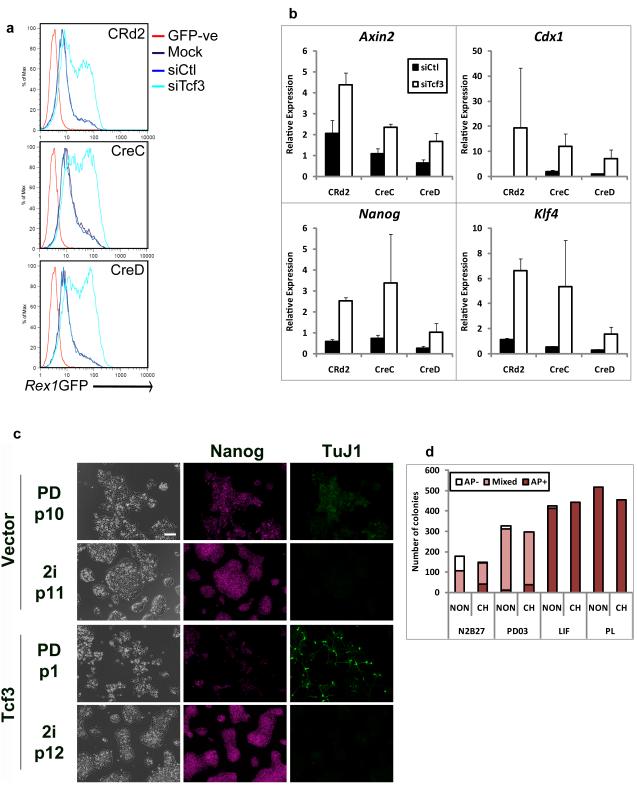
Figure 4. β-catenin functions by abrogating Tcf3 repression.
(a). Flow cytometry analysis showing the profile of Rex1GFP expression in βcatfl/− (CRd2) or βcatΔ/− (CreC and CreD) Rex1GFP reporter ES cells mock transfected (Mock) or transfected with control siRNAs (siCtl) or siRNA against Tcf3 (siTcf3) and cultured for 48 hours in N2B27 alone. GFP-ve: wild-type ES cells that do not express GFP.
(b). Histograms showing relative expression of Axin2, Cdx1, Nanog and Klf4 in βcatfl/− (CRd2) or βcatΔ/− (CreC and CreD) Rex1GFP reporter ES cells. Cells were cultured for 24 hours in N2B27 alone and were transfected with control siRNAs (siCtl) or siRNAs against Tcf3 (siTcf3). Expression is shown relative to untreated. Mean ± s.d. of three biological replicates.
(c). Phase contrast and fluorescent images showing Nanog and βIII-tubulin (TuJ1) expression in Tcf3-null ES cells stably expressing a randomly integrated Tcf3 transgene or the corresponding vector control cultured in N2B27 plus PD or plus 2i for the indicated number of passages (p). Scale bar, 100μm.
(d). Histogram showing number of undifferentiated (AP+), mixed and differentiated (AP-) colonies formed from 600 Tcf3-null ES cells in N2B27 alone or plus PD, LIF or PD+LIF in the presence or absence of CH
Tcf3-null ES cells18 self-renew robustly in LIF or PD alone and are insensitive to CH (Fig. 4c,d), suggesting that a requirement for Gsk3 inhibition is dictated by Tcf3. To test whether the effect of CH involves direct interaction of β-catenin with Tcf3 we established Tcf3-null ES cell lines expressing wild-type Tcf3 (Tcf3WT) or Tcf3 lacking the β-catenin binding domain (Tcf3ΔN) (Supplementary Information, Fig. S8b). Both Tcf3WT and Tcf3ΔN suppress activation of the TOPFlash reporter by CH (Fig. 5a). Similarly, both reduced expression of Cdx1 (Fig. 5b). Consistent with insensitivity to β-catenin, Tcf3ΔN was the more effective repressor of this endogenous Wnt target. In PD alone Tcf3-null ES cells efficiently formed undifferentiated colonies whereas both Tcf3WT and Tcf3ΔN transfectants behaved like wild type and failed to clone (Fig. 5c). Addition of CH restored colony forming efficiency by Tcf3WT cells. In contrast cells expressing Tcf3ΔN formed very few colonies. Importantly, however, they produced undifferentiated colonies at similar frequency to other cells in PD+LIF. Interestingly, the addition of CH in this condition increased colony numbers for both Tcf3WT and Tcf3ΔN cells. These data demonstrate that the response to CH is largely mediated by a direct interaction between Tcf3 and β-catenin but also confirm that additional effects downstream of Gsk3 inhibition promote maximal ES cell self-renewal.
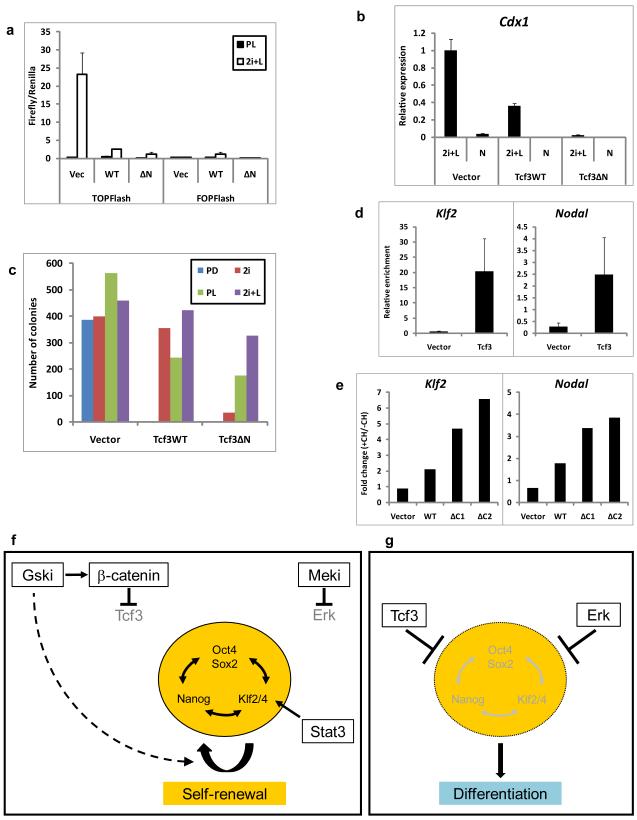
Figure 5. Gsk3 Inhibition Relieves the Core Pluripotency Network from Repression by Tcf3 and Complements Mek Inhibition and/or Stat3 Activation to Stabilise ES Cell Self-Renewal.
a) Histogram showing TOPFlash and FOPFlash activation in Tcf3-null ES cells stably expressing randomly integrated wild-type (WT) or N-terminal-deleted (ΔN) Tcf3 transgenes or the corresponding vector control (Vec) in PD+LIF(PL) or 2i+LIF (2i+L). Mean ± s.d. of three biological replicates is shown
b) Histogram showing relative expression of Cdx1 in Tcf3-null ES cells stably expressing randomly integrated wild-type (Tcf3WT) or N-terminal-deleted (Tcf3ΔN) Tcf3 transgenes or the corresponding vector control in 2i+LIF (2i+L) or N2B27 alone. Mean ± s.d. of three biological replicates is shown
c) Histogram showing number of undifferentiated colonies formed from 600 Tcf3-null ES cells stably expressing randomly integrated wild-type (Tcf3WT) or N-terminal-deleted (Tcf3ΔN) Tcf3 transgenes or the corresponding vector control in N2B27 plus PD, 2i, PD+LIF (PL) or 2i+LIF (2i+L). Mean of two biological replicates.
d) Histogram showing relative enrichment of Klf2 and Nodal promoter regions following chromatin immunoprecipitation for Tcf3 in Tcf3-null ES cells stably expressing a randomly integrated Tcf3 transgene or the corresponding vector control. Mean ± sd of three replicates is shown
e) Histogram showing response to CH of Klf2 and Nodal in βcatΔ/− cells expressing randomly integrated wild-type (WT) or C-terminal-deleted (ΔC) β-catenin transgenes cultured in N2B27 alone for 24 hours followed by 8 hours in N2B27 plus PD in the presence or absence of CH. Mean ratio (+CH/−CH) of two biological replicates.
f) In the presence of Gsk-3 inhibitor (Gski) and a mitogen activated protein kinase (Erk) kinase inhibitor (Meki), repressive effects on the pluripotent gene regulatory network are abolished. The pluripotent circuitry is also positively regulated by Stat3 acting primarily through Klf4. Any two of these three effects are sufficient to stabilise the network and sustain ES cell self-renewal. Gski generates intracellular β-catenin which interacts with Tcf3 and abolishes its repressor effect on multiple genes in the pluripotent network. Gski additionally supports ES cell propagation through stimulatory effects on metabolic and biosynthetic processes (dashed arrow).
g) In the absence of inhibitors, Tcf3 repression and activated Erk drive ES cells into differentiation.
When ES cells are maintained in serum using LIF without inhibitors, cultures are heterogeneous and metastable due to co-existence of states (f) and (g).
Tcf3 binds in proximity to many gene promoters that are also bound by Oct4 and has been proposed as a component of a recursive circuit at the core of the pluripotent transcriptome6, 35, 36. By chromatin immunoprecipitation (ChIP) we confirmed detection of Tcf3 at promoters of Klf2 and Nodal (Fig. 5d)36. In the presence of PD and LIF we found that these and other candidate Tcf3 targets showed no consistent transcriptional up-regulation on addition of CH (data not shown). However, CH has minimal functional impact in this context (Fig. 2a,b,c). Cells cultured for 24 hours without 2i+LIF alone down-regulate naive genes preparatory to commitment. In these conditions addition of CH reproducibly induced up-regulation of Klf2 and Nodal (Fig. 5e). This effect was β-catenin-dependent but was observed in RescueΔC cells indicating that it is not mediated by canonical activation. We also found that Tcf3 target genes6, 36 were up-regulated when CH was added to ES cells in LIF and serum (Supplementary Information, Fig. S8c).
Canonical Wnt signalling has been proposed to support pluripotency by converting Tcf3 complexes from repressors to activators or by displacing Tcf3 with other Tcfs through which β-catenin activates transcription6. However, a simpler model is indicated by the findings that: (i) loss of Tcf3 phenocopies inhibition of Gsk3 even in the absence of β-catenin; (ii) transcriptionally inactive β-catenin fully restores responsiveness to Gsk3 inhibition; and, (iii) full responsiveness to CH requires interaction between β-catenin and Tcf3. We propose that β-catenin abrogates Tcf3 repression and that this is sufficient to permit transcription at Tcf3 target genes that are also bound by pluripotency factors (Fig. 5f,g).
Recently Kelly et al37 proposed a model in which β-catenin acts to inhibit differentiation of ES cells independent of activation of Tcf/Lef targets. Interaction with Oct4 and modulation of Oct4 target genes is proposed to account for the effects of β-catenin stabilisation downstream of Gsk 3 inhibition. However, these authors did not consider Tcf3. The data they present are consistent with our findings and can be explained by β-catenin-mediated derepression of Tcf3 targets, including Oct436. Furthermore, the reported interaction between β-catenin and Oct437 could reflect recruitment of β-catenin by Tcf3 to promoter sites co-occupied by Oct4.
In the mouse embryo, genetic analyses have not revealed a function for canonical Wnt signalling in early epiblast cells21, although a later role in axis formation in vertebrate embryos is well-defined20, 21, 38. Wnt signalling has been shown to promote differentiation in ES cell-derived embryoid bodies39 and characterised targets induced by the canonical pathway in differentiating ES cells include mesodermal lineage specification genes such as brachyury30. Exit from naïve pluripotency and entry into differentiation may be associated with up-regulation of other Tcfs and a switch in the mode of action of β-catenin from derepression of Tcf3 to direct transcriptional activation. It should also be noted that the cell adhesion role of β-catenin is crucial for differentiation, as documented in the accompanying paper from Lyashenko et al.
Characterisation of β-catenin-null ES cells establishes that β-catenin is not essential for maintenance of the pluripotent ground state. However, β-catenin does mediate the additional resistance to differentiation conferred by inhibition of Gsk3. This facultative recruitment of β-catenin is only required if ES cells are cultured without LIF. Consistent with this, Stat3-null ES cells are dependent on inhibition of Gsk32. It is important to note, however, that although the two pathways can substitute for one another they also have independent effects because ES cells cannot be stably maintained by LIF or CH alone but thrive in the presence of both31. Furthermore, addition of LIF to 2i or of CH to PD+LIF increases clonogenicity and for some mouse strains and the rat the combination of all three is important for robust ES cell propagation31.
In summary, selective inhibition of Gsk3 consolidates the ES cell ground state primarily, although not exclusively, via stabilisation of intracellular β-catenin, which eliminates the repressive influence of Tcf3 on the pluripotency network (Fig. 5 f,g). This mechanism is not essential when the core pluripotency factors Oct4, Sox2, Nanog, Klf2 and Klf4 are robustly expressed, but can complement activation of Stat3 and/or inhibition of Mapk to stabilise transcription of these core factors and their targets.
Methods
ES cell culture
Cells were cultured without feeders or serum, unless specifically noted, in N2B27 medium prepared as described2 or preformulated (NDiff™ N2B27 base medium, Stem Cell Sciences Ltd, Cat. No. SCS-SF-NB-02) supplemented with Small molecule inhibitors PD0325901 (PD, 1 μM) and CHIRON99021 (CH, 3 μM) and LIF prepared in house (2i+LIF). Cells were routinely propagated on 0.1% gelatine-coated plastic and replated every 3 days at a split ratio of 1 in 10 following dissociation with Accutase (Gibco). Alternative Gsk3 inhibitors were provided by Pfizer: Compounds A (750 nM), B (50 nM), C (1 μM), D (250 nM), E (100 nM), F (150 nM) and G (50 nM) are described, see Supplementary Information, Figure 1. Colony forming assays were performed by plating 600 ES cells per well on laminin (Sigma) coated plates. Plates were fixed and stained for alkaline phosphatase (Sigma) according to the manufacturer’s protocol. Plates were scanned using a CellCelector (Aviso) and scored manually.
βcatfl/−, βcatΔ/− and Rex1GFPd2 ES cells
βcatfl/− (CBC32) contain one null allele and one allele with exons 2-6 flanked by loxP sites40. βcatΔ/− lines were established by transiently transfection using Lipofectamine2000 (Invitrogen) of CAG-IRES-GFP or CAG-IRES-Cherry, sorting GFP- or Cherry-positive cells by flow cytometry (see below) and seeding at low density to permit expansion of clonal lines. To generate Rex1GFPd2 reporter lines the entire Rex1 coding sequence was replaced with destabilised GFP (GFPd2) linked to a blasticidin resistance cassette by homologous recombination. Correct targeting was confirmed by genomic PCR.
Generation of β-catenin, β-cateninΔC and Tcf3-expressing ES cell lines
Restriction fragments containing wild-type β-catenin and C-terminal-deleted β-catenin (β-cateninΔC)5 were cloned into pCAG-IRES-Puro. The Tcf3 (Tcf7l1) open reading frame was amplified from ES cell cDNA by PCR (Primers: AttB1-kozak-Tcf3 5′-ggggacaagtttgtacaaaaaagcaggcttcaccatgccccagctcggtggtgg-3′; Tcf3-AttB2 5′-ggggaccactttgtacaagaaagctgggtcttagtgggcagacttggtga-3′). Tcf3ΔN was in turn amplified from this by PCR (Primers: AttB1-kozak-Tcf3ΔN 5′-ggggacaagtttgtacaaaaaagcaggcttcaccatggatgaggtcaagtcgtccct-3′, Tcf3-AttB2 (see above)). Amplified sequences were subsequently cloned into pCAG-IRES-Puro using the Gateway system (Invitrogen). βcatΔ/− cells were stably transfected with full-length β-catenin or β-cateninΔC expression vectors. Tcf3-null (HRG418) ES cells were stably transfected with Tcf3 or Tcf3ΔN vectors. Stable transfectants were isolated by electroporation with linearised plasmid DNA and selection in 0.5-0.75μg/ml puromycin.
siRNA knockdown
siRNAs were transfected at a final concentration of 20nM using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions. ES cells were transfected and maintained in 2i+LIF overnight. Cells were then replated at low density to assess colony forming efficiency (see above) or cultured for a further 48 hours under different culture conditions before assessing Rex1GFP by flow cytometry (see below). Gsk3α, Gsk3β and control siRNAs were obtained from Qiagen (GS606496, GS56637 and SI03650325 respectively). Tcf3 and control siRNAs were obtained from Dharmacon (TCF3 ON-TARGETplus SMARTpool, L-048614-01-0005; ON-TARGETplus Non-Targeting Pool, D-001810-10).
Flow cytometry
For Pecam1 analysis cells were dissociated with cell dissociation buffer (Gibco), resuspended in PBS/1% FCS and incubated with FITC-conjugated Pecam1 antibody (1:200). Pecam1-FITC and GFP were analysed on a CyAn (Beckman Coulter). Cell sorting experiments were performed on a MoFlo (DAKO). For both analysis and sorting TO-PRO-3 staining was used to exclude dead cells.
Luciferase Assays
105 cells per well were transfected with 0.8μg TOPFlash or FOPFlash (Upstate) and 0.04μg Renilla luciferase plasmids in 24-well plates. 24hrs later cells were lysed and analysed using the dual luciferase kit (Promega) according to the manufacturer’s protocol. For cotransfection experiments 1.2μg plasmid DNA was transfected together with TOPFlash.
Gene expression analysis by RT–qPCR
RNA was isolated using the RNeasy Kit (Qiagen), reverse transcribed using SuperScriptIII (Invitrogen) according to the manufacturer’s instructions and analysed by real-time PCR using TaqMan Fast Universal Master Mix and TaqMan probes (Applied Biosystems) or the Universal Probe Library (UPL, Roche) system. Primers and UPL probe numbers are detailed in Supplementary Information, Table 1. Technical replicates were carried out for all qPCR reactions. An endogenous control (β-actin or Gapdh) was used to normalise expression.
Table 1.
qRT-PCR primers and corresponding UPL probe numbers
| Forward | Reverse | UPLprobe | ||
|---|---|---|---|---|
| qPCR | Actb | ctaaggccaaccgtgaaaag | accagaggcatacagggaca | 64 |
| Axin2 | gcaggagcctcacccttc | tgccagtttctttggctctt | 50 | |
| Cdx1 | acgccctacgaatggatg | cttggttcgggtcttaccg | 70 | |
| Fgf5 | aaaacctggtgcaccctaga | catcacattcccgaattaagc | 29 | |
| Klf2 | ctaaaggcgcatctgcgta | tagtggcgggtaagctcgt | 48 | |
| Klf4 | cgggaagggagaagacact | gagttcctcacgccaacg | 62 | |
| Nanog | ttcttgcttacaagggtctgc | agaggaagggcgaggaga | 110 | |
| Nodal | ccaaccatgcctacatcca | cacagcacgtggaaggaac | 40 | |
| Nr0b1 | cgtgctctttaacccagacc | ccggatgtgctcagtaagg | 3 | |
| Pou5f1 | gttggagaaggtggaaccaa | ctccttctgcagggctttc | 95 | |
| Rex1 | tcttctctcaatagagtgagtgtgc | gctttcttctgtgtgcagga | 71 | |
| Sox3 | cgctggcttctgaccact | gcaaacaccacagcgattc | 106 | |
| Tcf3 (Tcf7l1) | ctgagcagcccgtacctct | aggggccatttcatctgtag | 22 | |
| ChIP | Klf2 | aggtgtggtgcaaatgagggg | ggctgcagccagacgatcaa | |
| Nodal | gtccggagggaattttgggg | ttgaggcttgaaccagcggtc | ||
| Control Promoter | ggtatttggaaacgtcccacactcactcg | gatggaagatgaaaaagaaattgcaaggatccc |
Western blot analysis and immunofluorescence
Western blots and immunofluorescence were carried out using the following antibodies at the indicated dilutions: Gsk3 (Biosource, 44-610, WB 1:1000); β-catenin (BD Biosciences, 610154, WB 1:2000, IF 1:400; Cell Signalling, 9581, IF 1:400); α-Tubulin (Abcam, ab7291, WB 1:5000); Oct3/4 (Santa Cruz, sc5279, WB 1:1000, IF 1:200); Nanog (eBiocsiences, 14-5761, WB 1:1000, IF 1:200); βIII-tubulin (Covance, MMS-435P, IF 1:500); Pecam1 (BD Biosciences, 553372, flow cytometry 1:200); active caspase 3 (R and D systems, AF835, IF 1:200)
Chromatin immunoprecipitation (ChIP)
ES cells were fixed for 10mins in 1% formaldehyde, harvested in cold PBS and incubated for 20mins on ice in swelling buffer (5 mM HEPES [pH 8.25], 85 mM KCl, and 0.5% NP-40). Nuclei were released with 60 strokes of a dounce homogenizer, pelleted in a microfuge and resuspended in lysis buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, and 1% SDS). Lysates were sonicated to obtain average DNA fragment size of ~500bp. Lysates were diluted 1:10 in ChIP dilution buffer (50 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.1% Triton X-100, and 0.11% Na Deoxycholate), pre-cleared for 2h at 4C with protein-G sepharose beads (Amersham), and incubated overnight at 4C with 4μg Tcf3 antibody (Santa Cruz, sc8635). Lysates were then incubated for 30 mins at 4C with blocked protein-G sepharose beads, beads were washed twice each in RIPA (Tris-HCl pH 8.0, 100mM, NaCl, 300mM, EDTA pH 8.0, 2mM, Triton X-100, 2%, SDS, 0.2%, Na Deoxycholate, 0.2%), RIPA/150mM NaCl and TE. Chromatin was eluted 30mins at room temperature in elution buffer (Tris-HCl pH 8.0, 100mM, NaCl, 300mM, EDTA pH 8.0, 5mM, SDS, 0.5%). Chromatin was analysed by SYBR green real-time PCR (see primers in Supplementary Information, Table 1). Enrichment was calculated relative to control region.
Supplementary Material
Acknowledgements
We thank Natalia Lyashenko and Christine Hartmann for discussion and exchange of unpublished data. We also thank Alfonso Martinez-Arias for comments on the manuscript. We are grateful to Brad Merrill for generously providing Tcf3 mutant ES cells and to Brad Doble and Jim Woodgett for Gsk3 mutant ES cells. We thank Rudolf Grosschedl for the β-cateninΔC construct. Gsk3 inhibitors, Compounds A-G, were provided by Pfizer. Rachael Walker and Peter Humphreys supported flow cytometry and imaging respectively. The study was funded by the Biotechnology and Biological Sciences Research Council and the Medical Research Council of the United Kingdom, the Wellcome Trust, and the European Commission FP7 project EuroSyStem. SGL was supported by a CONACYT Studentship. AS is a Medical Research Council Professor.
References
- 1.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 2.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartwright P, et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 4.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–86. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–18. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–55. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Behrens J, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 9.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–9. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 10.van de Wetering M, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–99. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun. 2006;343:159–66. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 12.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinfeld B, et al. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–6. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 14.Hanna J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–24. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 18.Pereira L, Yi F, Merrill BJ. Repression of nanog gene transcription by tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479–91. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo G, Huang Y, Humphreys P, Wang X, Smith A. A PiggyBac-based recessive screening method to identify pluripotency regulators. PLoS One. 2011 Apr 18;6(4) doi: 10.1371/journal.pone.0018189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haegel H, et al. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–37. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 21.Huelsken J, et al. Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol. 2000;148:567–78. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 23.Bain J, et al. The selectivity of protein kinase inhibitors; a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev Cell. 2007;12:957–71. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 26.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 27.Guo G, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–9. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soncin F, et al. Abrogation of E-cadherin Mediated Cell-cell Contact in Mouse Embryonic Stem Cells Results in Reversible LIF-independent Self-renewal. Stem Cells. 2009 doi: 10.1002/stem.134. [DOI] [PubMed] [Google Scholar]
- 29.Anton R, Kestler HA, Kuhl M. beta-Catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–54. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Arnold SJ, et al. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91:249–58. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- 31.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–32. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 32.Li X, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–5. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 33.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–18. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 34.Robson P, Stein P, Zhou B, Schultz RM, Baldwin HS. Inner cell mass-specific expression of a cell adhesion molecule (PECAM-1/CD31) in the mouse blastocyst. Dev Biol. 2001;234:317–29. doi: 10.1006/dbio.2001.0274. [DOI] [PubMed] [Google Scholar]
- 35.Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–60. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam WL, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–31. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly KF, et al. beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 8:214–27. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon RT, Kimelman D. From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus. Bioessays. 1998;20:536–45. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.ten Berge D, et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–18. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brault V, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–64. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.