Abstract
Vascular endothelial dysfunction underlies the genesis and progression of numerous diseases. Although the GATA transcription factor GATA-2 is expressed in endothelial cells and is implicated in coronary heart disease, it has been studied predominantly as a master regulator of hematopoiesis. Because many questions regarding GATA-2 function in the vascular biology realm remain unanswered, we used ChIP sequencing and loss-of-function strategies to define the GATA-2–instigated genetic network in human endothelial cells. In contrast to erythroid cells, GATA-2 occupied a unique target gene ensemble consisting of genes encoding key determinants of endothelial cell identity and inflammation. GATA-2–occupied sites characteristically contained motifs that bind activator protein-1 (AP-1), a pivotal regulator of inflammatory genes. GATA-2 frequently occupied the same chromatin sites as c-JUN and c-FOS, heterodimeric components of AP-1. Although all three components were required for maximal AP-1 target gene expression, GATA-2 was not required for AP-1 chromatin occupancy. GATA-2 conferred maximal phosphorylation of chromatin-bound c-JUN at Ser-73, which stimulates AP-1–dependent transactivation, in a chromosomal context-dependent manner. This work establishes a link between a GATA factor and inflammatory genes, mechanistic insights underlying GATA-2–AP-1 cooperativity and a rigorous genetic framework for understanding GATA-2 function in normal and pathophysiological vascular states.
Keywords: endothelium, genomics, genome-wide, transcriptome
An important approach to understanding the biology and pathophysiology of the vascular system involves dissecting transcriptional mechanisms underlying the development and function of endothelial cells. Transcription factors implicated in endothelial transcriptional mechanisms (1) include GATA binding protein 2 (GATA-2), which is required to generate and/or maintain multipotent hematopoietic precursors during embryogenesis (2, 3). GATA transcription factors function as master regulators of development and have important functions in differentiated cells (4). Although GATA-2 is expressed in endothelial cells (5, 6), the GATA-2–driven genetic network in endothelium and its role in vascular biology are unclear.
The pathophysiological importance of GATA-2 in the vascular system is supported by the finding that GATA2 polymorphisms correlate with coronary artery disease (7). GATA-2 regulates genes that encode important mediators of endothelial cell function, including angiopoietin-2 (8), matrix metalloproteinase-2 (9), and vascular cell adhesion molecule-1 (10). In vitro and in vivo studies indicate that GATA-2 functions in human umbilical vein endothelial cells (HUVEC) and retinal endothelial cells to mediate mechano-signaling–dependent angiogenesis, which involves GATA-2–dependent induction of VEGF receptor II (11). Despite this compelling evidence, Gata2-null mice die at embryonic day 10.5 (E10.5) because of severe anemia, although their vasculature appears to be morphologically normal (3). However, the absence of an overt vasculogenesis defect does not negate the importance of GATA-2 in adult endothelium.
We used ChIP sequencing (ChIP-seq) and expression profiling to describe a GATA-1– and GATA-2–driven genetic network in human K562 erythroleukemia cells, which resemble primitive erythroid cells (12). This dataset, which was validated in primary erythroid cells (12), lacked genes that impart a unique identity to endothelial cells. We hypothesize that the GATA-2 target gene ensembles in endothelial and hematopoietic cells differ considerably, but analogous to its hematopoietic functions, GATA-2 exerts important activities in endothelium.
Herein we use ChIP-seq and expression profiling in HUVEC to define a GATA-2–driven genetic network that differs greatly from that of K562 cells. The composition of the network indicated that GATA-2 functions in concert with activator protein-1 (AP-1), a pivotal regulator of inflammation (13, 14), thus linking GATA-2 with inflammatory processes. Given the dearth of mechanistic information regarding vascular functions of GATA-2 and the broad scope of pathophysiologies involving vascular dysfunction with associated inflammation (15), linking endothelial GATA-2 to inflammatory genes establishes an important framework for understanding the role of GATA-2 in normal and pathological vascular states.
Results and Discussion
Exquisite Cell Type-Specificity of GATA-2 Chromatin Occupancy.
To establish a framework for understanding GATA-2 function in endothelium, we used ChIP-seq to define the GATA-2 chromatin target sites in HUVEC. The ChIP assay was validated by quantitating GATA-2 occupancy at the GATA2 +9.5 kb enhancer (16), which activates a linked LacZ reporter in the vasculature of transgenic mouse embryos (17). GATA-2 occupancy at the +9.5 kb site was ∼15-fold greater than the preimmune control, whereas no significant occupancy was detected at the negative control necdin promoter (Fig. S1). The ChIP-seq analysis was conducted via the standard mode of analysis for the ENCODE Consortium. Immunoprecipitated DNA from two biological replicates was analyzed by ChIP-seq. Because the replicates met established criteria for reproducibility, the datasets were merged, and peaks were called again. Unique sequences were mapped to the genome (HG19). Replicates A and B yielded 9.9 million and 12.6 million unique sequences, respectively. Using a false-discovery rate of 0.0001, replicates A and B yielded 18,233 and 8,830 GATA-2–occupied loci (peaks), respectively. Comparison of replicates revealed major overlap, with 7,619 (86%) of the B peaks present in A. Merging the replicates yielded 15,529 peaks corresponding to 6,081 genes (Dataset S1). The mean peak height was 75 bp (range: 14–229 bp), and the mean width was 417 bp.
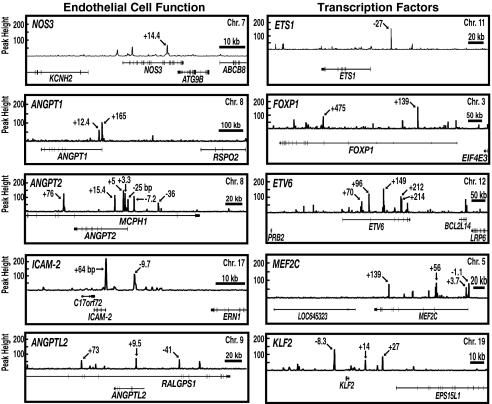
The GATA-2–occupied genes encode proteins known to be important for endothelial cell function, including endothelial NOS3, angiopoietins 1, 2, and 3, and intercellular adhesion molecule-2 (Fig. 1). Certain genes encoded transcription factors, including Kruppel-like factor-2 (KLF2) (Fig. 1). KLF2 is induced by shear stress, mediates shear stress signaling (18), determines vascular tone (19), mediates endothelial cell thrombotic function (20), and promotes vessel maturation (21). KLF2 is repressed by the proinflammatory cytokine IL-1β (18) and mediates the anti-inflammatory actions of statins (22). Additional transcription factors included the forkhead factor FOXP1 and multiple v-ets erythroblastosis virus E26 oncogene homolog (ETS)/ETS-variant (ETV) factors (Dataset S2), including ETV6, which is required for hematopoiesis and maintenance of the vascular network (23, 24). GATA-2 occupied myocyte-specific enhancer factor 2C (MEF2C), which is essential for vascular development (25). MEF2C contains an endothelial enhancer, corresponding to the GATA-2 peak at +139 kb, which directs endothelial cell-specific expression of a reporter in E7.5–E9.5 mice (26). The ChIP-seq analysis revealed a rich set of targets, including numerous genes not linked previously to endothelial cell biology.
Fig. 1.
Representative GATA-2 target genes in HUVEC. Signal maps of GATA-2 occupancy sites in HUVEC identified by ChIP-seq. Genes important for endothelial cell function and transcription factors are depicted. Arrows indicate ChIP-seq peak locations relative to the transcription start site of the respective GATA-2 target gene (kb).
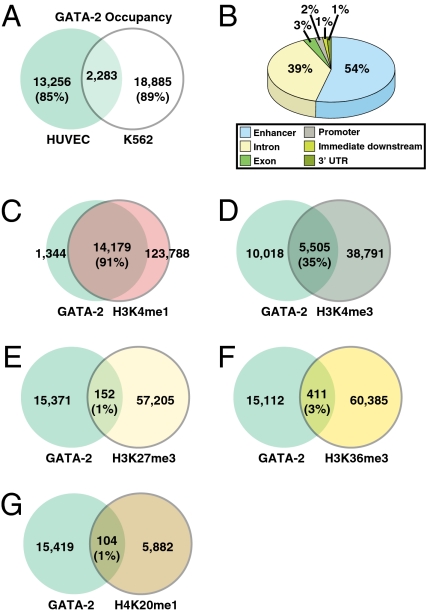
The majority of GATA-2–occupied peaks (13,256 peaks, 85%) were not occupied in our prior K562 cell analysis (12) (Fig. 2A). In HUVEC and K562 cells (12), the majority of peaks resided distal to the genes in putative enhancer regions (Fig. 2B). The unique chromatin-bound sites in HUVEC vs. K562 cells indicates that GATA-2 occupies a very small subset of genomic GATA motifs in a cell type-specific manner. To explore the relationship between GATA-2 occupancy and the epigenome, we evaluated histone marks in HUVEC at occupied sites. This analysis revealed that 91% of GATA-2–occupied sites overlap with H3K4me1 (Fig. 2C), a diagnostic enhancer mark (27). A smaller subset of occupied sites (35%) overlaps with H3K4me3 (Fig. 2D). As expected for a mark associated with transcription (28), 61% of these overlapping peaks reside in introns and exons. A small cohort of occupied sites overlaps with H3K27me3 (Fig. 2E), H3K36me3 (Fig. 2F), and H4K20me1 (Fig. 2G) (1%, 3%, and 1%, respectively).
Fig. 2.
Computational mining of ChIP-seq data. (A) Comparison of GATA-2–occupied peaks in HUVEC and K562 cells. (B) Locations of GATA-2–occupied peaks relative to nearest-neighbor genes determined with the Cis Element Annotation System (http://liulab.dfci.harvard.edu/CEAS/; http://ceas.cbi.pku.edu.cn/). (C–G) Comparison of GATA-2–occupied peaks and peaks of H3K4me1 (C), H3K4me3 (D), H3K27me3 (E), H3K36me3 (F), and H4K20me1 (G).
Linking GATA-2 with the Inflammatory Regulator AP-1.
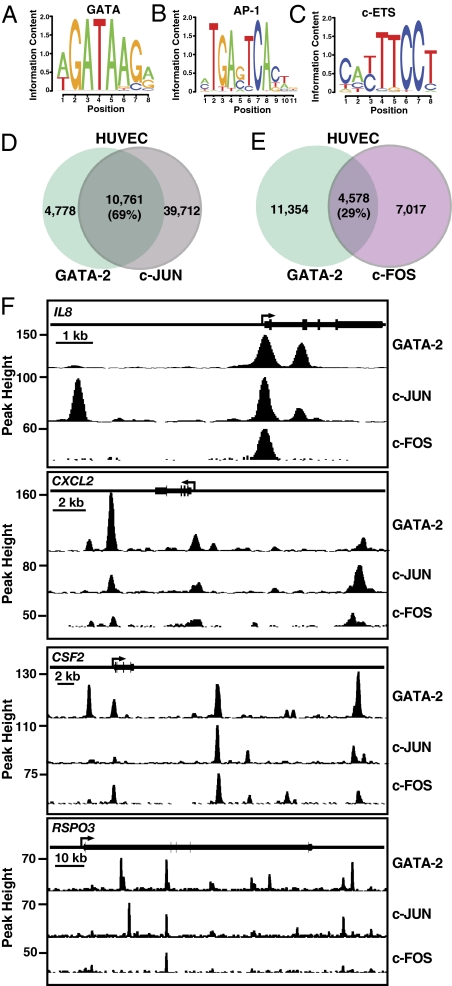
We assessed whether the GATA-2 cistrome in HUVEC resembles that of K562 cells, in which the majority of GATA-2–occupied peaks reside at sites containing the GATA motif (12). Of the 15,529 GATA-2 peaks, 12,930 (83%) contained at least one WGATAR motif. Of the 6,976,111 GATA motifs in the human genome, 25,823 (0.37%) resided within the 15,529 peaks. Constrained de novo motif finding with Cosmo (29) identified WGATAA that we defined previously for GATA-1 occupancy in K562 cells (Fig. 3A) (12). A broader GATA consensus, [CG][AT]GATAA[GAC][GAC], resided at 8,176 (43.3%) of the GATA-2–occupied K562 cell peaks and 4,698 (35.4%) of the HUVEC peaks.
Fig. 3.
Linking GATA-2 and AP-1 function. (A–C) Logos of overrepresented motifs from GATA-2–occupied ChIP-seq peaks: (A) GATA motif from constrained motif analysis and (B) AP-1 and (C) c-ETS motifs from de novo motif finding. (D and E) Comparison of GATA-2–occupied peaks and peaks of (D) c-JUN and (E) c-FOS occupancy in HUVEC. (F) Representative profiles demonstrating overlap among GATA-2, c-JUN, and c-FOS occupancy peaks.
De novo motif finding using MEME (30) to pinpoint cis-elements at GATA-2–occupied sites in HUVEC revealed canonical motifs for the inflammatory regulator AP-1 (14) (Fig. 3B) and c-ETS (Fig. 3C), which has important functions in endothelial cells (31). AP-1 motifs (TGA[G/C]TCA) (32) resided in 45.4% of the HUVEC GATA-2 peaks (7,055 of 15,523 peaks). This was highly significant (P < 0.01) relative to 16.3% of GATA-2 peaks in K562 cells (3,451 of 21,167 peaks). Because 69% of GATA-2–occupied peaks overlapped with those occupied by the jun proto-oncogene (c-JUN) component of AP-1 (Fig. 3D and Dataset S3), 29% overlapped with FBJ murine osteosarcoma viral oncogene homolog (c-FOS) (Fig. 3E and Dataset S4), and only 1% (137) overlapped with peaks of c-Myc occupancy (23,525 total c-Myc peaks), this GATA-2–AP-1 link has broad importance. The location with respect to neighboring genes of sites containing both GATA-2 and c-JUN did not differ significantly from the location of the complete cohort of GATA-2 occupancy sites. Examples of AP-1 target genes in which GATA-2 overlapped with c-JUN and c-FOS include IL-8 (33), CXCL2 (34), and CSF2 (35). GATA-2, c-JUN, and c-FOS overlapped at RSPO3, which is not known to be an AP-1 target (Fig. 3F). A high fraction of chromatin sites bound by both GATA-2 and c-Jun were enriched in H3K4me1 or H3K4me3 (76 and 83%, respectively).
We compared the frequency in which the c-ETS motif resided at GATA-2–occupied sites in HUVEC vs. K562 cells. In HUVEC, 5,446 (35.1%) of the 15,523 GATA-2–occupied sites contained c-ETS motifs. In K562 cells, 2,737 (12.9%) of the 21,167 GATA-2–occupied sites contained c-ETS motifs. Based on a binomial test for proportions, the differential frequency of c-ETS motifs at GATA-2–occupied sites in HUVEC vs. K562 cells was highly significant (P < 2.2e−16). Only 2,249 (14.5%) of the 15,523 GATA-2–occupied sites in HUVEC contained both c-ETS and AP-1 motifs. A GATA-2 peak was more likely to contain an AP-1 motif if it lacked a c-ETS motif (P < 0.001). Thus, it is likely that GATA-2 does not function frequently as a ternary complex with AP-1 and c-ETS in HUVEC.
Direct GATA-2 Target Gene Ensemble in Endothelium.
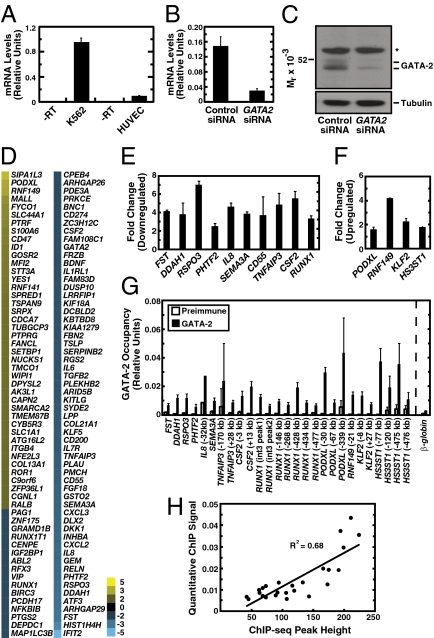
The frequent occurrence of AP-1 motifs at GATA-2–occupied sites and GATA-2 sharing of occupancy sites with a key component of AP-1 suggest that GATA-2 and AP-1 interact functionally. We used RNA interference to establish GATA-2 target genes in HUVEC. GATA2 mRNA was considerably lower in HUVEC than in K562 cells (Fig. 4A) (12), but endogenous GATA-2 in HUVEC was detectable by Western blotting (Fig. 4C). GATA2 siRNA significantly reduced GATA2 mRNA and protein in HUVEC vs. control siRNA-transfected cells (Fig. 4 B and C) without overtly affecting HUVEC morphology. Expression profiling of two biological replicates was conducted, and genes dysregulated upon GATA-2 knockdown were analyzed further. The direct GATA-2 targets were identified by comparing the differential expression with GATA-2 occupancy determined by ChIP-seq. This analysis revealed 116 direct targets differentially expressed by twofold or more upon GATA-2 knockdown (Fig. 4D). Because knockdowns do not eliminate expression, the analysis underestimates the target gene ensemble.
Fig. 4.
Direct GATA-2 target gene ensemble. (A) Comparison of GATA2 levels in HUVEC and K562 cells. (B) Quantitative RT-PCR analysis of GATA2 transcript levels after treatment with nontargeting control siRNA or GATA2 siRNA. (C) Western blotting of GATA-2 in HUVEC transfected with control or GATA2 siRNA. Whole-cell samples were analyzed. *, nonspecific band. (D) Heat map depicting the mean fold change of expression at GATA-2–occupied loci resulting from GATA2 knockdown (n = 2). (E and F) Quantitative RT-PCR validation of array results in GATA-2–activated (down-regulated with knockdown) (E) and repressed (up-regulated) genes (F). (G) Quantitative ChIP validation of GATA-2 occupancy at loci dysregulated by the knockdown. Data shown are mean ± SE; n = 3. (H) Correlation between height of ChIP-seq peak and quantitative ChIP signal.
The direct targets encode regulators of cell migration, including semaphorin 3A (36), and podocalyxin-like (37). GATA-2–activated direct targets encode proinflammatory mediators, including interleukins, the chemokine ligand CXCL2, and the cytokine CSF2 (GM-CSF). GATA-2 directly repressed KLF2, the key mediator of shear stress signaling in endothelium that opposes inflammation. GATA-2 activated RSPO3, which controls placental vascular plexus development and regulates angiogenesis during embryogenesis (38). Secreted RSPO3 activates wingless-type/β-catenin signaling, which exerts a proinflammatory function (38). GATA-2 induced expression of activating transcription factor 3 (ATF3), a stress-regulated transcription factor that protects endothelial cells from TNF-α–induced cell death (39) and negatively regulates allergic airway inflammation (40).
A subset of GATA-2–activated and –repressed targets was subjected to additional validation by real-time RT-PCR (Fig. 4 E and F) and quantitative ChIP analysis using the human β-globin (HBB) promoter as a negative control (Fig. 4G). ChIP-seq peak height correlated (R2 = 0.68) with the quantitative ChIP signal (Fig. 4H).
Our results indicate that GATA-2, c-JUN, and c-FOS commonly occupy the same chromatin sites containing GATA and AP-1 motifs. Taken together with the fact that AP-1 is a key regulator of inflammatory genes such as IL-8 and CSF2 that are GATA-2 targets (Fig. 4 D and E), this finding suggests the existence of a GATA factor–AP-1 regulatory network to control inflammatory genes. Ontological analysis of the direct GATA-2 target-gene ensemble revealed an enrichment of inflammatory genes, the majority of which are GATA-2–activated (Fig. 5A).
Fig. 5.
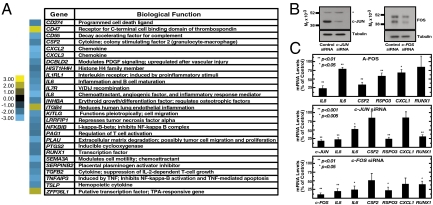
GATA-2/AP-1–dependent regulation of inflammatory genes. (A) Genes involved in inflammation and their expression changes with GATA-2 knockdown. Blue, down-regulated with GATA-2 knockdown; Yellow, up-regulated with GATA-2 knockdown. (B) Western blotting of endogenous c-JUN and c-FOS in HUVEC transfected with c-JUN or c-FOS siRNA, respectively. Transfected cells were boiled in SDS-sample buffer, and whole-cell samples were analyzed by Western blotting. *, nonspecific band. (C) Real-time RT-PCR analysis of gene expression in HUVEC transfected with a dominant-negative AP-1 antagonist (A-FOS), c-JUN siRNA, or c-FOS siRNA. The expression of empty vector-transfected cells (A-FOS) or nontargeting control siRNA-transfected cells (c-JUN and c-FOS) was designated as 100%, and expression for each gene is represented as a percentage of control expression. Data shown are mean ± SE; n ≥ 3.
GATA-2 Requirement for Assembly of Functional AP-1 Complexes on Chromatin.
In our loss-of-function analysis, HUVEC were cultured in medium containing FBS, EGF, and FGF2, which induce signaling that can activate AP-1 (13). Our results suggested that GATA-2 and AP-1 confer maximal AP-1 target gene transcription. We tested whether activity of presumptive AP-1 targets requires AP-1 in HUVEC. Transfection with dominant-negative c-FOS (A-FOS) (41, 42) significantly reduced IL-8 (P < 0.01), CSF2 (P < 0.01), and IL-6 (P < 0.01) expression (Fig. 5C). We tested genes that emerged from our direct GATA-2 target screen that were not established AP-1 targets. Expression of RSPO3 (P < 0.01), CXCL1, and RUNX1 was reduced by A-FOS. c-JUN, and c-FOS knockdowns confirmed AP-1 regulation of select GATA-2 targets; CSF2 and CXCL1 expression were not affected significantly (Fig. 5 B and C). Thus, GATA-2 and AP-1 ensure maximal activity of a cohort of the target genes.
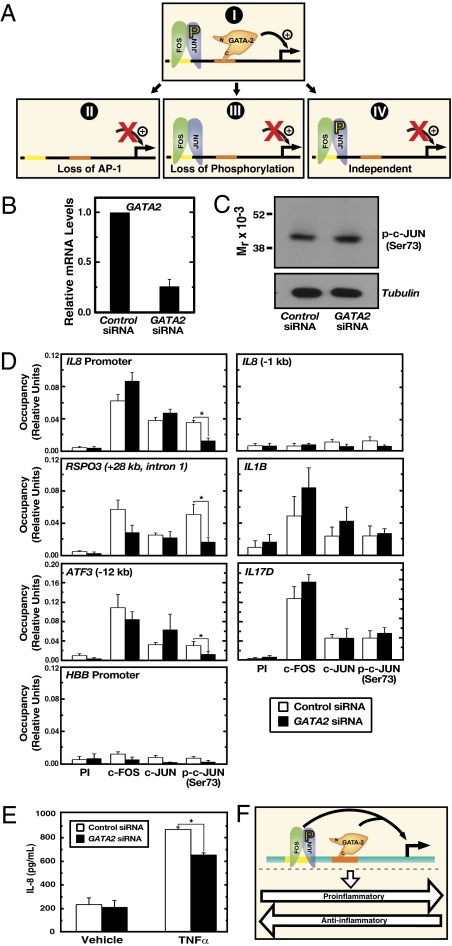
We considered potential mechanisms underlying the GATA-2–AP-1 cooperation (Fig. 6A). At target genes (Fig. 6AI), reduced GATA-2 occupancy might decrease AP-1 occupancy (Fig. 6AII), decrease AP-1 phosphorylation (Fig. 6AIII), or induce AP-1–independent molecular alterations (Fig. 6AIV). To distinguish among these possibilities, occupancy of c-FOS, c-JUN, and c-JUN phosphorylated at serine 73 was analyzed in control siRNA- and GATA2 siRNA-transfected cells. The analysis was conducted under conditions in which GATA2 expression was greatly reduced (Fig. 6B). Knocking down GATA-2 significantly reduced phosphorylated c-JUN occupancy (P < 0.05), but not total c-JUN and c-FOS occupancy, at the IL-8 promoter and distal sites at RSPO3 and ATF3 genes (Fig. 6D). Loss of chromatin-bound phosphorylated c-JUN was not associated with a change in total c-JUN (Fig. 6C).
Fig. 6.
GATA-2 requirement for assembly of functional AP-1 complexes on chromatin. (A) Models as described in Results and Discussion. (B) Real-time RT-PCR quantitation of GATA2 mRNA. (C) Western blotting of c-JUN phosphorylated at serine 73 [p-c-JUN (Ser73)] in HUVEC transfected with control or GATA2 siRNA. Whole-cell samples were analyzed by Western blotting. (D) Quantitative ChIP of preimmune control (PI), c-FOS, c-JUN, and p-c-JUN (Ser-73) occupancy at the IL-8 promoter, RSPO3, and ATF3, AP-1 targets that are GATA-2–independent (IL-1B, IL-17D), and negative controls (HBB and a site 1 kb upstream of the IL-8 promoter). Data shown are mean ± SE; n ≥ 3. (E) ELISA quantitation of IL-8 levels in medium from control siRNA- or GATA-2 siRNA-transfected cells treated with 10 ng/mL TNF-α or vehicle for 6 h. Data shown are mean ± SE; n = 6. (F) GATA-2–AP-1 coregulation of inflammatory genes in endothelium.
To assess the specificity of the GATA-2–dependent phosphorylated c-JUN occupancy, occupancy was analyzed at the GATA-2–insensitive IL-1B and IL-17D genes (Fig. 6D). GATA-2 knockdown did not affect phosphorylated c-JUN occupancy at these genes. Occupancy of all factors was low at the HBB promoter and an unoccupied site 1 kb upstream of the IL-8 promoter (Fig. 6D). The context-dependent GATA-2 requirement for phosphorylated c-JUN occupancy may explain why certain AP-1 target genes that are occupied by GATA-2 and AP-1 (Datasets S3 and S4) are insensitive to reducing GATA-2. In principle, GATA-2 may promote c-JUN phosphorylation indirectly or may promote phosphorylated c-JUN chromatin occupancy, and it has been reported that GATA-2 and c-JUN can interact (43). Coimmunoprecipitation analysis with HUVEC whole-cell lysates did not provide evidence for a stable complex containing GATA-2 and c-JUN.
We also tested whether c-JUN promotes GATA-2 chromatin occupancy, but knocking down c-JUN did not affect GATA-2 chromatin occupancy (Fig. S2).
To test whether GATA-2 facilitation of AP-1–mediated transcription has biological implications, we asked whether GATA-2 regulates secretion of the critical inflammatory mediator IL-8. GATA-2–knockdown HUVECs were treated with the proinflammatory factor TNF-α, which induced IL-8 secretion several fold as measured by ELISA (Fig. 6E). Knocking down GATA-2 significantly reduced IL-8 secretion (P = 0.0015), consistent with the mRNA expression data.
Linking GATA-2, AP-1, and Inflammatory Genes.
The results described herein, representing the genome-wide analysis of a GATA factor in endothelium, provide a rigorous genetic framework for understanding GATA-2 function in vascular biology. GATA-2 controls expression of genes involved in establishing endothelial cell phenotypes and inflammation. Analysis of 116 GATA-2–occupied genes differentially regulated upon knocking down GATA-2 using Ingenuity Pathways Analysis (http://www.ingenuity.com/products/pathways_analysis.html) revealed a complex network (Fig. S3) in which factors were knitted together based on established protein–protein interactions and abilities to regulate expression of other factors. The network highlights a GATA-2 function in regulating genes mediating inflammation (Fig. 6F). GATA-2 targets also included genes encoding anti-inflammatory factors, including TNFAIP3, which is induced by proinflammatory factors and suppresses proinflammatory signaling (44, 45) and ATF3 (40), as described above. Certain AP-1 targets are regulated by the proinflammatory factor NF-κB (46). Scanning the GATA-2 peaks with the NF-κB position/weight matrix from the JASPAR database using the FIMO tool (47) revealed that only 9.1% of the peaks match significantly (P ≤ 1e−4) to the matrix, a finding that is inconsistent with a frequent GATA-2–NF-κB association at HUVEC chromatin sites.
Because certain GATA-2 target genes with established roles in inflammation were co-occupied by GATA-2, c-JUN, and c-FOS, GATA-2 knockdown reduced phosphorylated c-JUN occupancy and IL-8 secretion, and AP-1 is a pivotal mediator of inflammation, it is attractive to propose that GATA-2–AP-1 cooperation is a common mode of orchestrating inflammatory processes. Vascular endothelial dysfunction involving inflammation underlies diseases including atherosclerosis, rheumatoid arthritis, and type II diabetes (15).
Given the biological importance of genes constituting the GATA-2 genetic network in HUVEC, why does the Gata2-knockout mouse appear to have morphologically normal vasculature, despite dying at E10.5 because of severe anemia (3)? Since multiple GATA factors are expressed in endothelium (48), a rigorous loss-of-function approach to ascertain GATA-2 function in the adult vasculature might require multigenic disruptions to reveal critical GATA factor-dependent pathways; alternatively, an important vascular activity of GATA-2 might not be apparent given the early embryonic lethality.
The direct regulation of inflammatory genes by GATA-2 has considerable biological and pathophysiological implications. It will be important to implement additional functional analyses and to determine whether endothelial cell subtypes differ in this mechanism. During the review of our manuscript, a genome-wide analysis of GATA-2 occupancy in human microvascular endothelial cells (HMVEC) was reported (49). Comparison of the HUVEC and HMVEC datasets will be informative.
Given the oncogenic activity of AP-1 subunits (50, 51), the role for GATA-3 in breast cancer (52), and links between GATA-2 dysregulation and leukemia (53), it will be important to determine whether GATA factors cooperate with AP-1 in diverse biological contexts and to what extent disruption of this mechanism underlies human pathologies. Because strong evidence implicates inflammation in a plethora of disease processes, including cancer, studies to evaluate GATA-2 function in endothelium in diverse disease models will be particularly instructive.
Materials and Methods
Cell Culture.
HUVEC were maintained in Medium 200 (Cascade Biologics) supplemented with Low Serum Growth Supplement (Cascade Biologics) and 1% antibiotic/antimycotic (Invitrogen). Cells were passaged at ∼70–80% confluence with 0.05% trypsin, and cells from passages four and six were used.
Antibodies.
Rabbit anti–GATA-2 polyclonal antibody was described previously (54). Anti–c-FOS (SC-253) and anti–c-JUN (SC-45) were from Santa Cruz Biotech. Anti–p-c-JUN (Ser73) was from Cell Signaling Technology (#9164). Mouse monoclonal α-tubulin (#CP06) was from Calbiochem.
Transfection and RNA Interference.
HUVEC were seeded 3–5 d before siRNA or plasmid transfection and grown to ∼70% confluence. After trypsinization, ∼106 cells per experiment were resuspended in HUVEC Nucleofector Solution (Lonza), and 0.45–0.60 nmol of siRNA, 5 μg A-FOS expression vector, or empty vector was transfected using the Nucleofector system (Lonza). siGENOME SMARTpool GATA2 siRNA, c-JUN siRNA, c-FOS siRNA, and control siRNA (SMARTpool #1) were from Dharmacon (Thermo Scientific). CMV-500-A-Fos and CMV-500 empty vector were provided by Charles Vinson (National Cancer Institute, Bethesda, MD). Cells were analyzed 24 or 40 h after siRNA or A-Fos transfection, respectively.
Generation of ChIP-Seq Data.
HUVEC histone ChIP-seq data were generated at the Broad Institute and by the B. Bernstein group (Massachusetts General Hospital, Boston, MA). HUVEC c-JUN ChIP-seq data were generated by D. Raha and Mike Snyder (Stanford University, Stanford, CA). HUVEC c-Myc ChIP-seq data were generated by Vishy Iyer (University of Texas at Austin, TX). HUVEC GATA-2, HUVEC c-FOS, and K562 GATA-2 ChIP-seq data were generated by the P. Farnham group. These data were collected as part of the ENCODE Consortium (http://www.genome.gov/10005107) and are available on the University of California, Santa Cruz Genome Browser.
Quantitative ChIP Analysis.
ChIP was conducted as described (55) using 1 × 108 HUVEC cells per condition for ChIP-seq, or 1 × 107 HUVEC cells per condition for quantitative ChIP.
ChIP-Seq Analysis.
HUVEC were crosslinked for 10 min with 1% formaldehyde, snap frozen, and stored at −80 °C. ChIP was conducted as described (http://www.genomecenter.ucdavis.edu/farnham/pdf/FarnhamLabChIP%20Protocol.pdf). Chromatin from 108 cells was diluted with three volumes of immunoprecipitation (IP) dilution buffer and incubated at 4 °C overnight with 60 μL of rabbit anti–GATA-2 antibody. StaphA cells were blocked with BSA. StaphA and Staph-Seq cells (SIGMA) were used. Blocked StaphA/Staph-Seq cells (100 μL) were added to the antibody/chromatin mixture and rotated for 15 min at room temperature. StaphA/Staph-Seq cells were washed twice with dialysis buffer and four times with polyclonal IP wash buffer. After reversal of crosslinks and RNase treatment, DNA was purified and analyzed as described with the Illumina GA2 platform (56). Short-sequence reads were aligned to the genome, and peaks were called using Sole-search (false-discovery rate, 0.0001; α value 0.001) with sequenced HUVEC input DNA as background (57). ChIP-seq data produced by the ENCODE Project Consortium were downloaded from the University of California, Santa Cruz browser and analyzed with Sole-search. Peak overlap was analyzed with Sole-search, allowing a distance of 200 bp between peaks.
Expression Profiling.
Control siRNA- and GATA2 siRNA-treated samples collected 24 h posttransfection were analyzed by hybridization to human 244k expression microarrays (Agilent Technologies). For each sample, 1.2 μg RNA was labeled with Cy-5 (control siRNA) or Cy-3 (GATA2 siRNA), and labeled cRNAs were combined and hybridized. Data were collected with an Agilent scanner. Two biological replicates for control siRNA/GATA-2 siRNA pairs were each hybridized in duplicate, yielding similar results, and average signals between the four replicates were used to establish fold-change cutoffs.
PCR Primers.
Primer sequences are indicated in Dataset S5.
Supplementary Material
Acknowledgments
We thank Dr. Charles Vinson (National Institutes of Health) for providing the A-Fos vector. ChIP-seq was supported by National Human Genome Research Institute funds. We acknowledge funding from National Institutes of Health Grants R21 HL091520 and DK068634 (to E.H.B.), HG003747 (to S.K.), and 1U54HG004558 (to P.J.F.). A.K.L. is an American Heart Association postdoctoral fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29531).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108440108/-/DCSupplemental.
References
- 1.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 3.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 4.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 6.Lee ME, Temizer DH, Clifford JA, Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J Biol Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]
- 7.Connelly JJ, et al. GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet. 2006;2:e139. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon MP, Tournaire R, Pouyssegur J. The angiopoietin-2 gene of endothelial cells is up-regulated in hypoxia by a HIF binding site located in its first intron and by the central factors GATA-2 and Ets-1. J Cell Physiol. 2008;217:809–818. doi: 10.1002/jcp.21558. [DOI] [PubMed] [Google Scholar]
- 9.Han X, Boyd PJ, Colgan S, Madri JA, Haas TL. Transcriptional up-regulation of endothelial cell matrix metalloproteinase-2 in response to extracellular cues involves GATA-2. J Biol Chem. 2003;278:47785–47791. doi: 10.1074/jbc.M309482200. [DOI] [PubMed] [Google Scholar]
- 10.Minami T, Aird WC. Thrombin stimulation of the vascular cell adhesion molecule-1 promoter in endothelial cells is mediated by tandem nuclear factor-kappa B and GATA motifs. J Biol Chem. 2001;276:47632–47641. doi: 10.1074/jbc.M108363200. [DOI] [PubMed] [Google Scholar]
- 11.Mammoto A, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 14.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 15.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 16.Grass JA, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wozniak RJ, Boyer ME, Grass JA, Lee Y-S, Bresnick EH. Context-dependent GATA factor function: Combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J Biol Chem. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- 18.SenBanerjee S, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Lin Z, et al. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96:e48–e57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J Biol Chem. 2008;283:3942–3950. doi: 10.1074/jbc.M707882200. [DOI] [PubMed] [Google Scholar]
- 22.Parmar KM, et al. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J Biol Chem. 2005;280:26714–26719. doi: 10.1074/jbc.C500144200. [DOI] [PubMed] [Google Scholar]
- 23.Wang LC, et al. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LC, et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q, et al. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 26.De Val S, et al. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 29.Bembom O, Keles S, van der Laan MJ. Supervised detection of conserved motifs in DNA sequences with cosmo. Stat Appl Genet Mol Biol. 2007;6(1) doi: 10.2202/1544-6115.1260. Article 8. [DOI] [PubMed] [Google Scholar]
- 30.Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan Y, et al. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. J Clin Invest. 2005;115:2508–2516. doi: 10.1172/JCI24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angel P, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 33.Yasumoto K, et al. Tumor necrosis factor alpha and interferon gamma synergistically induce interleukin 8 production in a human gastric cancer cell line through acting concurrently on AP-1 and NF-kB-like binding sites of the interleukin 8 gene. J Biol Chem. 1992;267:22506–22511. [PubMed] [Google Scholar]
- 34.Faouzi S, et al. Anti-Fas induces hepatic chemokines and promotes inflammation by an NF-kappa B-independent, caspase-3-dependent pathway. J Biol Chem. 2001;276:49077–49082. doi: 10.1074/jbc.M109791200. [DOI] [PubMed] [Google Scholar]
- 35.Wang CY, et al. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol Cell Biol. 1994;14:1153–1159. doi: 10.1128/mcb.14.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serini G, Valdembri D, Bussolino F. Integrins and angiogenesis: A sticky business. Exp Cell Res. 2006;312:651–658. doi: 10.1016/j.yexcr.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Larrucea S, et al. Expression of podocalyxin enhances the adherence, migration, and intercellular communication of cells. Exp Cell Res. 2008;314:2004–2015. doi: 10.1016/j.yexcr.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Kazanskaya O, et al. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development. 2008;135:3655–3664. doi: 10.1242/dev.027284. [DOI] [PubMed] [Google Scholar]
- 39.Kawauchi J, et al. Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-alpha-induced apoptosis through down-regulation of p53 transcription. J Biol Chem. 2002;277:39025–39034. doi: 10.1074/jbc.M202974200. [DOI] [PubMed] [Google Scholar]
- 40.Gilchrist M, et al. Activating transcription factor 3 is a negative regulator of allergic pulmonary inflammation. J Exp Med. 2008;205:2349–2357. doi: 10.1084/jem.20072254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olive M, et al. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 42.Gerdes MJ, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66:7578–7588. doi: 10.1158/0008-5472.CAN-06-1247. [DOI] [PubMed] [Google Scholar]
- 43.Kawana M, Lee ME, Quertermous EE, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol Cell Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper JT, et al. A20 blocks endothelial cell activation through a NF-kappaB-dependent mechanism. J Biol Chem. 1996;271:18068–18073. doi: 10.1074/jbc.271.30.18068. [DOI] [PubMed] [Google Scholar]
- 46.Natoli G. NF-kappaB: No longer an island, but a piece of a continent. EMBO Rep. 2010;11:246–248. doi: 10.1038/embor.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202-8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song H, et al. Critical role for GATA3 in mediating Tie2 expression and function in large vessel endothelial cells. J Biol Chem. 2009;284:29109–29124. doi: 10.1074/jbc.M109.041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanki Y, et al. Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J. 2011;30:2582–2595. doi: 10.1038/emboj.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 52.Kouros-Mehr H, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang SJ, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci USA. 2008;105:2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Im H, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Im H, et al. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 56.O’Geen H, Frietze S, Farnham PJ. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol Biol. 2010;649:437–455. doi: 10.1007/978-1-60761-753-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blahnik KR, et al. Sole-Search: An integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.