Abstract
Human cytomegalovirus induces and requires fatty acid synthesis. This suggests an essential role for lipidome remodeling in viral replication. We used mass spectrometry to quantify glycerophospholipids in mock-infected and virus-infected fibroblasts, as well as in virions. Although the lipid composition of mock-infected and virus-infected fibroblasts was similar, virions were markedly different. The virion envelope contained twofold more phosphatidylethanolamines and threefold less phosphatidylserines than the host cell. This indicates that the virus buds from a membrane with a different lipid composition from the host cell as a whole. Compared with published datasets, the virion envelope showed the greatest similarity to the synaptic vesicle lipidome. Synaptosome-associated protein of 25 kDa (SNAP-25) is a component of the complex that mediates exocytosis of synaptic vesicles in neurons; and its homolog, SNAP-23, functions in exocytosis in many other cell types. Infection induced the relocation of SNAP-23 to the cytoplasmic viral assembly zone, and knockdown of SNAP-23 inhibited the production of virus. We propose that cytomegalovirus capsids acquire their envelope by budding into vesicles with a lipid composition similar to that of synaptic vesicles, which subsequently fuse with the plasma membrane to release virions from the cell.
Human cytomegalovirus (HCMV) is a member of the enveloped herpesvirus family. It is a widespread pathogen that can cause disease in immunologically immature or compromised individuals (1). HCMV infection substantially alters the metabolism of cultured fibroblasts (2), inducing a marked increase in the flux of carbon from glucose into fatty acids (3). Consistent with the enhanced flux, inhibition of acetyl-CoA carboxylase, the first committed enzyme in the fatty acid biosynthetic pathway, reduces the yield of infectious virus. Thus, lipid synthesis is required during the HCMV replication cycle. Newly synthesized membranes likely accumulate in the assembly compartment (4–8), a virus-induced organelle with virus-coded proteins and cellular proteins that include markers of the exocytic and endocytic networks. This is the site at which viral capsids acquire an envelope (9–12).
Glycerophospholipids are major components of cellular membranes. This family of lipids consists of a glycerol backbone, a functional head group, and two fatty acids of varying length and unsaturation. Phosphatidic acid (PA) serves as the basic glycerophospholipid building block, with its phosphate group esterified to a head group to produce phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), or diphosphatidylglycerol. By using mass spectrometry, it is possible to identify and quantify numerous glycerolphospholipid species, differing in head group, acyl chain length, and degree of unsaturation (13–16). This quantitative lipidomics approach has been used to generate detailed descriptions of several viral envelopes, including those acquired by HIV (17), Semliki Forest virus and vesicular stomatitis virus (18), and hepatitis B virus (19).
In this study, we show that the glycerophospholipid composition of HCMV virions is markedly different from that of infected cells, arguing that the virus acquires its envelope at a specialized cellular membrane. By comparing these results with published datasets, we noted a striking similarity of the virion lipidome to that of synaptic vesicles (20). This similarity suggested that HCMV acquires its envelope by budding into exosome-like vesicles in fibroblasts, which then fuse at the plasma membrane to release mature virions from the cell. Synaptosomal-associated protein of 25 kDa (SNAP-25) is a component of the SNARE complex that mediates exocytosis of synaptic vesicles in neurons and exocrine cells, and it has been implicated in the exit of HSV-1 from neurons (21). SNAP-23 is a homolog of SNAP-25 (22) that functions in nonneuronal cells (23, 24). Using siRNA and shRNA specific for SNAP-23, we observed a role for SNARE machinery in the maturation of HCMV particles. This is consistent with a recent report showing that another component of the machinery, syntaxin 3, is needed (25). We propose a model in which HCMV induces the synthesis of membranes with a lipid composition similar to synaptic vesicles, which sponsor envelopment and egress of viral progeny.
Results
Modest Impact of HCMV on Total Cell Glycerophospholipids.
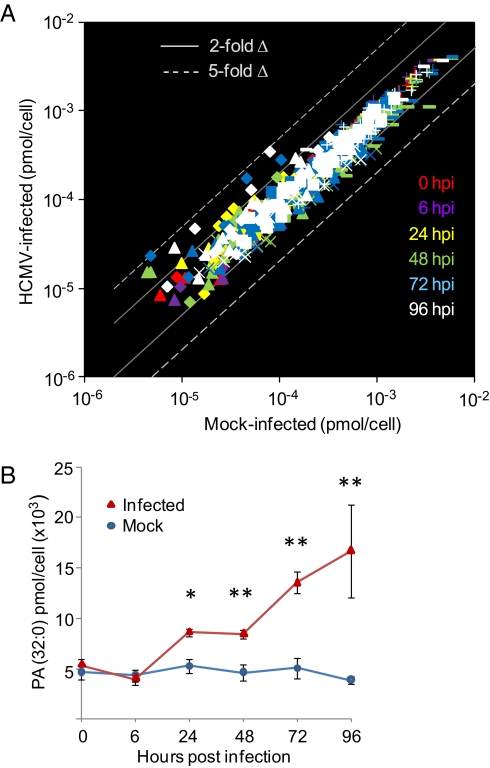
Using mass spectrometry, we monitored 146 unique glycerophospholipids, with acyl chains containing a total of 30–42 carbons, over the course of the HCMV replication cycle. Confluent fibroblast cultures were fed with serum-free medium at 24 h before infection and maintained in the absence of serum throughout the infection cycle. This treatment synchronized the cells in G0, reducing cell-to-cell variation in response to infection, and it removed a complex source of extraneous metabolites (2). A comparison of mock-infected vs. virus-infected fibroblasts is displayed in Fig. 1A, where each data point represents the average concentration of a single lipid at a specific time. Complete quantitative data for each lipid species is available in Dataset S1.
Fig. 1.
Glycerophospholipids in mock-infected vs. HCMV-infected fibroblasts. (A) Total glycerophospholipid levels change only modestly during infection. Glycerophospholipids were measured by liquid chromatography-mass spectrometry (LC-MS) profiling of mock vs. infected cells at 0 (red), 6 (purple), 24 (yellow), 48 (green), 72 (blue), and 96 (white) hpi. Gray solid lines and dashed lines indicate a twofold and fivefold change in amount in infected vs. mock, respectively. Lipids with different head groups are shown by different symbols: PA (◆); PE (■); PG (▲); PI (×); PS (▬▬▬); and PC (+). The number of individual lipid species measured was PA, 20; PE, 37; PG, 24; PI, 17; PS, 23; and PC, 25. (B) PA (32:0) levels increase in primary fibroblasts with time after infection with HCMV. Data are from Dataset S1. Red triangles, infected cells; blue circles, mock-infected cells. *P < 0.05 and **P < 0.01, Student t test. Error bars indicate SEM.
Glycerophospholipids in mock-infected samples exhibited variable but modest changes over the time course. The glycerophospholipids generally showed less than twofold changes in response to infection. The largest changes were increases in saturated and monounsaturated PA species; for example, PA (32:0) increased with time after infection, exhibiting an approximately fourfold increase relative to mock-infected cells at 96 hpi (P = 0.013; Fig. 1B). Multiple long chain PC species increased by typically 15–40% (e.g., at 96 hpi 38:6 P = 0.012, 40:6 P = 0.015), whereas several short chain PC species decreased ≈15–30% (e.g., at 96 hpi 32:0 P = 2.2E-4, 34:2 P = 0.0028). PI levels generally dropped over the course of infection, with PI (38:4) showing the largest reductions from basal levels (approximately twofold, P = 0.0054 at 96 hpi) during later stages of infection. Saturated and monounsaturated PG species generally increased with infection (e.g., 32:0, 32:1, 34:0, 34:1 all by more than 60% by 96 hpi, P < 0.05 by two-way ANOVA), whereas many polyunsaturated fatty acid species of PG and PS decreased. In summary, HCMV infection induced reproducible but generally modest changes in the levels of cellular glycerophospholipids.
Virion Glycerophospholipid Composition Is Different from That of Whole-Cell Lysates.
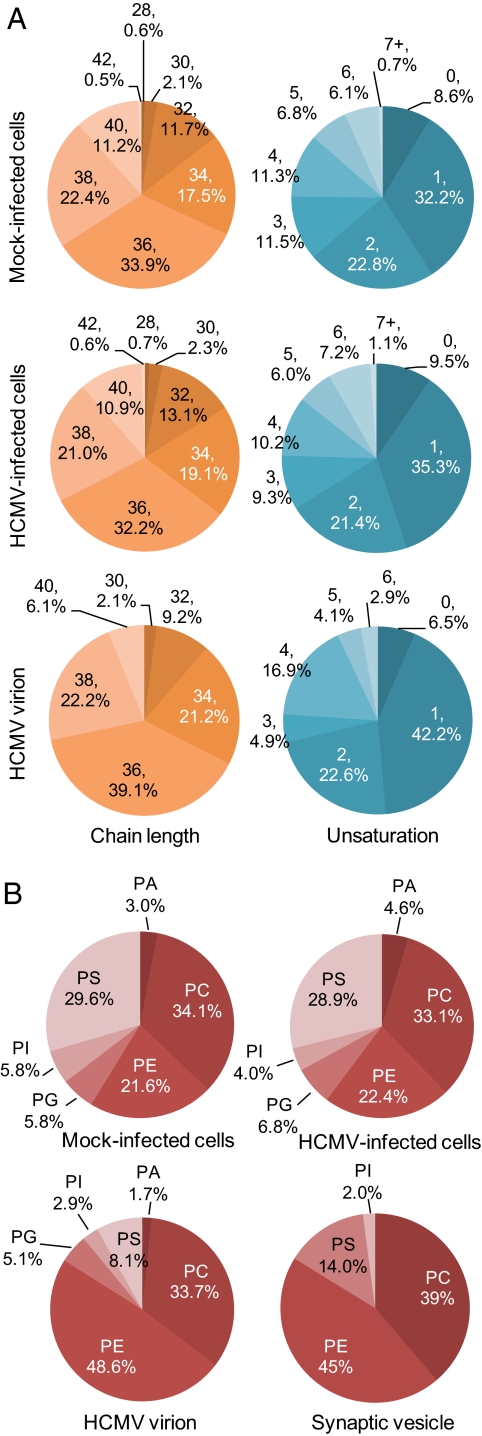
Virions were purified from the medium of infected fibroblast cultures by velocity sedimentation in glycerol-tartrate gradients (26), and judged to be substantially free of cellular membrane contaminants by electron microscopy (Fig. S1). Analyses of three independent virion preparations (Dataset S2) showed modest differences between virions and mock-infected or HCMV-infected cells in terms of acyl chain length (for measured lipids, which contained a total of 30–42 carbons in the fatty acid chains) or degree of unsaturation (Fig. 2A). The head group composition of the glycerophospholipids from the virion was, however, substantially different from that of the infected host (Fig. 2B). Whereas PE constituted ≈21% of glycerophospholipids assayed in whole-cell lysates, it was the dominant head group (≈48%) in purified virions. PS, PA, and PI were lower by factors of approximately 3, 2, and 2, respectively, whereas PC levels did not change the ratio. Interestingly, only one species each of PA and PG was detected in virions: PA (38:4) and PG (36:2). PA (38:4) was higher in infected compared to mock-infected cells by a factor of approximately 3.5, and PG (36:2) did not change significantly in infected cells through 96 hpi (Dataset S1).
Fig. 2.
Glycerophospholipids in virions compared with flbroblasts. (A) Modest differences between virions and mock-infected or HCMV-infected cells in terms of glycerophospholipid acyl chain length or degree of unsaturation. Acyl chain lengths were determined for lipids containing 30–42 total carbons in their two fatty acids combined. Percentages were calculated from Datasets S1 and S2. (B) HCMV virion and synaptic vesicle glycerophospholipids share similar head group composition. The percentage of each species in fibroblasts at 96 h after mock-infection or HCMV infection was calculated and compared with the virion lipidome. Synaptic vesicle lipidome data are from Takamori et al. (20).
Finally, the virion PE population contained multiple plasmalogen species that were found in lesser amounts or not detected in cell lysates. Plasmalogens are lipids in which the fatty acid linked to the first carbon of the glycerol backbone is attached through an ether or vinyl-ether bond rather than an ester bond. Although the fraction of PEs that were plasmalogens was lower in virions than in cells (19% and 28% of their total PE populations, respectively) (Fig. S2A), given the large increase in PEs in the virions, the representation of plasmalogen PEs as a percentage of the total glycerophospholipid pool was higher in virions (Fig. S2B). The distinct composition of the virion envelope relative to that of whole cells argues that the envelope is derived from a specialized cellular membrane.
SNAP-23 Is Required for the Efficient Production of HCMV Progeny.
By comparing the glycerophospholipid composition of HCMV virions with published datasets, we noted a resemblance to synaptic vesicles (20). HCMV virions and synaptic vesicles contain similar ratios of phospholipid head group classes (i.e., both have higher PE content and lower PS). In addition, both are enriched for PE plasmalogens. These similarities suggest that HCMV might acquire its envelope by budding into vesicles within fibroblasts that are similar in lipid composition to neuronal synaptic vesicles.
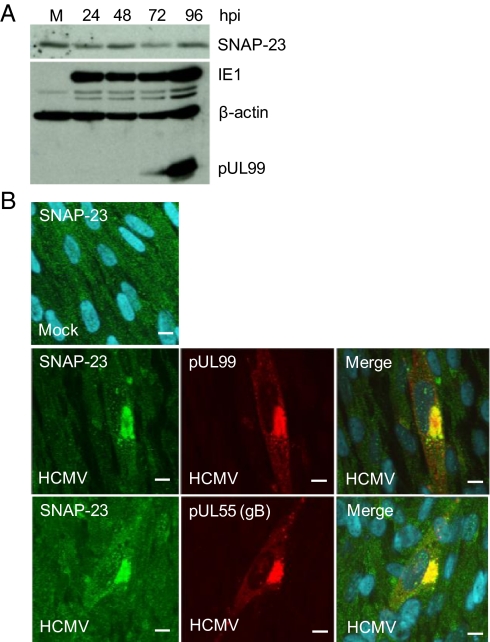
Synaptic vesicles are specialized trafficking vesicles that store neurotransmitters and undergo Ca2+-dependent exocytosis in response to an action potential. SNAP-25 is a key component of the SNARE complex that mediates exocytosis of synaptic vesicles in neurons and hormonal exocytosis in endocrine cells (reviewed in ref. 27). SNAP-23 is a SNAP-25 homolog that is expressed in many cell types (28), including primary human fibroblasts (29). Given the role of SNAP-25 in fusion of synaptic vesicles, the close relationship of SNAP-23 to SNAP-25, and the presence of SNAP-23 in fibroblasts, we tested the possibility that SNAP-23 was used by HCMV during its assembly/egress process. SNAP-23 was quantified by Western blot, and its levels did not change substantially with time after infection (Fig. 3A). Virus-coded IE-1 and pUL99 were assayed to confirm the progression of infection, and β-actin was used as a loading control. Immunofluorescent analysis (Fig. 3B) showed that SNAP-23 was spread through the cytoplasm in mock-infected cells, presumably owing to its association with vimentin filaments (29). In contrast, at 96 hpi, a substantial portion of SNAP-23 colocalized with pUL99 and pUL55 (gB), markers for the HCMV assembly compartment, the site at which the virion acquires an envelope (9–12).
Fig. 3.
SNAP-23 is relocalized to the assembly compartment after HCMV infection. (A) SNAP-23 protein levels remain nearly constant after infection. MRC5 fibroblasts were harvested at various times after infection at a multiplicity of 3 TCID50 units/mL, and SNAP-23 protein was assayed by Western blot. IE1, pUL99, and β-actin were monitored as controls. (B) SNAP-23 is colocalized with markers of the assembly compartment. MRC5 fibroblasts were analyzed by immunofluoresence at 96 hpi using antibodies to SNAP-23 (green) and pUL99 (red, upper infected panels) or pUL55 (red, lower infected panels).
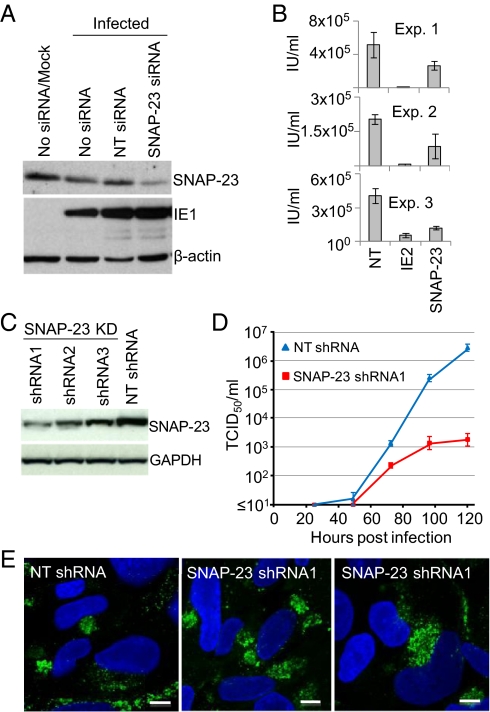
The functional significance of the relocalization was probed by performing a knockdown experiment with siRNA against SNAP-23 (30). A Western blot showed a substantial decrease in SNAP-23 levels relative to the nontargeting control using β-actin for normalization, and the cells remained competent to express the IE1 protein after infection with HCMV (Fig. 4A). However, three independent experiments showed an ≈50% decrease in the production of extracellular virus in knockdown compared with normal cells (Fig. 4B). In these experiments the siRNA was introduced into fibroblasts by transfection, and, consequently, some cells did not receive the siRNA and produced a normal yield of virus. Accordingly, we repeated the knockdown experiment using three different shRNAs expressed from a lentivirus vector. The most efficient knockdown occurred with shRNA1 (≈85%; Fig. 4C), and it was used to generate cultures in which SNAP-23 was reduced in all cells. Upon infection, these cells produced markedly less extracellular virus; the yield of infectious virus was reduced by a factor of approximately 1,000 at 120 hpi (Fig. 4D). Immunofluorescent analysis demonstrated that the HCMV pUL99 late protein accumulated and properly localized to the assembly compartment (Fig. 4E), arguing that the infection progressed to a very late stage but failed to generate normal levels of infectious progeny in the knockdown cells.
Fig. 4.
SNAP-23 is required for the efficient production of infectious virus. (A) siRNA knockdown of SNAP-23. Twenty-four hours before infection, MRC5 fibroblasts received no treatment (No siRNA) or were transfected with a nontargeting (NT) siRNA or with an SNAP-23–specific siRNA. Cells were infected at a multiplicity of 3 TCID50 units/mL and harvested at 72 hpi. Lysates were prepared and assayed by Western blot using antibodies to the indicated proteins. Mock-infected cells that were not treated with an siRNA were assayed as a control. (B) siRNA knockdown of SNAP-23 impairs HCMV replication. Cells were pretreated for 24 h with siRNA to HCMV IE2 or SNAP-23, infected at a multiplicity of 0.5 TCID50 unit/mL, supernatants were harvested at 72 hpi, and virus was quantified by assaying for IE1 expression by immunofluorescence. Error bars indicate SEM. (C) shRNA knockdown of SNAP-23. Cells containing lentiviruses expressing SNAP-23–specific shRNAs (shRNA1-3) or a nontargeting shRNA (NT shRNA) were assayed by Western blot using antibodies to the indicated proteins. (D) shRNA knockdown of SNAP-23 impairs HCMV replication. Fibroblasts expressing shRNA1 or NT siRNA were infected at a multiplicity of 3 pfu per cell, and virus from the media of infected cultures was quantified by TCID50 assay at various times after infection. Error bars indicate SEM. (E) shRNA knockdown of SNAP-23 does not interfere with accumulation or localization of pUL99. Infected cells were monitored for expression of pUL99 by immunofluorescence (green), and nuclei were stained with DAPI (blue).
Discussion
HCMV up-regulates much of central carbon metabolism, including the production of fatty acids (3); and, although the changes observed for total cellular glycerophospholipids assayed were modest (Fig. 1A and Dataset S1), virions exhibited a signature markedly distinct from the whole cell (Fig. 2B and Dataset S2). This suggests that the virus acquires its membrane from a discrete cellular compartment. There is strong evidence that the virion matures at the membrane-rich assembly compartment (9–12); so it is likely that the fatty acids synthesized during infection are used to produce membranes in this structure, supplying virion envelopes with a distinct lipid composition. The differential lipid composition of the virion might also reflect enrichment for certain lipids during the viral budding process.
The most substantial changes in infected cell glycerophospholipids were in PAs (Fig. 1B and Dataset S1). PAs serve a membrane structural role but also regulate diverse cell functions, including cell survival and proliferation, vesicular trafficking, cytoskeletal dynamics, oxidative bursts, etc. (31). Cells generate PAs from several sources, including diacylglycerols, another family of signaling molecules (32). PA (32:0) was elevated to the greatest extent within infected cells. This and other unsaturated and monounsaturated PAs induced during infection (Dataset S1) could accumulate in part through phospholipase D hydrolysis of PC (33), and likely have signaling roles (32). For example, unsaturated PAs can induce MAPK activity (34), and p38 MAPK is induced during HCMV infection in two waves, one of which seems to coincide with the elevation of PAs (35); and the interaction of PA with glucose transporter 4 (GLUT4) (36) could facilitate the massive induction in activity of this glucose transporter after infection (37). PA (38:4) was the only species identified in virions, and it was also elevated in infected relative to mock-infected cells. Polyunsaturated PAs are likely to be produced by phospholipase C catalyzed hydrolysis of PI-4,5 P2 followed by diacylglycerol kinase action (32). The differences in PA species caused by infection likely represent a combination of changes in metabolic, signaling, and structural lipid requirements that are needed to support virus replication and assembly.
Relative to infected cells, virions are enriched for PEs (Fig. 2B). PEs are “nonbilayer” lipids that form a liquid-crystalline phase when purified and hydrated under physiological conditions; “bilayer” lipids form bilayers under similar conditions (38). Nonbilayer lipids can impose curvature stress on bilayer membranes (39). This might favor formation of the compact spherical envelope of the virion, in particular its inner leaflet, which requires negative curvature to surround the HCMV tegument. Further, PEs promote membrane fusion in a variety of assays, including the fusion of Sindbis virus with model lipid membranes (40) and fusion in a reaction using pure yeast lipids and proteins (41). The reconstituted yeast system also revealed that PE is essential for SNARE complex assembly before fusion. The virion PE population contained plasmalogen species (Fig. S2A), consistent with the elevation of its dihydroxyacetone phosphate precursor in infected cells (3), and plasmalogens support fusion (42). Thus, the virion PE composition might facilitate the envelopment/egress process or the subsequent fusion of the virion with the plasma membrane at the start of a new round of infection.
Not surprisingly, the phospholipid composition of the HCMV virion is markedly different from that of viruses that bud from the plasma membrane, including HIV-1 (17), vesicular stomatitis virus, and Semliki Forest virus (18). It is noteworthy, however, that the envelope of HCMV also differs from that of another herpes virus, HSV-1 (43). HSV-1 is not known to induce a membranous organelle similar to the HCMV assembly compartment.
Although different from other studied viruses, the virion lipidome strongly resembles that of the synaptic vesicle (Fig. 2B), suggesting the virion envelope and synaptic vesicle are generated via similar mechanisms. SNAP-25 is a key component of the SNARE complex that mediates exocytosis of synaptic vesicles, and SNAP-23 is a SNAP-25 homolog expressed in many cell types, including fibroblasts. SNAP-23 is relocalized to the HCMV assembly compartment (Fig. 3), and depletion of SNAP-23 with siRNA or shRNA (Fig. 4) demonstrated that it is essential for the efficient production of HCMV progeny. Depletion of a second SNARE component, syntaxin 3, has also been found to reduce the yield of HCMV (25).
Like SNAP-25, SNAP-23-mediated fusion is Ca2+-dependent. SNAP-25 interacts with the synaptotagmin 1, which serves as a Ca2+ sensor (27) and requires relatively high Ca2+ (1 μM) for activation. SNAP-23 requires a lower Ca2+ concentration (100 nM), presumably because it uses different members of the synaptotagmin family. Importantly, HCMV infection elevates cytoplasmic Ca2+; mock-infected cells contain 30–100 nM Ca2+, and infected cells contain to 150–250 nM Ca2+ (44), providing an environment where SNAP-23 would be expected to function efficiently.
HCMV (45) and other herpesviruses (46–48) depend on elements of the endosomal sorting complex required for transport machinery, which mediates endosome sorting and multivesicular body formation (49). Perhaps the lipidome of endosomes and/or multivesicular bodies will prove to be similar to that of synaptic vesicles and the HCMV envelope. One model for synaptic vesicle biogenesis postulates that they are derived from endosomal intermediates (50).
Additional observations are consistent with the view that the virion envelope is acquired from synaptic vesicle-like membranes. Synaptic vesicle biogenesis has been linked to two clathrin-related pathways, one of which uses budding from endosomes and/or the Golgi complex and requires AP1 or AP3 clathrin adaptors along with the Ras-related GTPase ADP-ribosylation factor (ARF), which is sensitive to brefeldin A (51). Early endosome markers are at the center of the HCMV assembly compartment (5, 10), clathrin and ARF4 have been detected in the virion (52), and trafficking of the pUL99 virion protein is brefeldin sensitive (53). Further, a portion of the PS in the virion envelope has been inferred to be exposed on its outer leaflet, because PS serves as a cofactor for activation of prothrombinase, and HCMV virions are procoagulant (54). PS is normally located on the inner leaflet of the cellular plasma membrane, but synaptic vesicles contain similar amounts of PS on their two membrane leaflets (55). Our lipidomic observations suggest a unique framework for interpreting these data, wherein HCMV hijacks synaptic vesicle-like machinery to enable virion egress.
Materials and Methods
Virus Growth and Purification.
Primary human foreskin fibroblasts and MRC5 fibroblasts (ATCC) were cultured in medium containing 10% FBS. HCMV infections used the AD169 strain BADwt (56). Virus stocks were produced in fibroblasts by pooling cell-free and cell-associated virus, concentrated by centrifugation through sorbitol cushions, resuspended in serum-free medium, and titered by tissue culture infective dose (TCID50) or fluorescent focus assay.
Sample Preparation and Mass Spectrometry.
For analysis of cellular lipids, primary fibroblasts were serum starved for 24 h, infected at a multiplicity of 3.0 TCID50 units per cell, and then maintained in serum-free medium. To extract phospholipids, 2 × 106 cells were washed twice with ice-cold PBS, scraped into 800 μL of ice-cold 0.1 N HCl:CH3OH (1:1), and transferred into a cold microfuge tube. After addition of 400 μL of ice-cold CHCl3, samples were mixed by vortexing for 1 min at 4 °C, and phases were separated by centrifugation in a microfuge (18,000 × g for 5 min, 4 °C). The lower organic layer was isolated, synthetic odd-carbon phospholipids (four per each phospholipid class) were added as internal standards, and solvent was evaporated. The resulting lipid film was dissolved in 100 μL of isopropanol (IPA):hexane:100 mM NH4COOH(aq) 58:40:2 (mobile phase A) (14).
For analysis of virion lipids, virus was purified as previously described (26). Serum-starved, primary fibroblasts were infected at a multiplicity of 0.1 TCID50 units per cell, maintained in serum-free medium, and supernatant was harvested 6 d later. Cell debris was removed by centrifugation, bacitracin was added to prevent clumping, and virus particles were pelletted by centrifugation through the 20% sorbitol cushion at room temperature. Pellets were resuspended in buffered saline, and virions were resolved by centrifugation in sodium tartrate gradients. The virion band was removed with a syringe, diluted, and purified through a second round of centrifugation. Purified virions were resuspended in buffer containing 100 mM NaCl and 50 mM Tris·HCl, pH7.4. Infectivity of the product was quantified by TCID50 assay, and purity was assessed by electron microscopy (Fig. S1). Phospholipids were extracted from an average of 3.4 × 106 TCID50 of purified virions by using acidified methanol followed by chloroform extraction, as described for cells above.
Mass spectrometric analysis and quantitation were performed essentially as previously described (14). An MDS SCIEX 4000QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems) was used for the analyses. Coupled to it was a Shimadzu HPLC system (Shimadzu Scientific Instruments) consisting of an SCL 10 APV controller, two LC 10 ADVP pumps, and a CTC HTC PAL autosampler (Leap Technologies). Phospholipids were separated on a Phenomenex Luna Silica column (2 × 250 mm, 5-μm particle size) using a 20-μL sample injection. A binary gradient consisting of IPA:hexane:100 mM NH4COOH(aq) 58:40:2 (mobile phase A) and IPA:hexane:100 mM NH4COOH(aq) 50:40:10 (mobile phase B) was used for the separation. Instrumentation parameters and solvent gradient were as previously described (14). Data are presented as means plus SEs based on three independent experiments. Statistical analysis was performed by ANOVA. For time courses, a two-way ANOVA using factors of time and virus state (infected, uninfected) was used, with post hoc t test at individual time points.
Protein Analysis.
For Western blots, MRC5 cell pellets were resuspended in lysis buffer (150 mM NaCl, 1% nonidet-p40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, and Roche protease inhibitor mixture), sonicated, rocked for 30 min at 4 °C, and subjected to centrifugation in a microfuge to remove insoluble material. Equal amounts of protein were separated by electrophoresis in SDS-containing 12% polyacrylamide gels and transferred to nitrocellulose membranes, which were then probed with antibodies. For immunofluorescence, cells were plated on glass coverslips and mock infected or infected with HCMV. After various time intervals, cells were fixed and permeabilized with methanol. Cells were blocked and incubated with mouse or rabbit antibodies. The following antibodies were used: HCMV IE-1 [1B12 (57)], pUL99 [10B4-29 (9)], pUL55 gB (Goodwin Institute), SNAP-23 (111-202; Synaptic Systems), β-actin-HRP (ab49900; Abcam), and GAPDH (G8795; Sigma-Aldrich).
Knockdown of SNAP-23.
MRC5 fibroblasts were transfected with siRNA (10 nM) directed against SNAP-23 (5′CCAACAGAGAUCGUAUUGATT3′) (30), an siRNA specific for IE-2 (5′AAACGCAUCUCCGAGUUGGACTT3′) (58), or the Mission siRNA Universal Negative Control #1 (Sigma-Aldrich) by using Oligofectamine (Invitrogen) in Optimem (Gibco). After 6 h, transfection reagent was removed and replaced with medium containing 10% FBS. Twenty-four hours later, cells were infected and analyzed at 72 or 96 hpi. shRNAs in pLKO.1-puro (Sigma-Aldrich) also were used for knockdown of SNAP-23. MRC5 cells were infected with the lentivirus vector, selected with 500 μg/mL G418, and then used for analysis. shRNA1 (TRCN0000144789) was chosen for further study (5′CCGGGCAATGAGATTGATGCTCAAACTCGAGTTTGAGCATCAATCTCATTGCTTTTTTG-3′).
Supplementary Material
Acknowledgments
We thank L. Vastag and J. Hwang for helpful discussions and advice and M. Bisher and J. Goodhouse for technical support. This work was supported by National Institutes of Health Grants CA82396 (to T.S.), AI068678 (to J.D.R.), and U54 GM069338 (to H.A.B.). S.T.H.L. received fellowship support from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109796108/-/DCSupplemental.
References
- 1.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 2.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AuCoin DP, Smith GB, Meiering CD, Mocarski ES. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J Virol. 2006;80:8199–8210. doi: 10.1128/JVI.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, Vasanji A, Pellett PE. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol. 2007;81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krzyzaniak M, Mach M, Britt WJ. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J Virol. 2007;81:10316–10328. doi: 10.1128/JVI.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: Characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez V, Sztul E, Britt WJ. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J Virol. 2000;74:3842–3851. doi: 10.1128/jvi.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepeda V, Esteban M, Fraile-Ramos A. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol. 2010;12:386–404. doi: 10.1111/j.1462-5822.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 11.Homman-Loudiyi M, Hultenby K, Britt W, Söderberg-Nauclér C. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J Virol. 2003;77:3191–3203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Severi B, Landini MP, Govoni E. Human cytomegalovirus morphogenesis: An ultrastructural study of the late cytoplasmic phases. Arch Virol. 1988;98:51–64. doi: 10.1007/BF01321005. [DOI] [PubMed] [Google Scholar]
- 13.Milne S, Ivanova P, Forrester J, Alex Brown H. Lipidomics: An analysis of cellular lipids by ESI-MS. Methods. 2006;39:92–103. doi: 10.1016/j.ymeth.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 15.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreyev AY, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51:2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan R, et al. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalvodova L, et al. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol. 2009;83:7996–8003. doi: 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodgers MA, Saghatelian A, Yang PL. Identification of an overabundant cholesterol precursor in hepatitis B virus replicating cells by untargeted lipid metabolite profiling. J Am Chem Soc. 2009;131:5030–5031. doi: 10.1021/ja809949r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Miranda-Saksena M, et al. Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J Virol. 2009;83:3187–3199. doi: 10.1128/JVI.01579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- 23.Chen D, Whiteheart SW. Intracellular localization of SNAP-23 to endosomal compartments. Biochem Biophys Res Commun. 1999;255:340–346. doi: 10.1006/bbrc.1999.0173. [DOI] [PubMed] [Google Scholar]
- 24.Valdez AC, Cabaniols JP, Brown MJ, Roche PA. Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J Cell Sci. 1999;112:845–854. doi: 10.1242/jcs.112.6.845. [DOI] [PubMed] [Google Scholar]
- 25.Cepeda V, Fraile-Ramos A. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol. 2011;13:846–858. doi: 10.1111/j.1462-5822.2011.01583.x. [DOI] [PubMed] [Google Scholar]
- 26.Womack A, Shenk T. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio. 2010;1:e00282. doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang ZP, Südhof TC. Cell biology of Ca2+-triggered exocytosis. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 29.Faigle W, Colucci-Guyon E, Louvard D, Amigorena S, Galli T. Vimentin filaments in fibroblasts are a reservoir for SNAP23, a component of the membrane fusion machinery. Mol Biol Cell. 2000;11:3485–3494. doi: 10.1091/mbc.11.10.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okayama M, Arakawa T, Mizoguchi I, Tajima Y, Takuma T. SNAP-23 is not essential for constitutive exocytosis in HeLa cells. FEBS Lett. 2007;581:4583–4588. doi: 10.1016/j.febslet.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Hodgkin MN, et al. Diacylglycerols and phosphatidates: Which molecular species are intracellular messengers? Trends Biochem Sci. 1998;23:200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- 33.Pettitt TR, McDermott M, Saqib KM, Shimwell N, Wakelam MJ. Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. Biochem J. 2001;360:707–715. doi: 10.1042/0264-6021:3600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siddiqui RA, Yang YC. Interleukin-11 induces phosphatidic acid formation and activates MAP kinase in mouse 3T3-L1 cells. Cell Signal. 1995;7:247–259. doi: 10.1016/0898-6568(94)00083-n. [DOI] [PubMed] [Google Scholar]
- 35.Johnson RA, Ma XL, Yurochko AD, Huang ES. The role of MKK1/2 kinase activity in human cytomegalovirus infection. J Gen Virol. 2001;82:493–497. doi: 10.1099/0022-1317-82-3-493. [DOI] [PubMed] [Google Scholar]
- 36.Heyward CA, et al. An intracellular motif of GLUT4 regulates fusion of GLUT4-containing vesicles. BMC Cell Biol. 2008;9:25. doi: 10.1186/1471-2121-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruner SM. Intrinsic curvature hypothesis for biomembrane lipid composition: A role for nonbilayer lipids. Proc Natl Acad Sci USA. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q, Guo Y, Li L, Hui SW. Effects of lipid headgroup and packing stress on poly(ethylene glycol)-induced phospholipid vesicle aggregation and fusion. Biophys J. 1997;73:277–282. doi: 10.1016/S0006-3495(97)78068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheule RK. Fusion of Sindbis virus with model membranes containing phosphatidylethanolamine: Implications for protein-induced membrane fusion. Biochim Biophys Acta. 1987;899:185–195. doi: 10.1016/0005-2736(87)90399-3. [DOI] [PubMed] [Google Scholar]
- 41.Mima J, Wickner W. Complex lipid requirements for SNARE- and SNARE chaperone-dependent membrane fusion. J Biol Chem. 2009;284:27114–27122. doi: 10.1074/jbc.M109.010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glaser PE, Gross RW. Plasmenylethanolamine facilitates rapid membrane fusion: A stopped-flow kinetic investigation correlating the propensity of a major plasma membrane constituent to adopt an HII phase with its ability to promote membrane fusion. Biochemistry. 1994;33:5805–5812. doi: 10.1021/bi00185a019. [DOI] [PubMed] [Google Scholar]
- 43.van Genderen IL, Brandimarti R, Torrisi MR, Campadelli G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 44.Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci USA. 2006;103:19117–19122. doi: 10.1073/pnas.0609353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tandon R, AuCoin DP, Mocarski ES. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol. 2009;83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori Y, et al. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic. 2008;9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calistri A, et al. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J Virol. 2007;81:11468–11478. doi: 10.1128/JVI.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawliczek T, Crump CM. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol. 2009;83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurley JH, Boura E, Carlson LA, Różycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos MS, Li H, Voglmaier SM. Synaptic vesicle protein trafficking at the glutamate synapse. Neuroscience. 2009;158:189–203. doi: 10.1016/j.neuroscience.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shupliakov O, Brodin L. Recent insights into the building and cycling of synaptic vesicles. Exp Cell Res. 2010;316:1344–1350. doi: 10.1016/j.yexcr.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 52.Varnum SM, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 2010;9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pryzdial EL, Wright JF. Prothrombinase assembly on an enveloped virus: evidence that the cytomegalovirus surface contains procoagulant phospholipid. Blood. 1994;84:3749–3757. [PubMed] [Google Scholar]
- 55.Deutsch JW, Kelly RB. Lipids of synaptic vesicles: Relevance to the mechanism of membrane fusion. Biochemistry. 1981;20:378–385. doi: 10.1021/bi00505a024. [DOI] [PubMed] [Google Scholar]
- 56.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiebusch L, Truss M, Hagemeier C. Inhibition of human cytomegalovirus replication by small interfering RNAs. J Gen Virol. 2004;85:179–184. doi: 10.1099/vir.0.19453-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.