Abstract
BLOC-1 is one of four multi-subunit complexes implicated in sorting cargo to lysosome-related organelles, as loss of function of any of these complexes causes Hermansky-Pudlak syndrome. Eight subunits of BLOC-1 interact with each other, and with many other proteins. Identifying new interactors of BLOC-1 will increase understanding of its mechanism of action, and studies in model organisms are useful for finding such interactors. Psi-BLAST searches identify homologues in diverse model organisms, but there are significant gaps for BLOC-1, with none of its eight subunits found in Saccharomyces cerevisiae. Here we use more sensitive searches to identify distant homologues for three BLOC-1 subunits in S. cerevisiae: Blos1, snapin and cappuccino (cno). Published data on protein interactions show that in yeast these are likely to form a complex with three other proteins. One of these is the yeast homologue of the previously uncharacterized KxDL protein, which also interacts with Blos1 and cappuccino in higher eukaryotes, suggesting that KxDL proteins are key interactors with BLOC-1.
Keywords: endosome, endocytosis, Hermansky-Pudlak syndrome (HPS), BLOC1S1 (Blos1), cappuccino (cno), snapin, S. cerevisiae
Introduction
Traffic through the secretory pathway involves the action of many multi-molecular complexes. Adaptor protein (AP-3) complexes define a traffic step from early endosomes, or in yeast from the TGN (1), to later endocytic compartments (2,3). AP-3 dysfunction in humans leads to Hermansky-Pudlak Syndrome (HPS), characterized by oculocutaneous albinism and platelet dysfunction caused by defects in melanosomes and platelet dense granules respectively (1). These two endocytic compartments are lysosome-related organelles (LROs), a term that applies to specialized acidic organelles related to late endosomes or lysosomes (4,5).
Biogenesis of LROs relies not only on AP-3, but also three other multimeric complexes called BLOC-1/2/3 (for biogenesis of LRO complex) (6). Of these, BLOC-1 is the most complicated with eight subunits: Blos1, Blos2, Blos3 (also called BLOC1S1-3), cappuccino, dysbindin, muted, pallidin and snapin (6). HPS in humans results from mutations in two of the eight BLOC-1 subunits: HPS7 and 8 arise from mutations of dysbindin and Blos3 respectively. Mutations in three other BLOC-1 subunits (pallidin, muted and cappuccino) have been identified in mouse models of HPS (7,8). BLOC-1 has been shown to reside on endosomes (9,10), and is required for different aspects of cargo sorting in early endosomes (11,12). A similar function is conserved across evolution, as in flies and plants mutations in BLOC-1 subunits affect lysosome and LRO function (13,14) A wide variety of other proteins implicated in membrane trafficking have been identified as interactors of the eight BLOC-1 subunits, mainly by yeast two-hybrid analysis (15). Among the few interactors described to bind the intact octameric complex are AP-3 and BLOC-2 (9), as well as the SNAREs syntaxin-13 and SNAP-25 (16). Despite the large number of interactions known for BLOC-1, its mechanism of action is not known.
All of AP-3 and BLOC-1/2/3 are ubiquitously expressed in mammals (4,17), suggesting that they may affect the secretory pathway in non-specialized cells. In agreement with this, HPS affects not only LROs but also the lysosome itself, such that lysosomal proteins such as CD63 are misrouted to the cell surface (9), and lipofuscin accumulates in different cell types (18). This suggests that the mutations causing HPS might have effects in all cell types, even those lacking LROs. In the budding yeast S. cerevisiae, AP-3 mediates traffic of a subset of cargo to the vacuole, the terminal degradative organelle equivalent to the lysosome (1). Even though BLOC components are conserved widely throughout eukaryotic evolution (for example, the slime mold Dictyostelium has 11 of the possible 13), only two direct homologues to BLOC components have been found in any fungus: Blos1 and Blos2 in the oleaginous yeast Yarrowia lipolytica, but none have been identified in S. cerevisiae (19).
We have extended previous iterative psi-BLAST searches, using a more sensitive technique that combines structural predictions with the detection of sequence homology. This has identified putative homologues for three BLOC-1 subunits in S. cerevisiae: Blos1, snapin and cno. These form a complex with three other proteins, one of which is the yeast homologue of the previously uncharacterized KxDL protein. Database mining shows that KxDL proteins have conserved interactions with BLOC-1 in higher eukaryotes.
Results and Discussion
A Blos1 homologue in S. cerevisiae
A psi-BLAST search seeded with human Blos1 (125 aa) identified a yeast homologue in Y. lipolytica (113 aa) on the second iteration, but no homologue in S. cerevisiae was found (19). We reasoned that there might be a homologue of Blos1 in S. cerevisiae whose sequence has extensively diverged, that was missed because the set of sequences used by the iterative psi-BLAST model was too dominated by sequences closely related to human Blos1. Among the non-significant hits for Blos1 there are no S. cerevisiae proteins, but there are proteins from other yeast of the typical size of Blos1. For example, an ORF of 104 aa from Kluyveromyces lactis (a yeast that is more closely related to Saccharomyces than to Yarrowia) has an E-value = 0.09 (threshold from inclusion in psi-BLAST = 0.005, see Methods and Table S1). To test if this is a Blos1 homologue in K. lactis, we seeded psi-BLAST with the K. lactis sequence, which identified ORFs from six other yeast, including the S. cerevisiae ORF of unknown function YLR408Cp (122 aa). Although this psi-BLAST failed to expand, known Blos1 homologues appeared among the non-significant hits (e.g. Rana, E-value = 0.5; Yarrowia, E-value = 4, Table S1), which left open the possibility that the sequence in K. lactis is a Blos1 homologue, in which case YLR408Cp could also be a Blos1 homologue. To test this, we seeded a psi-BLAST search with YLR408Cp. At first this identified the same 7 yeast sequences; then the alignment expanded to include first the known fungal Blos1 homologues, then all other Blos1 sequences (Table S1). Thus, although human Blos1 could not detect YLR408Cp by psi-BLAST, the reverse search did establish the link, which strongly suggests that YLR408Cp is the S. cerevisiae homologue of Blos1.
We next enhanced homology detection for Blos1 by using HHpred, which supplements sequence alignment with structure prediction (20), to achieve far greater sensitivity (21). This takes advantage of the fact that proteins diverge in terms of structure much more slowly than in specific sequence, conservation of which may be undetectable. HHpred compares a structural model of the query sequence against a database of ~135,000 records that contains not only all solved structures, but also structural models of every ORF in human, fly, worm, plant and budding yeast (see Methods). HHpred searches include information not only on amino acid frequencies, but also on the position-specific probability for opening and closing gaps (20). HHpred seeded with human Blos1 identified YLR408Cp as the sole yeast homologue (E-value 0.005, Table S2), and in the reverse search, HHpred seeded with YLR408Cp identified Blos1 in human, fly, worm and plant (E-values 0.0001 – 0.01, Table S2). Thus, a search that includes structural information supports the psi-BLAST results that YLR408Cp is a Blos1 homologue.
We next extended sensitivity using HHsenser, which uses an alignment originating from HHpred to find distant homologues with high sensitivity and virtual absence of false positives (22). HHsenser combines the iterative approach of psi-BLAST with an “intermediate profile search”, whereby information obtained from a fixed number of iterations are then used to seed new (“intermediate”) searches (22). In addition, HHsenser compares profiles with profiles (not with sequences), which improves sensitivity (23). Submitting the HHpred alignment for Blos1 to HHsenser increased the significance of the alignment to YLR408Cp ten-fold (E-value 0.0005, Table S2). For the reverse search, HHsenser seeded with YLR408Cp produced highly significant alignments with Blos1 in higher eukaryotes (E-values < 10−26, Table S2). Variation in the significance of alignments depending on the initial seed is a known feature of HHsenser (22). These highly significant alignments strongly suggest that YLR408C can be assigned to be the Blos1 homologue in S. cerevisiae, so we provisionally suggest the gene name BLS1, for Blos1-homologue, although this needs to be confirmed by functional testing for protein sorting functions.
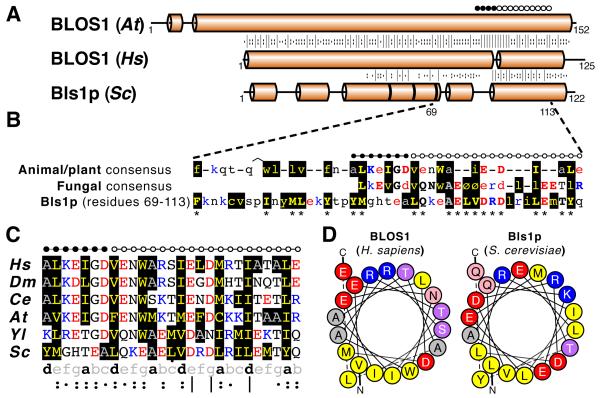
What are the conserved features that underlie the alignment of Blos1 with Bls1p? Blos1 homologues are predicted to be helical, as is Bls1p. Fig 1A shows that while human Blos1 shows great sequence homology with its plant homologue, there is only marginal sequence conservation with Bls1p. For all BLOC-1 subunits, short linear motifs have been identified from multiple sequence alignments, which can help identify divergent homologues (19). In Blos1 a conserved motif was identified at residues 87-93: “ALKEIGD” (Fig. 1B) (19), but Bls1p does not contain this, and its conservation with Blos1 is maximal in a region of 20 residues between the motif and the extreme carboxy-terminus (Fig. 1A). To analyze this conservation in Bls1p, previously known Blos1 homologues were divided into two groups: animal/plant and fungal, with consensus sequences being constructed for each group (Fig. 1B). The animal/plant Blos1 consensus shows maximal conservation at the motif and ~20 aa on either side (44 aa in total, Fig. 1B, top row). For Blos1 in fungi, conservation is confined to the motif and ~20aa downstream (26 residues in total, Fig. 1B, middle row). As expected from our psi-BLAST results, the sequence of Bls1p is closer to the fungal group than to the animal/plant group, and 13 out of 20 residues between 94-113 of Bls1p are shared with the fungal consensus (Fig. 1B, bottom row).
Fig. 1. Alignment of Blos1 homologues with Bls1p.
A. Secondary structural prediction for Blos1 homologues. Blos1 in H. sapiens (Hs) was aligned by HHpred with Blos1 in Arabidopsis thaliana (At) and Bls1p in S. cerevisiae (Sc). Cylinders represent alpha helices (no beta sheet predicted). Alignments are from HHsenser seeded with the human sequence, with asterisks indicating gaps of single residues, and conservation indicated by | : and · (excellent, good & moderate respectively). Black dots above indicated the motif at residues 87–93 of human Blos1 identified previously (19). White dots indicate residues 94–113 of Bls1p/YLR408Cp, the region that is most highly shared by Bls1p and fungal sequences (see B).
B. Alignment of the carboxy-terminal half of Bls1p with consensus sequences from known Blos1 homologues. The top two rows are consensus sequences created using MUSCLE (see Methods) from 62 previously identified Blos1 sequences, 35 from animals and plants, and 27 from fungi (see Table S3). All positions in these consensus sequences were scored as weakly, moderately or highly conserved (see Methods). The regions shown are those that are rich in highly conserved residues (indicated in upper case and bold). For animal/plant Blos1 (top row) the region is equivalent to residues 70–113 of human Blos1 (44 residues). For fungal Blos1 (middle row) the region is equivalent to residues 88-113 (26 residues). Conserved hydrophobicity is indicated by Ø. Residue colours are: basic – blue, acidic – red, hydrophobic – yellow on black, alanine – grey on black. The bottom row is the homologous region from Bls1p (residues 69–113). Here residues that are shared with either of the consensus sequences above are indicated by bold capitals and with asterisks. White dots above the sequence indicate the region of maximum identity between Bls1p (residues 94–113) and the fungal consensus. As in A, the motif described by Cheli et al. (19) is indicated by black dots.
C. Alignment of the most conserved regions of six key Blos1 homologues. Alignment of Blos1 sequences from human (Hs), fly (Dm), worm (Ce), plant (At), oleaginous yeast (Yl) and budding yeast (Sc) covering the conserved 27 residues indicated by black and white dots as in (B). Residue colours are as in (A). Heptad repeats with hydrophobic residues at positions “a” and “d” are shown below, together with the degree of conservation at each position as calculated by MUSCLE: excellent, good or moderate (indicated by | : and ·).
D. Helical wheel projections of 20 residues from Blos1 and Bls1p. Helical wheel projections for carboxy-terminal helices of human Blos1 and Bls1p (residues 94–113 for both). Circle colours: basic – blue, acidic – red, hydrophobic – yellow, alanine – grey, serine/threonine – purple, hydrophilic – pink.
In addition to specific amino acids, key properties of the helix are conserved from human to yeast (Fig. 1C and D). Viewed as a helical wheel, both human and yeast sequences form amphipathic helices with negative charges flanking a hydrophobic face, and with positive charges and other polar residues on the opposite face (Fig. 1D). This amphipathicity correlates with conservation of hydrophobic residues at positions “a” and “d” of a heptad repeating pattern (Fig. 1C). These regions are typically not predicted to form coiled coils (8), likely because most Blos1 homologues have only one leucine in this region (24). By comparison, fungal Blos1 homologues identified previously, as well as Bls1p, have 3-4 leucines here and so are predicted to form coiled-coils. The significance of this predicted difference between Blos1 higher eukaryotes is not known. Thus, identification of Bls1p as a Blos1 homologue is based on overall predicted structure, and sequence conservation with fungal Blos1 homologues that does not include the motif identified previously.
A possible snapin homologue in S. cerevisiae
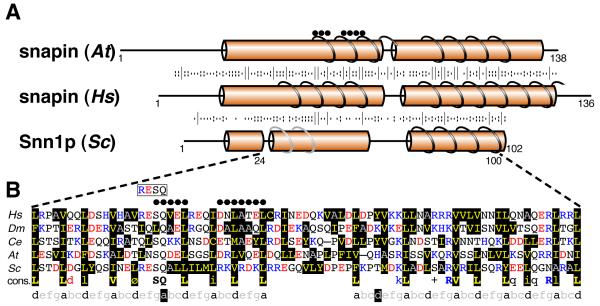
Continuing the search for different BLOC-1 subunits, we considered snapin (136 aa), where psi-BLAST has found homologues in metazoa, plants and protists, but not fungi (19). HHpred seeded with snapin weakly identified the yeast ORF of unknown function YNL086Wp (102 aa, E-value 0.3, Table S2), and HHsenser improved the alignment (E-value 0.003, Table S2). In reversed searches, psi-BLAST with YNL086Wp only identified homologues in closely related yeast (Table S1). HHpred with YNL086Wp showed weak homology to snapin from human, fly and worm (E-values 0.6 – 2, Table S2), and these alignments were improved by HHsenser (E-values 0.003 – 0.09, Table S2). Even though the snapin/YNL086Wp alignment is not strong, our findings suggest that YNL086Wp could be the snapin homologue in S. cerevisiae, and we propose that YNL086W be named SNN1 (from snapin), again pending confirmation by functional testing.
Snapin and its known homologues have two predicted helical regions, both with predicted coiled coils, and Snn1p is highly similar, aligning without gaps across a region of 77 residues (Fig. 2A and B). Two sequence motifs were identified previously in the amino-terminal helix of snapin: “SQxEL” and “DxLAxEL” at residues 50-54 and 59-66 respectively (Fig 2A and B) (19), the first of which is well conserved in Snn1p (Fig 2B). Other homology between snapin and Snn1p is distributed along their whole length (Fig. 2A). Within the predicted coiled-coils, key leucines in heptad repeats are conserved (Fig. 2B). Snapin and Snn1p also share the motif “RESQ” (Arg-Glu-Ser-Gln, Fig. 2B), the serine of which is phosphorylated by PKA (25). The finding that Snn1p shares functionally important sequence elements with snapin supports the notion that it may be a snapin homologue.
Fig. 2. Alignment of snapin homologues with Snn1p.
A. Secondary structural prediction for Snapin homologues. Human snapin was aligned with the plant homologue and with Snn1p, as in Fig. 1A. Motifs identified by Cheli et al. are indicated by black dots. Predicted coiled-coils are shown, with those near the amino-terminus of Snn1p in grey to indicate their prediction strength is just below the threshold of 0.5 (0.43).
B. Alignment of the central region of 77 amino acids from five key snapin homologues. Sequences from the central region (as indicated by the dotted lines) of snapin homologues from human (Hs: 33-109), fly (Dm: 34-110), worm (Ce: 21-96), plant (At: 45-120), and budding yeast (Sc: 24-100) were aligned, residue colours as in Fig 1B. Below is a consensus sequence made from 38 snapin homologues (see Table S3), with conserved hydrophobicity indicated by “Ø”, conserved K or R indicated by “+”, and the most conserved positions shown in capitals and bold. The bottom line shows heptad repeats with hydrophobic residues at positions “a” and “d”. In each of the two helices, one of the key “a” or “d” positions is not hydrophobic (shown as white “a” or “d” on black). Leucine is common at positions “a” and “d” (56%), explaining why these helices are predicted to form coiled-coils (24). Boxed “RESQ” above indicates the PKA phosphorylation site found in both human snapin and Snn1p. Black dots above are previously identified motifs, as in A.
A new family of cno-like proteins with a S. cerevisiae homologue
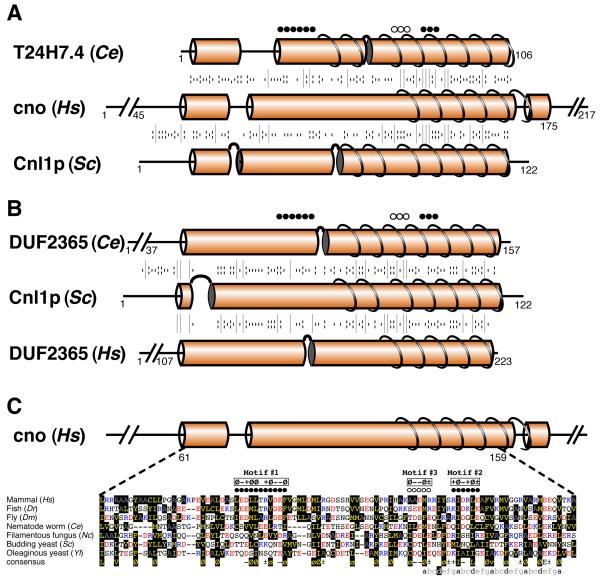
Psi-BLAST searches with cno (217 aa) have found homologues in mammals, fish, flies and a protist (slime mold), but not in nematode worms, plants or any fungi (19). We hypothesized that the species lacking cno may contain homologues that have diverged beyond the ability of psi-BLAST to recognize them, but which are detectable when structural information is included in alignment searches. HHpred seeded with the protist cno (the most divergent) identified human and fly homologues with E-values < 10−50. The next hits were distant matches to two proteins of unknown function (Fig. 3A): T24H7.4 in the nematode C. elegans (106 aa, E-value 0.2) and YDR357Cp in S. cerevisiae (122 aa, E-value 1), and similar results were obtained seeding HHpred with human cno (Table S2). Unlike for Blos1/Bls1p and snapin/Snn1p, cno alignments did not improve with HHsenser. However, searches seeded with these two new sequences did yield interesting information.
Fig. 3. Alignment of cno and newly described homologues.
A and B. Secondary structural prediction and alignment of cno and sequences identified by HHpred. Alignments of (A) Cno from human with T24H7.4 from C. elegans (Ce) and Cnl1p in S. cerevisiae (Sc); and (B) Cnl1p with DUF2365 proteins in C. elegans and human. Alignments shown as in Fig. 1A, but requiring gaps as indicated. Predicted helices and coiled-coils are indicated as in Fig. 2A. Black dots above indicate motifs identified by Cheli et al. (19) in cno. White dots above indicate a motif we identify to be shared between cno and Cnl1 sequences - see Fig. S1 and (C).
C. Alignment of the central region from cno homologues and cno-like homologues in fungi. The central region of cno in human (Hs: 61-159) was aligned to the homologous region from cno in zebrafish (Dr: 84-180), cno in fly (Dm: 5-102), T24H7.4 in worm (Ce: 14-103), and the cno-like proteins in the filamentous fungus Neurospora (Nc: 70-161), budding yeast (Sc: 21-114), and the oleaginous yeast Yarrowia (Yl: 21-113); residue colours as in Fig 1B. Dots above represent motifs (see text and Fig. S1). Below is a consensus sequence made from these seven sequences. Here and in the motifs above the alignment, conserved residues are: “Ø” = hydrophobic, “+” = K/R, “−” = D/E, “±” = K/R/D/E. Lower case “ø” indicates where hydrophobicity is weakly conserved. The bottom line shows heptad repeats underlying predicted coiled-coils at the carboxy-terminus, with hydrophobic residues at positions “a” and “d” (except for one “d” position shown as white “d” on black). Leucine is common at positions “a” and “d” (31%), explaining why these helices are predicted to form coiled-coils (24).
For T24H7.4, psi-BLAST only found a homologue in the closely related nematode C. briggsae. HHpred produced weak hits to YDR357Cp, fly cno and human cno (E-values 0.6,1 and 2 respectively, Table S2). While T24H7.4 is shorter than most cno homologues, cno in some insects is almost as short (e.g. beetle 125 aa). Therefore, we considered it possible that T24H7.4 is the missing nematode cno homologue.
Looking at YDR357Cp, psi-BLAST found a family of homologous sequences in fungi (one in each of 51 species, Table S1), but not outside fungi. HHpred with YDR357Cp (Fig. 3B) revealed weak homology both to a nematode ORF named systematically DUF2365, and to its relatives in human and fly (E-values 0.01, 0.2 and 0.4 respectively, Table S2). All that is known about the DUF2365 family (also systematically named c17orf59 from the location of the human gene) is that divergent eukaryotes have a single protein, typically of 125-325 residues, with a domain of unknown function (hence DUF) of approximately 95 residues identified by automated BLAST searches. A reciprocal HHpred search with nematode DUF2365 identified human, fly and plant DUF2365 proteins (E-value <10−10); the next hit was YDR357Cp (E-value 0.006, Table S2), which strongly indicates that YDR357Cp is the yeast member of the DUF2365 family. Since YDR357C is cno-like, we propose the name CNL1. While this role has not yet been tested, there is some functional evidence that this gene functions in vacuolar protein sorting (see Conclusion).
The similarities that are shared by cno, T24H7.4, Cnl1p, and DUF2365 proteins are distributed throughout the conserved domain of approximately 95 amino acids, which is predicted to be largely helical (Fig. 3A and B). Cno is predicted to form a coiled-coil at the carboxy-terminus of its region of shared homology (Fig. 3A) (26), and the same is true for DUF2365 proteins (Fig. 3B). At the level of primary sequence, two motifs were identified in cno (Fig. 3C) (19), but only one of these is conserved in T24H7.4 and Cnl1p (Fig. 3A and C). To visualize how elements of primary sequence are shared between cno and fungal cno-like proteins, we made a large alignment of cno homologues and fungal cno-like sequences (Fig. S1A). This showed that there is little conservation of the first motif at residues 89-99 (“Ø−+ØØx+Ø−−Ø”, where Ø/− /+ are hydrophobic, acidic and basic residues respectively). In contrast, the second motif at residues 134-139 (“+Ø−+ر”, where ± indicates charge of either type) is well conserved in fungal cno-like sequences. The alignment also revealed a third short motif (“Ø−−ر”, residues 125-129) in most fungal cno-like proteins (and all DUF2365, not shown) that is conserved to some extent in cno (Figs S1A and 3C). A tree of cno and fungal cno-like sequences shows that while they mainly divide into their two groups, there are exceptions: the cno homologue we have identified in worms (T24H7.4) and the cno homologue the choanoflagellate Monosiga brevicollis are intermediate between the two groups (Fig. S1B). These results support the idea that a common ancestor has diverged into the two groups of cno and cno-like proteins, the link being undetectable by conventional sequence alignment.
A yeast BLOC-1 complex that contains a KxDL protein
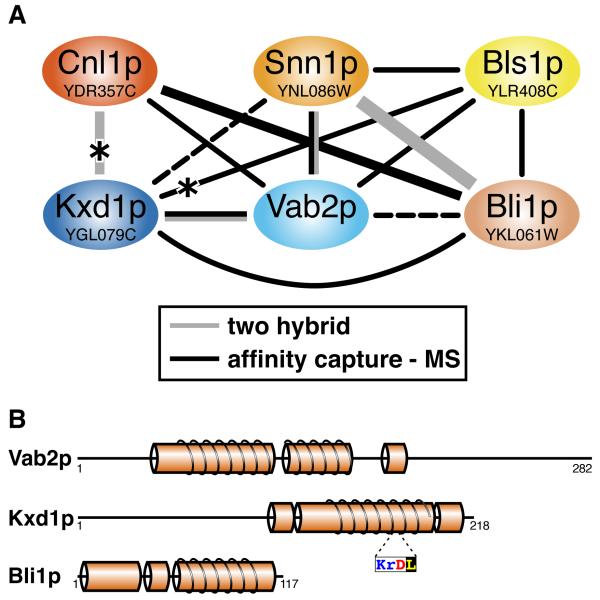
All BLOC-1 subunits are relatively small (between 125 and 351 residues), and are predicted to contain alpha helices (6), which raises the possibility that the yeast ORFs found by HHpred are false positives, identified solely because their simple helical structure is so common. However, additional evidence that Bls1p, Snn1p and Cnl1p are BLOC-1 components comes from published data on their physical interactions. All three have been found in a single multi-subunit complex among 546 such complexes identified in a genome-wide study of protein-protein interactions in S. cerevisiae (27). Among the 2,400 tagged proteins purified in this study, one was Vab2p, a 31 kDa cytoplasmic protein of unknown function originally identified as a binding partner for Vac8p (28,29), which is a peripheral vacuolar protein that co-ordinates multiple vacuolar functions (29,30). Affinity-purified Vab2p co-precipitated five other proteins: Bls1p, Cnl1p, Snn1p, and two proteins of unknown function: YGL079Wp and YKL061Wp (Table S4) (27,31). Precipitates of Bls1p, Cnl1p and YKL061Wp revealed five more interactions among Bls1p, Cnl1p, Snn1p, YGL079Wp and YKL061Wp (Table S4). Still more physical interactions within this group of proteins have been mapped: one affinity purification identified by its combination with similar levels of expression (32), and two interactions identified by two-hybrid studies (33-35) (Table S4). In total, 13 of the possible 15 pair-wise interactions have been detected between the six proteins Vab2p, Bls1p, Cnl1p, Snn1p, YGL079Wp and YKL061Wp (Fig. 4A). The high density of connections suggests that they form a single complex (27). Current data do not indicate that Vab2p or any other component protein is the “node” for the complex, which will have to be determined by examining pairwise interactions in strains missing other components of the complex.
Fig 4. A complex of six yeast proteins includes all three BLOC-1 subunits.
A. Known direct physical interactions between six proteins identified in Vab2p complexes. Black and grey lines represent interactions defined by affinity purification/mass spectrometry and yeast two hybrid, respectively. The Vab2p-Bli1p and Kxd1p-Snn1p interactions are indicated by dashed lines as they scored below the threshold defining core interactions (Table S4). Interactions obtained in two or three studies are indicated by correspondingly thicker lines. Of the possible 15 pair-wise interactions, 11 were found by direct interaction (27,32), and two by two hybrid. Asterisks indicate two interactions of KxDL proteins that are recapitulated in higher eukaryotes (see Conclusion).
B. Predicted secondary structure of three yeast proteins interacting with BLOC-1. The predicted helical nature of Vab2p, Bli1p and Kxd1p is shown, together with regions predicted to form coiled coils, as in Fig. 2A. The conserved KxDL motif at residues 175-8 of Kxd1p is indicated.
The three extra components that interact with BLOC-1 in yeast are similar to known BLOC-1 subunits in size and in predicted content of alpha-helices (with some coiled coil) but no beta-sheet (Fig. 4B). Only Vab2p has been studied previously, but apart from its interaction with Vac8p, no function is known (28,29). We next scanned the database for homologues of Vab2p, YGL079Wp and YKL061Wp using psi-BLAST (Table S1); no extra alignments arose from HHpred. Homologues for Vab2p were found in fungi only. For YKL061Wp, homologues were restricted to close relatives of S. cerevisiae (Table S1). Since the majority of physical interactions documented for YKL061W are with BLOC-1 subunits (27,31,33-35), we propose the name BLI1, for BLOC-1 interactor.
Unlike Vab2p and Bli1p, YGL079Wp is in a conserved protein family, with a single identifiable member in most eukaryotes, from mammals to plants (Table S1). However, none of the homologues have been characterized. Automated database curation has designated them as IPR019371, Pfam10241, or DUF-KxDL. They are all short proteins, defined by a conserved helical region of 90 residues that includes a “KxDL” (Lys-x-Asp-Leu) motif near the carboxy-terminus (Fig. 4B). Since YGL079W is the yeast KxDL homologue, we propose the name KXD1.
The multiple interactions between all of Bls1p, Snn1p, Cnl1p, Vab2p, Bli1p and Kxd1p indicate that the latter three might also be subunits of BLOC-1 in yeast. In support of this, high-throughput analysis of localization ascribes all six ORFs to a punctate distribution, in some cases reported to co-localize with endosomes (36).
Conclusions
Including structural information in homology searches identified potential yeast homologues for Blos1 and snapin, and a cno-like protein. The same approach found no homologues in yeast for the five remaining BLOC-1 subunits (Blos2, Blos3, muted, dysbindin, and pallidin), but did identify a new homologue of muted in C. elegans (C34D4.13, not shown). This indicates that whatever the function may be of the yeast complex we have highlighted, it may be only distantly related to BLOC-1 in mammals. Nevertheless, there is some evidence that the yeast proteins are performing a function that parallels BLOC-1 in mammals, since a genome-wide screen that identified ~200 genes functioning in vacuolar protein sorting included both CNL1 and KXD1, as well as all four sub-units of AP-3, but none of AP-1,-2 and clathrin (37). The inclusion of Kxd1p in complexes containing yeast BLOC-1 homologues is particularly significant because, although KxDL proteins are completely uncharacterized, in higher eukaryotes they recapitulate some if the interactions with BLOC-1 subunits (Fig. 4): in D. melanogaster and C. elegans, the KxDL protein interacts with Blos1 (38); in C. elegans, the KxDL protein interacts both with the newly discovered cno (T24H7.4), and with the DUF2365 homologue (39). Conservation of interactions in yeast, worms and flies suggests that KxDL proteins may be key interactors with BLOC-1 in mammals.
Although it has not been shown that any of the interactions of the yeast complex occur simultaneously, the density of interactions among the six subunits is predicted to generate a multimeric complex (27). Would this be similar to BLOC-1 in higher eukaryotes, which, by a combination of size exclusion chromatography and velocity sedimentation, has been estimated to be an asymmetric complex of 200 kDa (40)? For most of the eight subunits, a large proportion co-migrates and co-precipitates in these large complexes (8,26,41). However purification of the complex has not yet been achieved, so its complete composition is not known. Our results predict that BLOC-1 might include KxDL or DUF2365 proteins. Knowing conserved components and interactions in yeast, it will now be possible to study ancestral functions of BLOC-1 in a genetically tractable model organism.
Methods
Psi-BLAST: The psi-BLAST tool examining the non-redundant protein database at NCBI. E-values returned here (and by other tools, below) give the average number of false positives expected to randomly produce an alignment as good as this. An E-value of 10 means that 10 wrong hits are expected to occur with the extent of alignment observed, while numbers very much smaller than one indicate likely significance. We used a threshold E-value of 0.005 for inclusion of alignments from one iteration of psi-BLAST in the next iteration. Also searches were limited to eukaryotes, and sequences masked for the lookup table only. Psi-BLAST was iterated until new sequences added to the list were larger proteins including other domains of known function. Sequences from transcripts with frame shift errors were excluded.
HHpred: The HHpred tool (20), which is part of the MPI bioinformatics toolkit (http://toolkit.tuebingen.mpg.de), was used to search against the protein databank of structures clustered at 70% sequence identity (PDB70), and to examine the genomes of five phylogenetically diverse organisms: H. sapiens, D. melanogaster, C. elegans, A. thaliana, and S. cerevisiae, with a total of ~135,000 records in total. Alignments were carried out with default settings in the local mode. E-values returned by HHpred and reported here are based on sequence alone, excluding secondary structural similarity, so hits can be significant even when the E-value is ~ 1 (20). Some HHpred alignments were submitted to HHsenser, using default settings (22). Coiled-coils were analyzed at www.ch.embnet.org/ using default settings; positions where any of the three windows scores ≥ 0.5 were considered positive. Structural predictions were made by Psi-Pred 3.0. Consensus sequences for Blos1 and snapin were made by MUltiple Sequence Comparison by Log-Expectation (MUSCLE) at EBI. For each position in the consensus, conservation was scored by comparison to maximum consensus strength: <50% weak, 50-75% moderate, and ≥75% high. The alignment for Fig. S1 was made by Kalign at EBI, and coloured with the Clustalx scheme.
Supplementary Material
Acknowledgements
Funding: Wellcome Trust (grant #082119).
Abbreviations
- AP-3
adaptor protein complex-3
- BLOC
biogenesis of lysosome-related organelle complex
- cno
cappuccino
- DUF
domain of unknown function
- HPS
Hermansky-Pudlak syndrome
- LRO
lysosome-related organelle
- ORF
open reading frame
References
- 1.Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: a coat of many colours. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- 2.Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theos AC, Tenza D, Martina JA, Hurbain I, Peden AA, Sviderskaya EV, Stewart A, Robinson MS, Bennett DC, Cutler DF, Bonifacino JS, Marks MS, Raposo G. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacino JS. Insights into the biogenesis of lysosome-related organelles from the study of the Hermansky-Pudlak syndrome. Ann N Y Acad Sci. 2004;1038:103–114. doi: 10.1196/annals.1315.018. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell’Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Falcon-Perez JM, Dell’Angelica EC. The pallidin (Pldn) gene and the role of SNARE proteins in melanosome biogenesis. Pigment Cell Res. 2002;15:82–86. doi: 10.1034/j.1600-0749.2002.1r082.x. [DOI] [PubMed] [Google Scholar]
- 8.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 9.Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell’Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, Wainer BH, Falcon-Perez JM, Dell’Angelica EC, Peden AA, Werner E, Faundez V. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, Dell’Angelica EC, Raposo G, Marks MS. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheli VT, Daniels RW, Godoy R, Hoyle DJ, Kandachar V, Starcevic M, Martinez-Agosto JA, Poole S, DiAntonio A, Lloyd VK, Chang HC, Krantz DE, Dell’Angelica EC. Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum Mol Genet. 2010;19:861–878. doi: 10.1093/hmg/ddp555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y, Li X, Chen Q, He X, Yang Q, Zhang A, Yu X, Chen H, Liu N, Xie Q, Yang W, Zuo J, Palme K, Li W. BLOS1, a putative BLOC-1 subunit, interacts with SNX1 and modulates root growth in Arabidopsis. J Cell Sci. 2010;123:3727–3733. doi: 10.1242/jcs.069732. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Fernandez IA, Dell’Angelica EC. A data-mining approach to rank candidate protein-binding partners-The case of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Inherit Metab Dis. 2009;32:190–203. doi: 10.1007/s10545-008-1014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, Malvar JS, de Vellis J, Sabatti C, Dell’Angelica EC. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol Psychiatry. 2010;15:204–215. doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dell’Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. Embo J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shotelersuk V, Gahl WA. Hermansky-Pudlak syndrome: models for intracellular vesicle formation. Mol Genet Metab. 1998;65:85–96. doi: 10.1006/mgme.1998.2729. [DOI] [PubMed] [Google Scholar]
- 19.Cheli VT, Dell’Angelica EC. Early origin of genes encoding subunits of biogenesis of lysosome-related organelles complex-1, -2 and -3. Traffic. 2010;11:579–586. doi: 10.1111/j.1600-0854.2010.01044.x. [DOI] [PubMed] [Google Scholar]
- 20.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battey JN, Kopp J, Bordoli L, Read RJ, Clarke ND, Schwede T. Automated server predictions in CASP7. Proteins. 2007;69(Suppl 8):68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 22.Soding J, Remmert M, Biegert A, Lupas AN. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006;34:W374–378. doi: 10.1093/nar/gkl195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 25.Chheda MG, Ashery U, Thakur P, Rettig J, Sheng ZH. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat Cell Biol. 2001;3:331–338. doi: 10.1038/35070000. [DOI] [PubMed] [Google Scholar]
- 26.Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1) Blood. 2003;101:4402–4407. doi: 10.1182/blood-2003-01-0020. [DOI] [PubMed] [Google Scholar]
- 27.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 28.Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang F, Peng Y, Nau JJ, Kauffman EJ, Weisman LS. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 2006;7:1368–1377. doi: 10.1111/j.1600-0854.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Chin CH, Chen SH, Ho CW, Ko MT, Lin CY. A hub-attachment based method to detect functional modules from confidence-scored protein interactions and expression profiles. BMC Bioinformatics. 2010;11(Suppl 1):S25. doi: 10.1186/1471-2105-11-S1-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 37.Norambuena L, Zouhar J, Hicks GR, Raikhel NV. Identification of cellular pathways affected by Sortin2, a synthetic compound that affects protein targeting to the vacuole in Saccharomyces cerevisiae. BMC Chem Biol. 2008;8:1. doi: 10.1186/1472-6769-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 39.Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falcon-Perez JM, Starcevic M, Gautam R, Dell’Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem. 2002;277:28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- 41.Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O’Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell’Angelica EC, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.