Abstract
Detection of recurrent somatic rearrangements routinely allows monitoring of residual disease burden in leukemias, but is not used for most solid tumors. However, next-generation sequencing now allows rapid identification of patient-specific rearrangements in solid tumors. We mapped genomic rearrangements in three cancers and showed that PCR assays for rearrangements could detect a single copy of the tumor genome in plasma without false positives. Disease status, drug responsiveness, and incipient relapse could be serially assessed. In future, this strategy could be readily established in diagnostic laboratories, with major impact on monitoring of disease status and personalizing treatment of solid tumors.
INTRODUCTION
For the clinical oncologist, it has become increasingly urgent to have access to accurate and sensitive methods for quantifying response to cancer therapy, assessing residual disease burden, and predicting impending relapse. In hematological malignancies, highly sensitive assays for monitoring minimal residual disease (MRD) have become standard practice in several disorders, allowing individualized therapeutic choices. For example, serial quantification of the BCR-ABL1 fusion gene in chronic myeloid leukemia can predict relapse on targeted therapy, enabling early prophylactic intervention (Branford, 2007); residual levels of the IGH rearrangement at milestone time-points in acute lymphoblastic leukemia chemotherapy can guide choice of subsequent treatment (Flohr et al., 2008); and monitoring the monoclonal protein in multiple myeloma allows duration of induction therapy to be individualized to achieve maximum benefit without over-treatment (Barosi, et al., 2004).
The routine clinical application and high sensitivity and specificity of these assays depend on the recurrent, somatically acquired genomic rearrangements found in leukemias. In these cancers, breakage and subsequent fusion of two genes to form a chimeric cancer gene commonly occurs, with the particular genes involved driving the type of leukemia that develops. At diagnosis, assays for specific fusion genes or rearrangements can be performed on the leukemic cells and, when positive, can be used during treatment to track small numbers of residual circulating cells carrying the rearrangement. This is achieved by the relatively straightforward process of PCR across the rearrangement junction, a procedure within the technical repertoire of many diagnostic laboratories. The high sensitivity and specificity is conferred by the design of the PCR assay, which only successfully connects the two abnormally joined segments of genome present in cancer cells, and fails to amplify DNA or RNA from normal cells.
This powerful approach has, in general, not been implemented in the routine clinical management of solid tumors (such as breast, ovarian, colorectal, and lung cancers) because recurrent somatic rearrangements have not been identified in most of these malignancies. Although a small fraction of solid tumors do carry recurrent genomic rearrangements, such as ETS fusion genes in prostate cancer (Tomlins et al., 2005, 2007) and Ewing’s sarcoma (Futreal et al., 2004; Vermeulen et al., 2006) and the EML4-ALK fusion in nonsmall cell lung cancer (Soda et al., 2007), these represent only a small fraction of all tumors. Nevertheless, the genomes of most solid tumors do contain rearrangements that are unique to each case. Until recently, mapping rearrangements in individual cancers to base-pair resolution has been impracticable in a clinical setting. However, we and others have recently shown that next-generation sequencing technologies allow genome-wide catalogues of somatic rearrangements to be generated cost-effectively in a clinically relevant time-frame (Campbell, et al., 2008; Mardis, et al., 2009; Stephens, et al., 2009; Stratton, et al., 2009; Pleasance, et al., 2010a,b). Moreover, it is well recognized that cells from many classes of solid tumor release naked DNA fragments into plasma (Nawroz, et al., 1996; Diehl, et al., 2005,2008; Yung, et al., 2009). In this report, we provide proof-of-principle that genomic rearrangements mapped with next-generation sequencing in individual cancers enable the development of straightforward, sensitive and specific assays to quantify disease burden in serial blood samples. This fundamental concept is potentially applicable across most types of solid tumor and in diverse therapeutic settings.
MATERIALS AND METHODS
Case Studies
We studied two patients with breast cancer and one with osteosarcoma. Patient 1 presented in 2002 at Age 35 with node-positive, oestrogen receptor-positive, invasive ductal breast cancer. Despite surgery, adjuvant radiotherapy, chemotherapy (doxorubicin and cyclophosphamide), and anti-oestrogen treatment (tamoxifen then goserelin), she relapsed with bony metastases in 2008. Patient 2 was a 46-year old who presented with a node-negative, oestrogen receptor-positive, invasive ductal breast carcinoma. She relapsed 17 months later with bony metastases despite surgery, radiotherapy and tamoxifen. Patient 3 is a 56-year-old woman who presented with a poorly differentiated, multifocal sarcoma involving the spine, pelvis, ribs, and femurs. She underwent extensive radiotherapy to T3-8 vertebrae, femurs, right tibia, and pubic bone (Month 2), followed by nine cycles of ifosfamide and doxorubicin (Months 3–10). Bortezomib was then administered (Months 10–13), but after the fourth cycle, the patient experienced rapid onset of thoracic back pain. CT scan showed a small, soft-tissue deposit around T9-10. After further radiotherapy (Month 14), weekly cycles of paclitaxel were given (Months 14–16). Despite this, the patient relapsed with widespread soft tissue metastases soon after stopping therapy (Month 17).
Methods
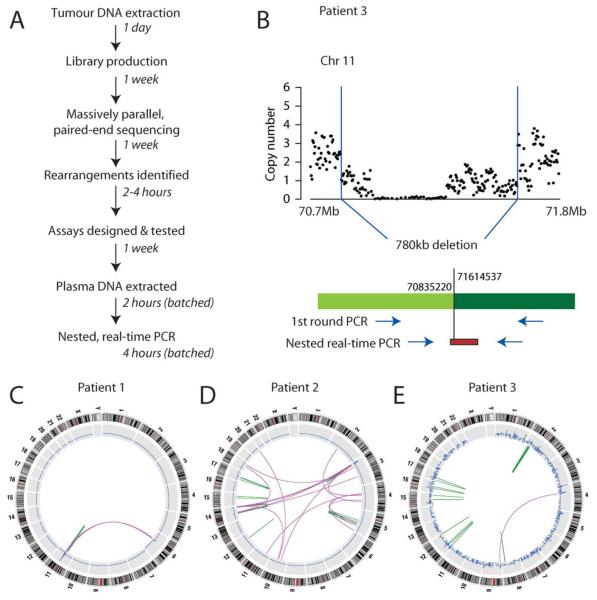
Samples of genomic DNA were extracted from fresh-frozen primary tumor material and studied by massively parallel, paired-end sequencing (Campbell, et al., 2008) (Fig. 1A). In brief, ~100 million fragments of DNA 400–500 bp in size were sequenced with paired 37 bp reads from either end of each fragment. Putative genomic rearrangements were identified as clusters of discordantly mapping read-pairs, namely sequence pairs which did not map back to the reference genome within ~400–500 bp of each other in the correct orientation. Rearrangements were then confirmed as real and somatic by PCR and sequencing across the rearrangement junction in tumor and germline DNA from each patient. This allowed the annotation of tumor-specific rearrangements to base-pair resolution. Nested, real-time PCR assays were then designed for several rearrangements per patient, as described (Morley et al., 2009). Two criteria were used to prioritize rearrangements for assay design. First, variants causing a definite copy number change were preferred (Fig. 1B), since these must be found in the overwhelming majority of cells and are therefore likely to be present in relapsing cells. Second, only breakpoints occurring in unique DNA sequence were used, to maximize specificity of the assay. The maximum product size in these assays was kept <200 bp, since circulating tumor DNA is highly fragmented (Diehl, et al., 2005, 2008).
Figure 1.
Protocol and rearrangement screens. (A) Outline of protocol and current time-frames for each step. (B) Example of a deletion mapped to base-pair resolution from the osteosarcoma sample (Patient 3). Using knowledge of the breakpoint, a nested PCR assay can be designed, with a fluorescent probe used for the second round real-time PCR reaction. (C) Genome-wide rearrangement screen for Patient 1, showing eight somatically acquired genomic rearrangements, including interchromosomal (purple arcs) and intrachromosomal (green arcs) variants. Copy number is shown in blue. (D) Genome-wide rearrangement screen for Patient 2. (E) Genome-wide rearrangement screen for Patient 3.
Free DNA was extracted from 2 ml plasma (Patients 1–2) or serum (Patient 3). For the two patients with breast cancer, these were taken at relapse with metastatic disease and for the patient with osteosarcoma, multiple samples during treatment were taken. Controls included tumor DNA (positive); normal DNA and water (negative); dilutions of tumor DNA in either normal DNA or water (for standard curves); and primers for nonrearranged regions of the genome (to quantify total plasma DNA). A detailed, step-by-step protocol for sequencing informatics, assay design and testing, plasma DNA extraction and analysis is provided in Supporting Information.
RESULTS
To provide proof-of-principle that tumor-specific genomic rearrangements can be detected in plasma, we first undertook massively parallel paired-end sequencing of genomic DNA from the primary tumors of two patients with metastatic breast cancer. From these data, we characterized 8 and 48 somatically acquired genomic rearrangements, respectively to base-pair resolution (Figs. 1B–1E, Supporting Information Table 1). Nested, real-time PCR assays were designed to amplify across three rearrangement junctions per patient with high sensitivity and specificity.
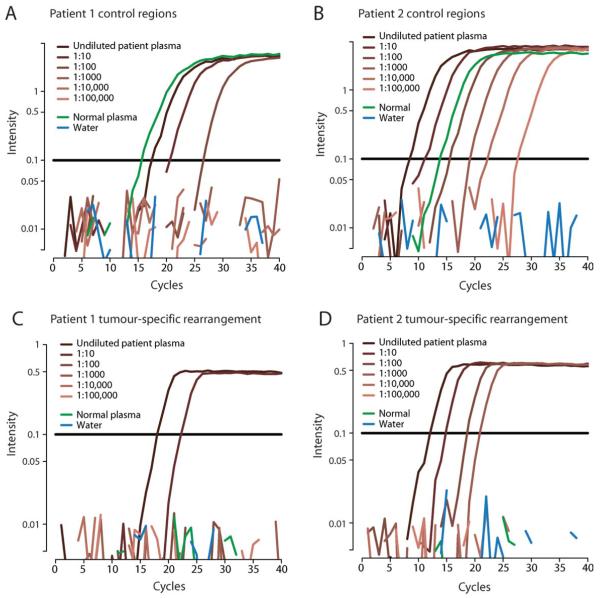
Free DNA was extracted from plasma samples taken at relapse with metastatic disease 17 months and 6 years, respectively after first presentation. Using control assays targeted against nonrearranged regions of the genome, we confirmed that amplifiable DNA was present in each sample even when diluted in water 100× (Patient 1, Fig. 2A) or 10,000× (Patient 2, Fig. 2B). The assays gave robust and reproducible quantification. We then proceeded to assay tumor-specific rearrangements in the two patients (Figs. 2C and 2D). Plasma DNA from a normal individual remained negative throughout the PCR for all of the rearrangements assessed, confirming the high specificity of the assay for the cancer genome. However, reactions containing free DNA from the patients’ plasma were strongly positive in both cases, confirming that tumor-specific genomic rearrangements found in the primary tumor can be detected in plasma at relapse with metastatic disease. Indeed, reactions remained positive even using a tenth of the template for Patient 1 and a thousandth for Patient 2, the latter being equivalent to detecting a signal in just 2 μl of plasma. Results from the other two rearrangements for each patient were very similar (data not shown). These data demonstrate that multiple rearrangements identified in the primary breast cancer were robustly detectable at relapse with distant metastatic disease years after initial surgery.
Figure 2.
Analysis of plasma DNA from two patients with breast cancer (Patients 1 and 2). (A,B) Results of a nested, real-time PCR from a control region of the genome performed on undiluted (dark brown) and serial 10-fold dilutions of DNA extracted from plasma of the two patients (lighter shades of brown). Results for plasma DNA from a normal individual are also shown (green). (C,D) Results of a nested, real-time PCR from a tumor-specific rearrangement performed on serial 10-fold dilutions of DNA extracted from plasma of the two patients (brown) and a normal control (green). The horizontal black lines represent the fluorescence threshold at which a reaction is deemed to become positive. The x axis denotes the number of cycles of PCR, with more strongly positive reactions crossing the threshold at an earlier cycle number.
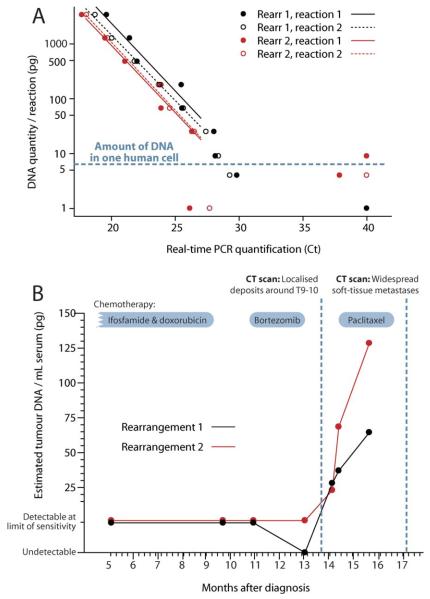
We next explored the utility of this approach to quantify tumor burden in a therapeutic, minimal disease setting. Somatically acquired rearrangements were identified in genomic DNA from the tumor of a patient with osteosarcoma (Fig. 1E), and assays designed as above. To generate a standard curve for quantification, tumor DNA was serially diluted in normal DNA such that each PCR reaction contained a defined amount of tumor DNA. Results from two replicates of two rearrangements demonstrate several favorable features of our protocol (Fig. 3A). First, the replicates show strong concordance with one another and with the other rearrangement for a given amount of tumor DNA, suggesting the assay has high reproducibility. Second, the standard curves are linear through a thousand-fold range of DNA levels, confirming the quantitative accuracy of the nested, real-time approach. Third, the assays remained consistently positive down to 25 pg of tumor DNA. At levels of 10 and 5 pg per reaction, we observed an on–off phenomenon, in that the assay was either positive or completely negative. Since a single human cell contains 6.75 pg DNA (Morton, 1991), reactions containing 5–10 pg DNA might have 0, 1, or ≥2 actual copies of the rearrangement (Stenman et al., 1999)—presumably the negative reactions we observed contained no amplifiable targets. These data imply that the nested, real-time reaction is sufficiently sensitive to detect a single copy of the cancer genome in many milliliters of plasma.
Figure 3.
Analysis of serial samples from a patient with osteosarcoma (patient 3). (A) Analysis of two rearrangements in duplicate reactions across a dilution series of tumor DNA into normal DNA. When the number of cycles to reach positivity (Ct) is ≤27, the absolute amount of tumor DNA can be estimated from the line of best fit. For reactions in which the number of cycles to reach positivity (Ct) > 27, disease can only be classified as detectable or undetectable. (B) Estimated amount of tumor DNA per ml of serum from seven samples collected at milestone time points in the patient’s clinical course.
We studied serum samples taken serially during the patient’s treatment for osteosarcoma, starting halfway through her first-line chemotherapy (Fig. 3B). Throughout first and second-line chemotherapy, residual disease was detectable by the assay, at the limits of sensitivity (1–2 tumor genomes/ml serum). In Month 14, we observed a distinct increase in circulating tumor DNA, which correlated with the clinical occurrence of thoracic back pain associated with localized disease progression. Over the next 2 months, circulating tumor DNA levels continued to increase, despite the patient showing clinical improvement on salvage chemotherapy. In Month 17, however, the patient relapsed with widespread soft-tissue metastases.
These data illustrate three key findings, each with significant implications for personalising drug therapy for cancer. First, at all stages during the treatment course at least one assay was positive. Since the half-life of plasma DNA is just 30 min (Lo et al., 1999) it seems that residual cancer cells were present throughout the patient’s therapy. Collected in real clinical time, such data could prospectively identify patients who are demonstrably not cured, leading to improved and individualized therapeutic decisions. Second, circulating tumor DNA levels actually rose rather than decreased during salvage treatment with a potentially toxic chemotherapeutic agent (paclitaxel). Although increased plasma DNA levels can be observed transiently with treatment-related tumor kill (Rago et al., 2007), the sustained nature of the increase observed here (6 weeks) would be more consistent with the conclusion that the cancer was progressing despite this treatment. This illustrates the potential for such an assay to identify failing therapies early, allowing quicker transfer to alternative therapeutic regimens. Finally, the results clearly demonstrate that rising levels of circulating tumor DNA can portend clinical progression. In this case, the increases were readily detectable 2–3 months before the widespread soft tissue deposits became clinically overt. In some clinical settings, such information might enable salvage therapy to commence while disease is in a less bulky state and before the development of complications associated with relapse.
DISCUSSION
A major goal of cancer medicine in the next decade is to progress from fixed, off-the-peg treatment regimens to bespoke therapy that is tailored to a patient’s tumor. Quantifying disease burden to monitor response to anti-cancer therapy has a direct intuitive appeal, with many potential applications for individualizing treatment choices. Unfortunately, for many solid tumors, such methods show poor sensitivity and specificity. Radiological imaging can only detect lesions >0.5 cm in size, already representing many millions of cancer cells. Serum markers, such as PSA for prostate cancer, can be helpful, but are not available for many tumor types and frequently lack specificity. Immunological detection of circulating tumor cells only identifies cells present in the blood and can result in false positive calls from nonmalignant cells expressing the marker of interest (Maheswaran, et al., 2008; Pantel, et al., 2008).
Tumor-specific genetic variants unequivocally identify cancer cells. For this reason, there has been considerable interest in using point mutations in circulating plasma DNA as a biomarker of disease burden (Nawroz, et al., 1996; Diehl, et al., 2005, 2008; Yung, et al., 2009). Early studies have shown that results correlate well with clinical status (Diehl, et al., 2008). However, sensitive and specific detection of a mutated base in a vast excess of normal DNA requires specialized techniques currently beyond the scope of many diagnostic laboratories (Diehl, et al., 2005, 2008; Yung, et al., 2009). The revolution in sequencing technology now makes it possible to identify all genetic variants of all classes in a given patient’s cancer. While next-generation sequencing is currently outside the scope of most diagnostic laboratories, it is anticipated that these technologies will become increasingly integrated into clinical practice. For monitoring disease burden, assays for genomic rearrangements have several practical advantages over point mutations in ease of implementation, accuracy of quantification, sensitivity and specificity and this is reflected in their routine implementation for many years in the management of hematological malignancies.
One interesting difference between our approach and minimal residual disease monitoring in leukemia is that most of the rearrangements for which we have developed patient-specific assays are probably passenger rearrangements, not causally implicated in the biology of the cancer. Provided that the relapsing cells also carry the rearrangement, this is of no consequence, but it would be possible for a relapsing clone to have lost the rearrangement and still be malignant. To minimize potential false negative results from this possibility, we recommend using a panel of assays against different rearrangements and choosing rearrangements in which copy number changes suggest the variant is present in nearly all of the tumor cells (see Supporting Information).
While this manuscript was in preparation, another group published similar paired-end rearrangement screens in six patients at diagnosis of colorectal cancer, with studies of plasma DNA in two patients (Leary et al., 2010). As we do, this article shows that tumor-specific rearrangements can be sensitively and specifically detected in plasma, with serial studies in one patient showing that levels correlated with decreasing disease burden during treatment. Our study adds to these findings by extending the proof-of-principle into two other tumor types, by showing that assays are positive at disease relapse as well as initial diagnosis and that impending relapse can potentially be identified. Other groups have studied differentially expressed transcripts, such as microRNAs, in the plasma of patients with cancer (Ng et al., 2009), but such assays are unlikely to have the sensitivity and specificity that can be achieved with genomic rearrangements.
To date, we have completed screens for somatically acquired genomic rearrangements in >100 solid tumor samples, including breast, pancreatic, ovarian, bone, and lung cancers. In all but one of these samples, we have successfully identified rearrangement breakpoints, and ~85% of samples have >10 rearrangements (Campbell et al., 2008; Mardis et al., 2009; Stephens et al., 2009; Stratton et al., 2009; Pleasance et al., 2010a,b). This underscores the widespread applicability of the approach described here for patients with solid tumors. Protocols have been developed that allow massively parallel sequencing to be performed on formalin-fixed, paraffin-embedded pathology blocks (Schweiger et al., 2009), which further broadens accessibility since most patients undergo biopsy or resection of their cancer. The primary paired-end screen for rearrangements requires only low coverage sequencing of the cancer cell genome, with subsequent validation of likely somatic rearrangements by PCR in cancer and normal DNAs. Currently, the screen takes a week for sequencing and a week for computer processing and can be performed for about US$4,000. It is likely that these timescales and costs will drop substantially over the next few years. Since genomic breakpoints are not generally recurrent across different patients [indeed, no recurrent fusion genes were identified in this screen or 24 other breast cancer samples (Stephens et al., 2009)], rearrangement screens must be performed separately for each patient. Once rearrangements have been ascertained in the primary cancer, monitoring of serial plasma samples would be within the purview of most molecular pathology laboratories, and could be achieved with a one-day turn-around.
Such a protocol could be adapted to numerous and diverse clinical settings. Examples include monitoring of tumor response in real time in patients on experimental, costly, or toxic therapies; identifying molecular relapse with the potential to initiate preemptive treatment; personalizing duration of therapy; using molecular response as a surrogate marker of cell kill in early phase clinical trials; and choosing the intensity of adjuvant therapy based on better risk stratification. The widespread implementation of analogous minimal residual disease assays in hematological malignancies testifies to both the clinical utility such measures provide and the ease and flexibility with which they can be integrated into disparate clinical care pathways.
ACKNOWLEDGMENTS
The authors thank Aarno Palotie and Leena Peltonen for the personal sacrifices they made to enhance the success of the project.
Supported by: Wellcome Trust, Grant number: 077012/Z/05/Z; Wellcome Trust Senior Clinical Fellowship, Grant number: 088340/Z/09/Z.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Barosi G, Boccadoro M, Cavo M, Corradini P, Marchetti M, Massaia M, Merlini G, Tosi P, Tura S. Management of multiple myeloma and related-disorders: Guidelines from the Italian Society of Hematology (SIE), Italian Society of Experimental Hematology (SIES), and Italian Group for Bone Marrow Transplantation (GITMO) Haematologica. 2004;89:717–741. [PubMed] [Google Scholar]
- Branford S. Chronic myeloid leukemia: Molecular monitoring in clinical practice. Am Soc Hematol Educ Program. 2007;1:376–383. doi: 10.1182/asheducation-2007.1.376. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohr T, Schrauder A, Cazzaniga G, Panzer-Grumayer R, van der Velden V, Fischer S, Stanulla M, Basso G, Niggli FK, Schafer BW, Sutton R, Koehler R, Zimmermann M, Valsecchi MG, Gadner H, Masera G, Schrappe M, van Dongen JJ, Biondi A, Bartram CR. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22:771–782. doi: 10.1038/leu.2008.5. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley AA, Latham S, Brisco MJ, Sykes PJ, Sutton R, Hughes E, Wilczek V, Budgen B, van Zanten K, Kuss BJ, Venn NC, Norris MD, Crock C, Storey C, Revesz T, Waters K. Sensitive and specific measurement of minimal residual disease in acute lymphoblastic leukemia. J Mol Diagn. 2009;11:201–210. doi: 10.2353/jmoldx.2009.080048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton NE. Parameters of the human genome. Proc Natl Acad Sci USA. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordonez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, Campbell PJ. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, Samuels Y, Velculescu VE, Vogelstein B, Kinzler KW, Diaz LA., Jr. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, Zatloukal K, Lehrach H. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One. 2009;4:e5548. doi: 10.1371/journal.pone.0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Stenman J, Finne P, Stahls A, Grenman R, Stenman UH, Palotie A, Orpana A. Accurate determination of relative messenger RNA levels by RT-PCR. Nat Biotechnol. 1999;17:720–722. doi: 10.1038/10942. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R, Sieuwerts AM, Martens JW, Silver DP, Langerod A, Russnes HE, Foekens JA, Reis-Filho JS, van’t Veer L, Richardson AL, Borresen-Dale AL, Campbell PJ, Futreal PA, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Vermeulen J, Ballet S, Oberlin O, Peter M, Pierron G, Longavenne E, Laurence V, Kanold J, Chastagner P, Lejars O, Blay JY, Marec-Berard P, Michon J, Delattre O, Schleiermacher G. Incidence and prognostic value of tumour cells detected by RT-PCR in peripheral blood stem cell collections from patients with Ewing tumour. Br J Cancer. 2006;95:1326–1333. doi: 10.1038/sj.bjc.6603438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]