Abstract
Background. Excessive inflammatory host response increases morbidity and mortality associated with seasonal respiratory influenza, and highly pathogenic virus strains are characterized by massive infiltration of monocytes and/or macrophages that produce a storm of injurious cytokines.
Methods. Here, we examined the role in respiratory influenza of serpinB1, an endogenous inhibitor of the serine proteases elastase, cathepsin G, and proteinase-3, increasingly recognized as regulators of inflammation.
Results. After challenge with high-dose surfactant protein-D (SP-D)–sensitive influenza A/Philadelphia/82 (H3N2), serpinB1−/− mice died earlier and in greater numbers than did wild-type mice. Sublethally infected animals suffered increased morbidity, delayed resolution of epithelial injury, and increased immune cell death. Viral clearance and SP-D/SP-A upregulation were unimpaired and so were early virus-induced cytokine and chemokine burst and influx of large numbers of neutrophils and monocytes. Whereas initial cytokines and chemokines rapidly cleared in wild-type mice, TNF-α, IL-6, KC/CXCL1, G-CSF, IL-17A, and MCP-1/CCL2 remained elevated in serpinB1−/− mice. Monocyte-derived cells were the dominant immune cells in influenza-infected lungs, and those from serpinB1−/− mice produced excessive IL-6 and TNF-α when tested ex vivo. Pulmonary γδ T-cells that produced IL-17A were also increased.
Conclusions. Because viral clearance was unimpaired, the study highlights the critical role of serpinB1 in mitigating inflammation and restricting pro-inflammatory cytokine production in influenza infection.
The serine proteases elastase, cathepsin G, and proteinase-3 are increasingly recognized as modulators of inflammation (reviewed in [1]). These enzymes, commonly called neutrophil serine proteases (NSPs), enter the lungs of infected mice as components of recruited monocytes and neutrophils. At the molecular level, they are related enzymes with distinct but, to a large extent, overlapping activities; they have broad capacity to regulate protective immune functions but, when neutrophils or monocytes are in excess, are key agents of inflammatory damage. We characterized SerpinB1 as a specific and efficient inhibitor of elastase, cathepsin G, and proteinase-3, all 3 of the NSPs [2]. We also deleted the murine gene encoding this common inhibitor to create a model in which the activities of the 3 NSPs would be coordinately increased [3]. Study of the serpinB1−/− mice makes it possible to identify pathological parameters of an infection that have their basis in exaggerated NSP action.
Innate immune cells, particularly myeloid cells, are extremely important in containing influenza [4]. Neutrophils can clear the virus in vitro and in vivo [5, 6]. Depletion of either alveolar macrophages or neutrophils before sublethal infection with a pandemic virus resulted in uncontrolled viral replication and increased mortality [7, 8]. However, myeloid cells can be detrimental to the host, contributing to morbidity and mortality through inflammatory injury [7, 9, 10]. Infection by highly pathogenic strains, such as 1918 H1N1 and avian H5N1, caused massive recruitment of neutrophils and monocytes and/or macrophages [11, 12]. These cells interfere with air exchange and rapidly release high levels of proinflammatory cytokines, including TNF-α, IL-1β, and IL-6, as part of the cytokine storm [7, 10, 13] associated with severe lung pathology in patients infected with avian H5N1 [11, 14].
A previous study of gene-targeted mice revealed a critical role for serpinB1 in protecting pulmonary antibacterial defense [3]. In response to Pseudomonas aeruginosa infection, serpinB1−/− mice showed defective clearance of bacteria, increased neutrophil death, elevated proinflammatory cytokines, and depletion by proteolysis of surfactant protein D (SP-D). This pulmonary collectin (collagenous lectin) opsonizes bacteria, neutralizes strains of influenza virus bearing high mannose carbohydrates on hemagglutinin, and maintains the anti-inflammatory environment of the lung [15–17]. In the present study, we evaluated the role of serpinB1 in influenza infection with the SP-D sensitive strain A/Philadelphia/82 (H3N2) (Phil/82), a human isolate that replicates injurious features of highly pathogenic influenza, including high-level recruitment of neutrophils and monocytes.
MATERIALS AND METHODS
Virus
Influenza A Philadelphia/82 (Phil/82) was grown in chorioallantoic fluid of 10-day-old embryonated hen’s eggs; it was purified on sucrose gradients, dialyzed against phosphate-buffred saline (PBS), and stored in aliquots at −80°C [18]. To determine infectivity, confluent MDCK cells in 96-well plates were infected with dilutions of the virus for 45 minutes at 37°C and evaluated 7 hours later using monoclonal antibody A-3 against influenza A nucleoprotein (provided by Dr. Nancy Cox; Centers for Disease Control and Prevention); the results were expressed as fluorescent focal counts (ffc) [19]. Phil/82 is a reassorted strain in which envelope proteins are from the human strain and internal proteins from PR/8, which allows strong growth in eggs [19]. The virus was given to the Hartshorn laboratory by Dr. Margot Anders in 1993 and, since then, has been propagated exclusively in eggs.
Animals and Infection Models
SerpinB1-deficient (serpinB1−/−) mice were generated in 129S6/SvEv/Tac (129S6) background [3]. Unchallenged serpinB1−/− mice were healthy with normal growth, reproduction, and tissue morphology. Wild-type 129S6 mice from Taconic Labs were maintained with serpinB1−/− mice in Immune Disease Institute animal facility for at least several weeks. Age- and sex-matched mice were sedated with 100 mg/kg ketamine and 10 mg/kg xylazine and were intranasally inoculated by applying 15 μL of inoculum onto each nare. Unless otherwise indicated, the dose (sublethal dose) was 2 × 106 ffc/mouse. In survival studies, mice were monitored at least twice daily for up to 20 days. Body weights were recorded daily. Animal studies were approved by the Institutional Animal Care and Use Committee of the Immune Disease Institute.
Lung Leukocyte and Cytokine Analysis
Groups of infected mice were sacrificed at indicated times to quantify leukocyte subsets. Excised lungs were enzymatically digested [12] (details in Supplementary Data). After red blood cell lysis and washing, cells were suspended in PBS with 2% FCS for cell counting (Coulter Ac·T diff2; Beckman Coulter) and staining with fluorochrome-conjugated antibodies (BD Biosciences; BioLegend). For viability testing of bronchoalveolar cells, tracheas were exposed and cannulated with an 18-gauge angiocath. Lungs were lavaged with 0.8 mL cold sterile PBS, and cells were washed with PBS, counted, and incubated for 30 minutes at 4°C with viability dye (ViD; Invitrogen). For cytokine synthesis assays, excised lungs were enzymatically digested as described above except that digestion time was 35 minutes and collagenase was Type 2 (Worthington) at 180 U/mL [20]. Harvested cells were suspended in complete DMEM (10% FCS, penicillin, streptomycin, and mercaptoethanol), and aliquots of 106 cells/mL were cultured for 4–6 hours with brefeldin A (1 μg; BD Biosciences; GolgiPlug) and either PMA (50 ng/mL) and ionomycin (0.5 μg/mL) for analysis of lymphocytes or LPS (Sigma L8643; P. aeruginosa; 2μg/mL) for analysis of myeloid cells. The cells were pelleted, resuspended in PBS with 2% FCS, stained with surface marker antibodies, fixed and permeabilized with BD Cytofix/Cytoperm, and stained intracellularly with indicated antibodies. Data were acquired on a Canto II cytometer (BD Biosciences) and were analyzed using FlowJo software (Tree Star).
Lung Homogenates, Enzyme-Linked Immunosorbent Assay (ELISA), Western Blots, and Real-Time PCR
Dissected lungs from infected mice were homogenized in PBS with leupeptin (50 μg/mL) and PMSF (2 mM), and aliquots were stored at –80°C. Cytokines and chemokines were measured in duplicate with use of ELISA (BioLegend; R&D Systems). Lung homogenates were used for real-time PCR and Western blots, which are detailed in Supplementary Data.
Histology
Lungs were fixed in Bouin's fixative and processed for hematoxylin and eosin staining with use of the Rodent Histopathology Core Facility, Harvard Medical School (Boston, MA).
Statistical Analysis
Data are expressed as mean ± standard error of the mean and were analyzed using the unpaired Student's t test or ANOVA with use of Prism (GraphPad). Kaplan-Meier survival curves were analyzed using the log-rank test. P < .05 was considered to indicate statistical significance.
RESULTS
Increased Mortality and Morbidity of SerpinB1-Deficient Mice After Influenza Infection
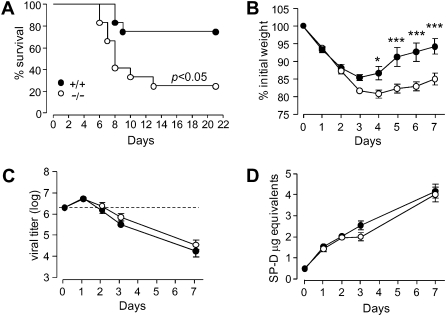
To test for a role for serpinB1 in respiratory influenza, serpinB1−/− and wild-type (129S6) mice were inoculated intranasally with a high dose of influenza Phil/82 (H3N2) virus, and the mice were monitored for survival and body weight for up to 20 days after infection. SerpinB1−/− mice had a significantly lower survival probability and a shorter median survival time, compared with wild-type mice (Figure 1A). Mean body weight was not different between the genotypes through day 5, after which the data were subject to survivor bias (data not shown). The significantly lower survival probability indicates that serpinB1 is critical to successful host response to influenza infection.
Figure 1.
Survival, morbidity, viral clearance and lung collectin levels of serpinB1−/− mice after influenza challenge. Wild type (+/+) and serpinB1 deficient (-/-) mice were inoculated with (A)107 ffc/mouse or (B-D) 2 ×106 ffc/mouse of Phil/82. A, Kaplan-Meier survival curves for 12 each wild type and serpinB1−/− mice. B, Mean body weights were significantly different between genotypes by 2-way ANOVA with Bonferroni’s posttest (*, P < .05; ***, P < .001). Ten mice/genotype were evaluated of which 5 randomized mice/genotype were sacrificed after day 5. Data are representative of two experiments. C, Viral titers were determined by real time PCR in lung homogenates of mice sacrificed on the indicated days post-infection. The dotted line represents the inoculum. D, SP-D levels in lung homogenates were detected by western blot, quantified by densitometry and expressed as “μg equivalents” using recombinant rat SP-D as internal standard. Mean ± SEM for 4-5 mice/group, and the data were analyzed by 2-way ANOVA.
To determine how serpinB1 alters survival in influenza, additional groups of serpinB1−/− and wild-type mice were inoculated with a sublethal dose of Phil/82. Body weight, pulmonary viral load, and surfactant proteins SP-D and SP-A were measured at various times after infection. Morbidity (body weight loss) was significantly increased in serpinB1−/− mice (Figure 1B). Body weight decreased rapidly for wild-type mice, reached its nadir (15% decrease) on day 3, and then rapidly recovered. In contrast, serpinB1−/− mice continued to lose weight through day 4 (20% total decrease), and recovery was protracted, so that significant differences of body weight were found after 4–7 days of infection. Viral burden was measured by quantitative real-time PCR based on amplification of the matrix gene [21]. The viral titer was increased on day 1 and decreased thereafter and did not differ between the genotypes (Figure 1C). We next examined SP-D, the collectin molecule that neutralizes influenza strains including Phil/82 and protects against inflammation; SP-D was of particular interest because of its depletion by proteolysis in bacterially infected serpinB1−/− mice [3]. Evaluation of lung homogenates by quantitative Western blot revealed substantial increase of SP-D levels beginning on day 1 and continuing through day 7 of infection for both wild-type and serpinB1−/− mice (Figure 1D). Moreover, the SP-D species was full-length monomer with no detectable proteolytic fragment (Supplemental Figure 1A; available online). SP-A, another pulmonary collectin that shares properties of SP-D [17], was also increased after infection and did not differ between the genotypes (Supplemental Figure 1B; available online). Real-time PCR revealed increased message for both SP-D and SP-A on day 1 of infection (Supplemental Figure 1C and 1D; available online). Thus, serpinB1 protects against mortality and attenuates the morbidity associated with influenza infection but is not required for viral clearance or maintenance of SP-D and SP-A levels in the lungs.
Increased Lung Injury and Cell Death in Infected SerpinB1−/− Mice
To understand the basis of the differential mortality and morbidity, the severity of lung injury was evaluated in histological sections. A large influx of neutrophils localized in airway lumen and in peribronchial areas was observed in both wild-type and serpinB1−/− mice on day 1 (not shown). In wild-type mice, maximal airway epithelial damage was observed on day 2, and phagocytic macrophages with large cytoplasms were observed in airways and alveolar lumen on day 3. In serpinB1−/− mice, epithelial injury appeared to be sustained on day 3, with accumulation of necrotic cell fragments and relatively lower proportion of phagocytic macrophages than in wild-type mice (Figure 2A). These findings indicate sustained injury and altered morphology of recruited monocytes and/or macrophages in serpinB1−/− mice. Consistent with the histological findings, neutrophil influx, measured by myeloperoxidase (MPO) activity in lung homogenates, increased on day 1 and decreased subsequently (Figure 2B). MPO activity did not statistically differ between the genotypes. To examine cell death, bronchoalveolar lavage cells were evaluated by flow cytometry using a fixation-compatible dye ViD [22]. Consistent with the lung histopathology, dye-permeable ViDbright dead leukocytes were significantly increased in serpinB1−/− mice compared with wild-type mice (Figure 2C).
Figure 2.
Lung histopathology, MPO activity, and death of recruited cells of influenza-infected mice. Wild type (+/+) and serpinB1 deficient (-/-) mice were infected with Phil/82 (2 × 106 ffc/mouse) and were sacrificed on the indicated days. A, Hematoxylin and eosin-stained lung sections on day 2 and 3. Note that inflammatory cells with segmented and doughnut shaped nuclei characteristic of neutrophils and monocytes (arrowheads) are abundant in airways and peribronchiolar areas on day 2 in mice of both genotypes, together with substantial cellular debris in airways (single arrows). Monocytic cells with bean-shaped or round nuclei with a large cytoplasm (double arrows) suggestive of resolution were observed in airways and alveolar spaces of wild type mice on day 3 but were not observed in serpinB1−/− mice, where the histological picture remains with similar features as on day 2. Areas of the top panels (40x objective) were enlarged and are shown in the lower panels. Histology is representative of at least 3 mice/genotype/time point. B, MPO activity in lung homogenates. Mean ± SEM is shown for 4-5 mice/group, and the data were analyzed by 2-way ANOVA. C, ViDbright (dead) leukocytes (CD45+) in BAL. Mean ± SEM is shown for 4-5 mice/group, and data were analyzed by unpaired t test (*, P < .05).
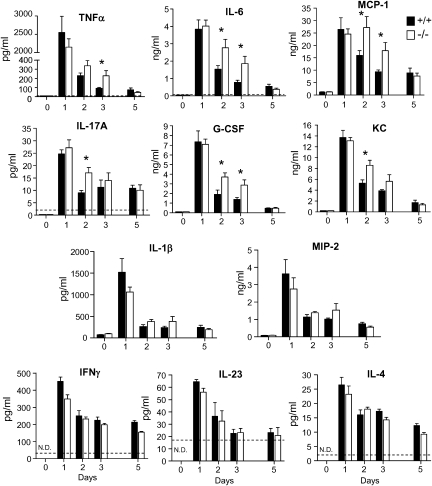
Enhanced and Prolonged Production of Proinflammatory Cytokines in SerpinB1−/− Mice
To determine whether the inflammatory response is abnormal in infected serpinB1−/− mice, we evaluated cytokines and chemokines, considered the double-edged sword of influenza because they are critical for antiviral protection but a source of inflammatory pathology if not well regulated. On day 1 of infection, lungs of serpinB1−/− and wild-type mice had comparable bursts of cytokines and chemokines tested. Whereas levels of these mediators decreased sharply thereafter in wild-type mice, the levels in serpinB1−/− mice remained elevated for TNF-α, IL-6, MCP-1, IL-17A, G-CSF, and KC (Figure 3), and levels of IL-1β and MIP-2 (CXCL2) trended higher. The increase in levels of proinflammatory cytokines and chemokines in serpinB1−/− mice on infection days 2 and 3 were reflected, to a large extent, in corresponding mRNAs (Supplemental Figure 2; available online), suggesting that the prolonged presence of these mediators is the result of failure of the serpinB1−/− mice to downregulate production. Of importance, prolonged production of cytokines was not a global cytokine defect, because the day 2 and 3 levels of IL-23, IFN-γ, and IL-4 in serpinB1−/− mice were not different from those in wild-type mice (Figure 3, bottom row).
Figure 3.
Cytokines and chemokines in lungs of influenza-infected mice. Wild type (+/+) and serpinB1−/− mice were inoculated with sublethal dose Phil/82 and were sacrificed on the indicated days. Lung homogenates were assayed by ELISA for the indicated cytokine/chemokine. Mean ± SEM is shown for 3-5 mice/group, and data were analyzed by unpaired t test (*, P < .05). N.D., not detectable
Altered Cytokine-Producing Cells From Infected SerpinB1−/− Mice
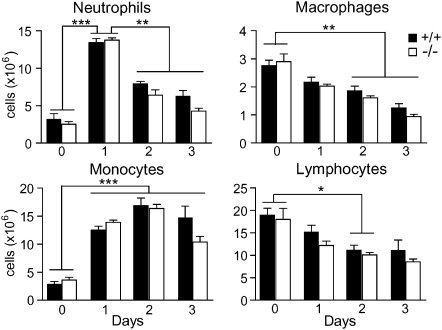
To identify the serpinB1−/− cells that produce excessive cytokines, we first quantified total immune cell subsets in the lungs of infected mice. The numbers of cells of each leukocyte subset were substantially altered during the course of infection, but no difference was observed between the 2 genotypes (Figure 4). Consistent with the histological data, levels of neutrophils peaked on day 1, whereas monocytes reached highest levels on day 2. A noteworthy feature of Phil/82 infection was the number of lung monocytes (>15 million), surprisingly high relative to the number of monocytes in blood of naive mice (∼0.5 million) and the absence of a substantial bone marrow reservoir. Levels of lung macrophages, including alveolar macrophages, decreased over time of infection for both genotypes (Figure 4), consistent with reports that lung macrophages undergo apoptosis on infection with influenza virus [23].
Figure 4.
Leukocyte subsets in lungs of influenza-infected mice. Wild type and serpinB1−/− mice were infected with sublethal dose Phil/82 and sacrificed on the indicated days. Subsets of leukocytes (CD45+) were quantified after enzymatic digestion of excised lungs. Shown are neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6Gneg), macrophages (CD11bnegCD11c+ highly autofluorescent cells) and lymphocytes (low side scatter, CD11bneg). The number of cells for wild type and serpinB1−/− mice at each time point were analyzed by the unpaired t test; there were no differences between the genotypes. Changes of cell numbers over time were analyzed by one-way ANOVA followed by Tukey post test (*, P < .05; **, P < .01; ***, P < .0001). Mean ± SEM is shown for 4 mice/genotype/time point. Data are representative of 2–3 experiments.
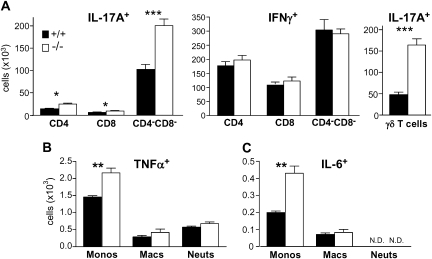
On the basis of these findings, we examined cytokine synthesis with use of lung cells from infection day 2 when immune cell density is highest and preceding the peak of morbidity. Ex vivo activation and intracellular cytokine staining revealed a dramatic increase in IL-17A–producing lymphocytes from virus-infected lungs of serpinB1−/− mice, compared with wild-type mice (Figure 5A, left). IL-17, a mediator of host defense to various infections and a pathogenic agent of autoimmune diseases [24], is not an established component of the cytokine storm of highly pathogenic influenza. Cell surface staining identified the predominant IL-17A–producer cells as CD4negCD8neg lymphocytes (Figure 5A, left). IL-17A–producing CD4+ and CD8+ cells were also present but at very low frequency. We also quantified IFN-γ–synthesizing lymphocytes and, consistent with the ELISA results for IFN-γ, the count of IFN-γ–producing lymphocytes was not different between the genotypes (Figure 5A, middle panel). The predominant IL-17A–producing CD4negCD8neg cells were identified as γδ T cells (Figure 5A right). The >3-fold increased count of IL-17A–producing γδ T cells from infected serpinB1−/− mice, compared with wild-type mice, reflects both an increased number of γδ T cells (2.2-fold) and an increased percentage of IL-17A producers among γδ T cells (70% for serpinB1−/− mice vs 45% for wild-type mice). These findings suggest that γδ T cells are the dominant IL-17–producing cells during this phase of influenza Phil/82 virus infection, and IL-17–producing γδ T cells are substantially more frequent in infected serpinB1−/− mice.
Figure 5.
IL-17A, IFN-γ, TNF-α, and IL-6 synthesizing lung leukocytes of infected mice. Wild type and serpinB1−/− mice were sacrificed on day 2 of sublethal Phil/82 infection. A, Single cell suspensions from excised lungs were cultured with PMA, ionomycin and brefeldin A and stained with the indicated antibodies and intracellularly for IL-17A and IFN-γ. IL-17A and IFN-γ producing cells were gated first on lymphocytes (low side scatter, CD45+CD11bneg). B, C, Single cell suspensions were incubated with LPS and brefeldin A and stained as in Figure 4 for monocytes, macrophages and neutrophils and intracellularly for (B) TNF-α or (C) IL-6. Mean ± SEM is shown for 4 mice/group, and data were analyzed by the unpaired t test (*, P < .05; **, P < .01; ***, P < .001; N.D., not detectable).
Among lung myeloid cells of Phil/82-infected mice, the predominant TNF-α–producing (Figure 5B) and IL-6–producing cells (Figure 5C) were monocytes (monocyte-derived cells) rather than lung macrophages or neutrophils. This was true for both genotypes. Of importance, the numbers of TNF-α–producing monocytes and IL-6–producing monocytes were markedly increased for serpinB1−/− mice, compared with wild-type mice (Figure 5B and C). These findings implicate monocytes (monocyte-derived cells) of infected serpinB1−/− mice as overproducers of proinflammatory cytokines.
DISCUSSION
Through the use of infection with Phil/82, we showed that deficiency of serpinB1 decreases the survival probability and increases the morbidity associated with murine pulmonary influenza. The greater morbidity among infected serpinB1−/− mice was associated with enhanced injury of lung epithelial structures, increased death of recruited immune cells, and failure of timely downregulation of proinflammatory cytokine production, hallmarks of exaggerated inflammation. Viral clearance was normal.
The substantial negative impact of overproduction of pro-inflammatory cytokines has been established by its association (along with high viral load) with fatal outcome of human influenza [14]. Specific features of the current infection model that may have influenced outcome are the use of a collectin-sensitive highly glycosylated strain and of high-dose inoculation. Highly glycosylated strains, unlike poorly glycosylated strains, such as PR/8, are more efficient in their ability to infect macrophages, induce cytokines, and recruit and activate leukocytes [25]. The choice of high-dose inoculation was based on a comparative study, which showed that the outcome of lethal/sublethal infection correlates with the initial tempo of virus replication, such that increasing the inoculum dose of a conventional strain mimics the effect of infection with a virus with increased virulence [26]. In another study, delay of antiviral treatment until 48 h after infection with high-dose H5N1 suppressed viral replication, but pro-inflammatory cytokines, lung damage, and interstitial inflammatory infiltration were similar to untreated mice [27], thus also highlighting the role of initial viral dose in driving the subsequent pro-inflammatory pathologic process. Consistent with these findings, high-dose Phil/82 infection reproduced select features of highly pathogenic influenza strains, including infiltration of very high numbers of monocytes. Moreover, many of the cytokines increased in Phil/82-infected serpinB1−/− mice are characteristic of highly pathogenic virus infections, for example, MCP-1, MIP-2, IL-1β, IL-6, and G-CSF, were increased in mice infected with the reconstructed 1918 H1N1 or highly pathogenic avian H5N1 virus [12, 28].
The collectin SP-D is thought to be protective in influenza, both for neutralizing the virus and maintaining anti-inflammatory properties of macrophages [15–17]. Contrary to anticipated findings, Phil/82 infection, rather than leading to proteolytic depletion of SP-D in serpinB1−/− mice, induced pronounced up-regulation of SP-D and SP-A to similar levels in both genotypes. The high level of SP-D is consistent with and may have contributed to the comparable clearance of virus in wild-type and serpinB1−/− mice. However, the pro-inflammatory cytokine production in lungs of infected serpinB1−/− mice occurred despite any putative opposing effects of SP-D.
Delays were noted in the effects of serpinB1 deletion in virus infection. The greater body weight loss of serpinB1−/− mice was not apparent until wild-type mice began to recover, and the appearance of increased levels of pro-inflammatory cytokines was also delayed. In contrast, the earliest burst of cytokines and chemokines was not altered by serpinB1 deletion, a finding likely explained because the responsible cells, the lung macrophages and epithelial cells [29], are lacking NSPs. Subsequent increased production of pro-inflammatory cytokines in the infected serpinB1−/− mice occurred after recruitment to the lungs of inflammatory neutrophils and monocytes that carry high levels of NSPs.
Several findings implicate monocytes (monocyte-derived cells) as key dysregulated responder cells in infected serpinB1−/− mice. Mobilization of monocytes to the infected lung has been shown to require a CCR2 dependent response to MCP-1 (CCL2) [30], and the Ly6C+ (Gr1+) subset, which is CCR2+, is preferentially recruited [30–32]. Infiltrating monocytes can differentiate into exudate macrophages and monocyte-derived dendritic cells, including cells that produce TNF-α and inducible NO synthase and stimulate proliferation of CD8 effectors for virus elimination [32, 33]. However, monocyte-derived cells have also been linked with tissue injury and morbidity associated with influenza [30, 32–34]. In lungs of Phil/82-infected mice, monocytes were most prevalent on infection days 2 and 3, when pro-inflammatory cytokines were increased in serpinB1−/− mice. Consistent with prior studies, monocytes in lungs of Phil/82-infected mice were primarily Ly6Chigh (data not shown). The numbers of monocytes in the lung were not different in serpinB1−/− and wild-type mice. When examined ex vivo, monocytes rather than lung macrophages were the predominant myeloid producers of IL-6 and TNF-α on day 2 of infection, and these cytokine-producing monocytes were increased in frequency in lungs of serpinB1−/− mice.
How serpinB1 regulates cytokine production remains to be elucidated, but the present study shows that lung γδ T cells and monocytes are severely skewed toward pro-inflammatory phenotypes when examined ex vivo. Whether there is interaction between the monocytes and IL-17+ γδ cells also needs to be addressed. Precedent exists for interaction of human Vγ9/δ2 cells [35], a minor population in blood with innate specificity for a microbial pyrophosphate metabolite, which are corecruited with monocytes to sites of infection. On coculture, HMB-PP–dependent crosstalk of Vγ9/δ2 cells and monocytes induces early inflammatory cytokines (including IL-6, TNF-α, and MCP-1/CCL2) and subsequent monocyte differentiation to antigen-presenting dendritic cells.
A putative role for γδ T cells and IL-17 in the increasingly pathogenic innate immune response of infected serpinB1−/− mice is consistent with known properties of IL-17. IL-17 stimulates granulopoiesis via G-CSF and recruits neutrophils by inducing MIP-2 and KC [36]. A recent study of influenza PR/8 infection also found that γδ T cells were the primary producers of IL-17 and that IL-17RA contributed to pathogenicity upstream of IL-6, KC, and G-CSF production, but was not required for viral clearance [37]. γδ T cells were recognized early as regulators of pulmonary influenza [38]. They are now recognized as innate immune responder cells (reviewed in [39, 40]), which unlike IL-17 producing adaptive T cells (Th-17 cells), display an activated phenotype, express IL-23R, CCR6, and transcription factor RORγT and are able, without the need for TCR engagement [41], to rapidly produce IL-17 on stimulation with IL-23 and IL-1β [42, 43]. Although IL-1β and IL-23 were elevated in influenza-infected serpinB1−/− mice, neither was detectably different from wild type at the time points tested. With exceptions, IL-17 has been historically considered as a Th-17 cell product in the adaptive immune response, and IL-17 was not included in evaluations of highly pathogenic influenza virus. However, a recent study of early immune responses to new variant H1N1 virus found elevated IL-17 levels exclusively in the more severely infected (hospitalized and critical care) patients [44]. A crosstalk between IL-17–producing γδ T cells and pro-inflammatory monocytes is suggested by the findings but remains to be established.
In conclusion, our findings indicate a special role for monocytes along with γδ T cells and IL-17 in the inflammatory pathology of influenza and indicate a critical regulatory function for serpinB1. The collective evidence strongly suggests that the function of serpinB1 in influenza infection is to protect the host by dampening, but not eliminating, the inflammatory innate immune response.
Supplementary Data
Supplementary data are available at The Journal of Infectious Diseases online.
Funding
This work was supported by National Institutes of Health (R21 AI-072552 to E. R. O. and C. B., R01 HL-066548 to E. R. O., RO1 AI-8322 to K. L. H., and RHL069031 to K. L. H.) and Parker B. Francis Fellowship in Pulmonary Research (to C. B.).
Acknowledgments
We thank Dr Sadis Matalon and Dr Erika Crouch,for reagents; J Michael Stolley, for overseeing the mouse colony; Jessica Cooley, for assistance with figures; and Dr Erika Crouch, Dr Heinz Remold, and Dr. Maziar Divangahi, for critical evaluation of the manuscript.
References
- 1.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–50. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 2.Cooley J, Takayama TK, Shapiro SD, Schechter NM, Remold-O'Donnell E. The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry. 2001;40:15762–70. doi: 10.1021/bi0113925. [DOI] [PubMed] [Google Scholar]
- 3.Benarafa C, Priebe GP, Remold-O'Donnell E. The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J Exp Med. 2007;204:1901–9. doi: 10.1084/jem.20070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGill J, Heusel JW, Legge KL. Innate immune control and regulation of influenza virus infections. J Leukoc Biol. 2009;86:803–12. doi: 10.1189/jlb.0509368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tecle T, White MR, Gantz D, Crouch EC, Hartshorn KL. Human neutrophil defensins increase neutrophil uptake of influenza A virus and bacteria and modify virus-induced respiratory burst responses. J Immunol. 2007;178:8046–52. doi: 10.4049/jimmunol.178.12.8046. [DOI] [PubMed] [Google Scholar]
- 6.Fujisawa H. Neutrophils play an essential role in cooperation with antibody in both protection against and recovery from pulmonary infection with influenza virus in mice. J Virol. 2008;82:2772–83. doi: 10.1128/JVI.01210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumpey TM, Basler CF, Aguilar PV, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 8.Tate MD, Deng YM, Jones JE, Anderson GP, Brooks AG, Reading PC. Neutrophils ameliorate lung injury and the development of severe disease during influenza infection. J Immunol. 2009;183:7441–50. doi: 10.4049/jimmunol.0902497. [DOI] [PubMed] [Google Scholar]
- 9.Kash JC, Tumpey TM, Proll SC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–81. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobasa D, Takada A, Shinya K, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–7. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y, Poon LL, Cheung CY, et al. H5N1 influenza: a protean pandemic threat. Proc Natl Acad Sci U S A. 2004;101:8156–61. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung CY, Poon LL, Lau AS, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–7. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 14.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wert SE, Yoshida M, LeVine AM, et al. Increased metalloproteinase activity, oxidant production, and emphysema in surfactant protein D gene-inactivated mice. Proc Natl Acad Sci U S A. 2000;97:5972–7. doi: 10.1073/pnas.100448997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawgood S, Brown C, Edmondson J, et al. Pulmonary collectins modulate strain-specific influenza a virus infection and host responses. J Virol. 2004;78:8565–72. doi: 10.1128/JVI.78.16.8565-8572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pastva AM, Wright JR, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4:252–7. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartshorn KL, Collamer M, Auerbach M, Myers JB, Pavlotsky N, Tauber AI. Effects of influenza A virus on human neutrophil calcium metabolism. J Immunol. 1988;141:1295–301. [PubMed] [Google Scholar]
- 19.Hartshorn KL, White MR, Voelker DR, Coburn J, Zaner K, Crouch EC. Mechanism of binding of surfactant protein D to influenza A viruses: importance of binding to haemagglutinin to antiviral activity. Biochem J. 2000;351:449–58. [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med. 2009;15:277–84. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward CL, Dempsey MH, Ring CJ, et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29:179–88. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perfetto SP, Chattopadhyay PK, Lamoreaux L, et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann PM. Susceptiblity of mononuclear phagocytes to influenza A virus infection and possible role in antiviral response. J Leukoc Biol. 1997;61:408. doi: 10.1002/jlb.61.4.408. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–67. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reading PC, Whitney PG, Pickett DL, Tate MD, Brooks AG. Influenza viruses differ in ability to infect macrophages and to induce a local inflammatory response following intraperitoneal injection of mice. Immunol Cell Biol. 2010;88:641–50. doi: 10.1038/icb.2010.11. [DOI] [PubMed] [Google Scholar]
- 26.Legge KL, Braciale TJ. Lymph node dendritic cells control CD8+ T cell responses through regulated FasL expression. Immunity. 2005;23:649–59. doi: 10.1016/j.immuni.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Zheng BJ, Chan KW, Lin YP, et al. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci U S A. 2008;105:8091–6. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szretter KJ, Gangappa S, Lu X, et al. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81:2736–44. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julkunen I, Sareneva T, Pirhonen J, Ronni T, Melen K, Matikainen S. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 2001;12:171–80. doi: 10.1016/s1359-6101(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 30.Dawson TC, Beck MA, Kuziel WA, Henderson F, Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am J Pathol. 2000;156:1951–9. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold S, von Wulffen W, Steinmueller M, et al. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J Immunol. 2006;177:1817–24. doi: 10.4049/jimmunol.177.3.1817. [DOI] [PubMed] [Google Scholar]
- 32.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–72. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 33.Aldridge JR, Jr, Moseley CE, Boltz DA, et al. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A. 2009;106:5306–11. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herold S, Steinmueller M, von Wulffen W, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205:3065–77. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberl M, Roberts GW, Meuter S, Williams JD, Topley N, Moser B. A rapid crosstalk of human gammadelta T cells and monocytes drives the acute inflammation in bacterial infections. PLoS Pathog. 2009;5:e1000308. doi: 10.1371/journal.ppat.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarzenberger P, La Russa V, Miller A, et al. IL-17 stimulates granulopoiesis in mice: use of an alternate, novel gene therapy-derived method for in vivo evaluation of cytokines. J Immunol. 1998;161:6383–9. [PubMed] [Google Scholar]
- 37.Crowe CR, Chen K, Pociask DA, et al. Critical role of IL-17RA in immunopathology of influenza infection. J Immunol. 2009;183:5301–10. doi: 10.4049/jimmunol.0900995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carding SR, Allan W, Kyes S, Hayday A, Bottomly K, Doherty PC. Late dominance of the inflammatory process in murine influenza by gamma/delta + T cells. J Exp Med. 1990;172:1225–31. doi: 10.1084/jem.172.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapsenberg ML. Gammadelta T cell receptors without a job. Immunity. 2009;31:181–3. doi: 10.1016/j.immuni.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Bonneville M, O'Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–78. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 41.Jensen KD, Su X, Shin S, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–41. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Ribot JC, Chaves-Ferreira M, d'Orey F, et al. Cutting edge: adaptive versus innate receptor signals selectively control the pool sizes of murine IFN-gamma- or IL-17-producing gammadelta T cells upon infection. J Immunol. 2010; 185:6421–5. doi: 10.4049/jimmunol.1002283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bermejo-Martin JF, Ortiz de Lejarazu R, Pumarola T, et al. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit Care. 2009;13:R201. doi: 10.1186/cc8208. [DOI] [PMC free article] [PubMed] [Google Scholar]