Abstract
The lengths of orthologous protein families in Eukarya are almost double the lengths found in Bacteria and Archaea. Here we examine protein structures in 745 genomes and show that protein length differences between superkingdoms arise as much shorter prokaryotic nondomain linker sequences. Eukaryotic, bacterial, and archaeal linkers are 250, 86, and 73 aa residues in length, respectively, whereas folded domain sequences are 281, 280, and 256 residues, respectively. Cryptic domains match linkers (P < 0.0001) with probabilities ranging between 0.022 and 0.042; accordingly, they do not affect length estimates significantly. Linker sequences support intermolecular binding within proteomes and they are probably enriched in intrinsically disordered regions as well. Reductively evolved linker sequence lengths in growth rate maximized cells should be proportional to proteome diversity. By using total in-frame coding capacity of a genome [i.e., coding sequence (CDS)] as a reliable measure of proteome diversity, we find linker lengths of prokaryotes clearly evolve in proportion to CDS values, whereas those of eukaryotes are more randomly larger than expected. Domain lengths scarcely change over the entire range of CDS values. Thus, the protein linkers of prokaryotes evolve reductively whereas those of eukaryotes do not.
Keywords: protein domain, evolutionary constraint, intrinsic disorder
The lengths of proteins are subject to systematic variation that relates to the cellular context in which they function. So, randomly chosen proteins from Archaea and Bacteria tend to be only two thirds as long as those chosen from Eukarya (1–3). Likewise, mean values for 300,000 protein sequences arranged in 18,000 orthologous families average 508, 309, and 311 aa for Eukarya, Bacteria, and Archaea, respectively (4).
These characteristic differences in protein lengths are thought to reflect the degree to which reductive pressure is expressed in the three superkingdoms (4). The length or mass of proteins is a potentially important characteristic because, for growth rate-optimized cells, there is always an advantage for proteins to be as small as possible to increase their mass-normalized kinetic efficiencies (4–6). Shorter proteins that retain maximum rates of function are expected to support faster growth rates of cells than longer proteins that have the same kinetic characteristics. However, the intensity of this reductive pressure will decrease as its cellular mass fraction decreases (4). As the complexity of the cellular proteomes of eukaryotes is, in general, much greater than that of prokaryotes (7, 8), an average individual eukaryotic protein will be present at a much lower mass fraction than its orthologues in Archaea and Bacteria. For this reason alone, we expect the selective pressure constraining a representative protein sequence length to be weaker in eukaryotes, which may account for the observation that the standard deviations (SDs) for orthologous protein lengths are twice the magnitude in eukaryotes compared with those for Archaea and Bacteria (4).
Proteins are highly structured and contain one or more regions that are generally recurrent and that fold into compact 3D molecular arrangements, the protein domains (9, 10). Multidomain proteins are usually described as tandem lengths of folded domains alternating with nondomain regions that are not folded. The distribution of domains in superkingdoms and in the Protein Data Bank is quite biased, with a significant number of domains being unique to Eukarya (10). Transmembrane domains are also underrepresented compared with those of globular proteins. Some nondomain sequences have been identified as intrinsically unstructured or natively unfolded sequences (11, 12). These lack a stable tertiary structure, but they do have a dynamic range of conformations that distinguishes them from the loops and diverse sequences that lack regular secondary structures but may also be classified as nondomain sequences (11, 12). Regions that are intrinsically disordered have been widely studied in proteins. They are much more enriched in eukaryotes compared with prokaryotes and are usually involved in binding and molecular recognition (13, 14). Intrinsically disordered sequences also have significantly different amino acid preferences (13). In the present study, we group all sequences without a stable tertiary fold as nondomain sequences. We note that domains are most often the catalytically active regions of proteins, but they may also support linkage to other proteins as well as nucleic acids and diverse macromolecules (12, 15). Nondomain regions in the form of terminal tails and internal linkers have been associated primarily with interactions that stabilize diverse macromolecular complexes, as well as the linking together of domains into higher-order tertiary folds (3, 11, 16–20). Here, for simplicity, we use the terms “domain” and “linker” to refer to folded domains and nondomain regions, respectively, but we recognize a degree of functional overlap between these two sorts of protein substructures.
The present data show that the evolutionary trajectories of domains and linkers are distinguishable by the fact that reductive constraints on protein lengths are preferentially expressed in linker sequences. In contrast, domain sizes are more stable throughout the three superkingdoms. Furthermore, a monotonic correlation between linker lengths and the total in-frame coding capacity [i.e., coding sequence (CDS)] of genomes is observed among archaeal and bacterial microbes. This correlation suggests that variation in the CDS values typical of prokaryotes evolving under growth rate optimization constraints drives the evolution of minimal linker lengths. In contrast, eukaryote linker lengths exceed expected minimal lengths, suggesting that, in these cells, reductive pressure from the putative growth rate optimization is vitiated or absent.
Results
Structural Assignments.
A large data set of structures assigned to protein sequences using hidden Markov models (HMMs) of structural recognition in SUPERFAMILY (15) was analyzed in 745 genomes, including 215 Eukarya, 478 Bacteria, and 52 Archaea genomes (Table S1). Our analysis assumes that all genes are accurately predicted in the genomes. The averages tabulated describe complex populations of proteins including substantial minorities of multidomain proteins that make up 26% to 32% of the proteins in the three superkingdoms (Table 1). Here, the composite lengths of both domains and linkers are described so that, for all proteins, these lengths are the sums of multiple domains and multiple linkers when applicable. Fig. S1 shows whisker plots for average protein, domain, and linker lengths of representative sets of genomes, describing the typical dispersion of length expected in the genomes that were sampled. We note that the possible existence of unsolved structures (21, 22) and the limitations of the HMM method cannot guarantee that all structures in a sequence will be uncovered. This might introduce some uncertainties to length estimates. However, careful analysis of the overall distribution of domains and linkers segments, domains that overlap, and HMM performance within families of orthologues suggest that this is a negligible problem (SI Materials and Methods). We find that HMMs detect superfamilies equally well in the three superkingdoms, and they do not show preferences for homologues of the seed sequences of the models (Table S2) or for superkingdom-specific sequences. These results are consistent with previous observations (15) and suggest that domain boundaries of remote homologues of the seed sequence are reliable. Moreover, if we assume that cryptic domains in linkers follow the same length distribution of domains detected by current HMMs, the data show that few of the putative domains could be accommodated in linkers, even if they were overlapped (Figs. S2–S4). In fact, comparison of frequency distributions of domains and linkers reveals that, on average, cryptic domains match linkers with probabilities in the range 0.022 to 0.042 (nonparametric Mann–Whitney test, P < 0.0001). Given these estimates, the maximum cumulative influence of such cryptic domains on average linker length would be, at most, approximately 22% in each superkingdom (Table S3). In summary, the reliability of the protein length patterns recovered in the present study have been tested and found to be robust.
Table 1.
Summary of lengths of proteins, protein domains, and linkers in 745 organisms belonging to the three superkingdoms of life
| Descriptor | All proteins | One-domain proteins | Two-domain proteins | Three-domain proteins | Four-domain proteins | Five-domain proteins | More than five domains |
| Eukarya (n = 215)* | |||||||
| Fraction of sequences, % | 100 | 68 | 19 | 6.1 | 2.5 | 1.1 | 2.1 |
| No. of sequences | 1,906,851 | 1,305,530 | 375,823 | 117,232 | 47,154 | 20,734 | 40,378 |
| Mean protein length ± SD | 532 ± 484 | 423 ± 337 | 623 ± 425 | 836 ± 512 | 981 ± 591 | 1,083 ± 719 | 1,509 ± 1,482 |
| Median protein length | 414 | 512 | 512 | 1,040 | 1,262 | 1,373 | 1,713 |
| Mean length ± SD | |||||||
| Domain | 281 ± 247 | 213 ± 134 | 338 ± 155 | 465 ± 207 | 566 ± 240 | 604 ± 269 | 943 ± 998 |
| Linker | 250 ± 372 | 210 ± 315 | 285 ± 383 | 371 ± 454 | 415 ± 533 | 479 ± 638 | 566 ± 757 |
| Median domain/linker length | 241/125 | 279/149 | 322/162 | 663/332 | 816/359 | 879/ 420 | 1,085/473 |
| Bacteria (n = 478)* | |||||||
| Fraction of sequences, % | 100 | 68.8 | 22.8 | 5.5 | 1.7 | 0.6 | 0.7 |
| No. of sequences | 1,008,634 | 694,039 | 229,892 | 55,208 | 17,003 | 5,590 | 6,902 |
| Mean protein length ± SD | 365 ± 258 | 304 ± 187 | 424 ± 219 | 583 ± 269 | 751 ± 292 | 916 ± 336 | 1,400 ± 1,105 |
| Median protein length | 473 | 270 | 564 | 795 | 716 | 1,263 | 1,604 |
| Mean length ± SD | |||||||
| Domain | 280 ± 181 | 225 ± 123 | 338 ± 139 | 480 ± 179 | 606 ± 173 | 731 ± 202 | 1,068 ± 691 |
| Linker | 86 ± 165 | 80 ± 147 | 86 ± 158 | 103 ± 190 | 145 ± 237 | 185 ± 279 | 332 ± 623 |
| Median domain/linker length | 368/45 | 206/27 | 474/47 | 678/64 | 572/77 | 1,026/149 | 1,364/243 |
| Archaea (n = 52)* | |||||||
| Fraction of sequences, % | 100 | 73.7 | 19.1 | 5.1 | 1.4 | 0.4 | 0.4 |
| Numbers of sequences | 68,350 | 50,378 | 13,027 | 3,496 | 951 | 247 | 251 |
| Mean protein length ± SD | 329 ± 217 | 275 ± 158 | 419 ± 207 | 574 ± 270 | 661 ± 308 | 866 ± 366 | 1,317 ± 765 |
| Median protein length | 437 | 375 | 381 | 822 | 638 | 792 | 1,080 |
| Mean length ± SD | |||||||
| Domain | 256 ± 158 | 209 ± 113 | 336 ± 134 | 476 ± 168 | 543 ± 189 | 669 ± 151 | 980 ± 551 |
| Linker | 73 ± 131 | 66 ± 113 | 83 ± 143 | 98 ± 197 | 118 ± 197 | 197 ± 301 | 337 ± 371 |
| Median domain/linker length | 342/39 | 292/35 | 325/28 | 689/77 | 525/51 | 642/136 | 879/234 |
| H value† | |||||||
| Domain | 4,081 | 9,140 | 31 | 368 | 604 | 1,163 | 1,345 |
| Linker | 376,492 | 202,523 | 121,933 | 40,633 | 9,781 | 2,781 | 1,914 |
Statistical tests of the reliability of these averages are described in SI Materials and Methods.
*An average of 55.02% ± 10.79 (SD), 65.52% ± 5.48, and 61.94% ± 3.88 of sequences had structural assignments in Eukarya, Bacteria, and Archaea, respectively.
†Kruskal–Wallis one-way ANOVA on ranks. The differences in all of the median lengths of domains or linkers among Eukarya, Bacteria, and Archaea are statistically significant (P ≤ 0.001).
To determine if linker sequences are enriched in intrinsically disordered regions, we studied the amino acid composition of linkers and domains. We found that linkers and domains showed significantly distinct amino acid preferences (paired t test, P < 0.01) in the three superkingdoms (Fig. S5). These preferences included known disorder-promoting residues (e.g., S, R, P, Q, E) in linkers that are typical of intrinsically disordered regions and known order-promoting residues (e.g., C, I, F, V, L, H) in domains typical of 3D structure (13). As cryptic domains match linkers with low probability, it seems reasonable to infer that intrinsically disordered regions are widely present in linkers. In fact, we analyzed the entire set of confirmed intrinsically disordered proteins (n = 643 in total) and found that 63 entries contained domains and had fewer than 30% structurally undetermined regions. Within these, 88.6% of residues in linker regions were disordered (only 1.8% were ordered), suggesting that intrinsically disordered regions can be substantial in linkers.
Domain and Linker Lengths Among the Three Superkingdoms.
The data summarized in Table 1 and Fig. 1 show that the mean values of protein lengths in Archaea are very roughly 10% smaller for single-domain as well as for multiple-domain proteins compared with those of Bacteria. Much more pronounced differences are found in comparisons with eukaryotes. Here, the proteins of Archaea and Bacteria tend to be less than two thirds the lengths of proteins in Eukarya, which confirms earlier observations (1–4).
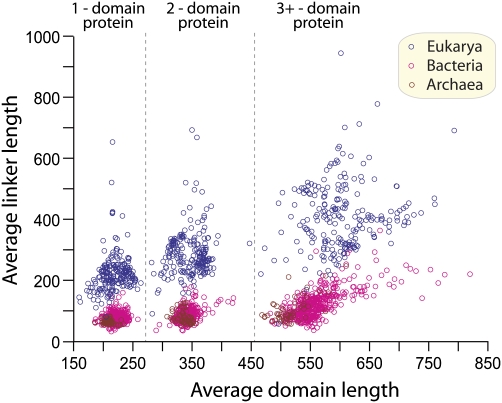
Fig. 1.
Plot shows average lengths (amino acid numbers) of domains and linkers in 745 genomes, including 215 Eukarya (blue circles), 478 Bacteria (pink circles), and 52 Archaea (brown circles). Mean values of proteins with different domain numbers within the same genome could be separated well (dash lines) because of the increasing aggregate lengths.
The immediate impression from the data summarized in Table 1 is that the most significant differences between overall lengths of proteins from Eukarya, and also those from Archaea and Bacteria, primarily result from systematic differences in the lengths of linker sequences. On average, the Eukarya have more than threefold greater linker lengths than Archaea and Bacteria (Table 1 and Fig. 1). The mean lengths of the linkers are 250, 86, and 73 aa for Eukarya, Bacteria, and Archaea, respectively.
In contrast, proteins in the three superkingdoms have similar domain lengths, with mean values of 281, 280, and 256 aa for Eukarya, Bacteria, and Archaea, respectively (Table 1). In addition, there is a characteristic scattering of domain and linker lengths from the three superkingdoms that is particularly evident in one- and two-domain proteins (Fig. 1). Here, the small cluster of archaeal domains and linkers overlaps the smaller end of the bacterial cluster. In contrast, the length of domains in multidomain proteins of Archaea overlaps but is generally smaller than in Bacteria (Fig. 1). Both Archaea and Bacteria are well segregated according to their shorter linker lengths from the Eukarya cluster. In general, the scattering of linker and domain lengths for Eukarya is greater than that for Bacteria and Archaea.
Multidomain Proteins.
An enhanced scattering of domain lengths and linker lengths in all multidomain categories parallels the progressive increase of their cumulative lengths with increasing domain number (Fig. 1). Furthermore, the relative numbers of sequences with three or more domains is approximately 30% greater in eukaryotes than in Archaea and Bacteria (Table 1).
The multidomain proteins also reveal distinctive size distributions (Fig. S4). The fractions of proteins with increasing numbers of domains decrease very rapidly in all three superkingdoms, whereby proteins with one or two domains constitute approximately 90% of all of sequences (Table 1). There is a marked tendency for the domains to shrink in size as their numbers per protein increase; this is most pronounced for Eukarya. Consequently, the aggregate domain lengths of the largest multidomain proteins among Eukarya may be as much as 20% shorter than their counterparts among Archaea and Bacteria (e.g., average domain lengths of 604 and 731 aa residues in five-domain proteins for Eukarya and Bacteria, respectively). Likewise, the cumulative linker sequences of the eukaryotic multidomain proteins do not increase at the same rate as the cumulative domain lengths do with increasing domain numbers. The archaeal multiple domain proteins, as a whole, are marginally shorter than their bacterial counterparts, which are significantly shorter than those of Eukarya. Nevertheless, the most striking discrepancy is that Eukarya proteins in all categories are equipped with linkers that are approximately two to three times longer than those of Archaea and Bacteria.
Continuous Variation of Linker Lengths.
We compared in greater detail the linker lengths for proteins in cells from the three superkingdoms and some phyla. Briefly, N-terminal linkers (79 and 25 residues for Eukarya and Archaea, respectively) are generally slightly shorter than C-terminal linkers (87 and 28 residues for Eukarya and Archaea, respectively) in Archaea and Eukarya, but not in Bacteria, in which there is the opposite inequality. In addition, the N-terminal linkers in almost all animals and fungi are shorter than C-terminal linkers with a small number of exceptions. In most Bacteria and Archaea, the lengths of their internal linkers are shorter than N-terminal or C-terminal linkers. Average lengths from animals, fungi, plants, and protists suggest that plants have the shortest linkers and that fungal linkers are the most narrowly distributed, whereas protists have the longest and most dispersed linker sequences. All this detailed variability needs further study. At this point, these data suggest that different phylogenetic groups of organisms have evolved linker lengths in idiosyncratic ways.
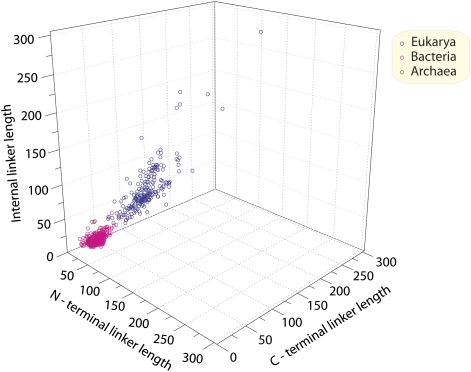
The present data contradict the expectation (3) of discrete differences between eukaryote linker lengths and prokaryote linker lengths. Fig. 2 is a 3D plot that highlights the aggregate linker lengths of all proteins compounded from their N-terminal, C-terminal, and, if present, internal linker sequences. We observe a continuous variation of the aggregate linker length vector with all the proteins of the three superkingdoms. Thus, linker lengths for all proteins of the three superkingdoms can be sorted in a continuous series rather than in discontinuous blocks characterized by discrete sizes.
Fig. 2.
Three-dimensional plot illustrates distribution of average lengths of corresponding N-terminal, C-terminal, and internal linker sequences in every genome analyzed in Fig. 1. The coordinates of every genome (circle) were determined by plotting average lengths of linkers of all protein sequences in a genome.
Linker and Domain Length Correlations with Proteome Diversity.
We recall that it is primarily linker lengths that vary between proteomes and not domain lengths (as detailed earlier). Accordingly, we might speculate that, if diversity of proteomes is approximately proportional to genome size, this might be reflected in plots of average linker lengths against genome sizes for cells of the three superkingdoms (4–7, 23). Although we observed a gradual increase of prokaryote linker lengths along with the corresponding genome lengths, eukaryote linker lengths were much longer than expected and approximately randomly distributed around a higher average value that did not correlate with genome length. The interpretation of these results was complicated by the potential influence of uncorrelated amounts of noncoding repetitive genome sequences.
To circumvent an influence of repetitive noncoding sequences on the analysis, we plotted average linker lengths against total CDS lengths. The CDS is the sum of codons in ORFs in a genome and is accordingly a measure of the total coding capacity for amino acid sequences of a genome, i.e., the diversity of the corresponding proteome. The CDS is also a measure of DNA and mRNA sequence heterogeneity for the corresponding genome. When the CDS values are plotted against domain diversity represented as the numbers of different domains at the fold superfamily (FSF) level of structural classification, significant linear correlations (R2 = 0.609–0.874; P < 0.001) are obtained (Fig. S6). In brief, the CDS is a useful measure of the potential proteome diversity of a particular cell, but it does not take into account variations of expression levels. When we plot average linker lengths against total CDS for individual genomes, similar results to those obtained with uncorrected genome lengths are obtained (Fig. 3).
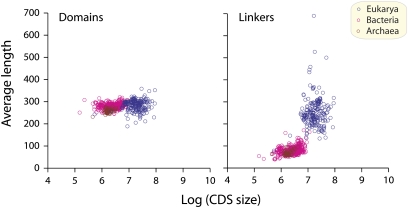
Fig. 3.
Average domain and linker length plotted against CDS length for individual genomes.
Fig. 3 describes the domain length and linker length distributions as a function of aggregate CDS for the genomes analyzed. The interpretation of this plot is based on the simplistic assumption that the combined coding capacity of a cell, as represented by its CDS value, is a valid measure of the diversity of that cell's proteome. Two convincing correlations surface in Fig. 3. First, the domain lengths of proteins from all three superkingdoms are relatively stable throughout the range of variation of CDS; the net increase over that range is less than 5%. In contrast, the lengths of linkers increase approximately twofold over the same range of CDS values for Archaea and Bacteria. In other words, the average lengths of the corresponding linker sequences are correlated with the molecular diversity encoded in the prokaryote genomes. This correlation suggests that reductive pressures are constraining the variation of linker lengths in the archaeal and bacterial populations (4–7, 23).
Here, one additional factor that may generate the observed scatter of these points is the dependence of the selective pressure for minimal protein lengths on the expression levels of individual proteins (4). The individual expression levels of proteins vary enormously within and between different cellular proteomes.
The results for the eukaryote cohort are much more chaotic. Thus, the plots of linker lengths versus CDSs for Eukarya reveal substantial random variations of linker lengths around a relatively constant average length (Fig. 3). This average length is greater than would be expected from an extrapolation of minimal lengths expected for eukaryotes based on the prokaryote relationship between linker length and CDS in Fig. 3. In other words, the best-fit simulation constructed for the Eukarya suggests that the average length of linkers within that cluster is not responding systematically to substantial increases in the molecular diversity (i.e., CDS) of the corresponding genomes (Fig. 3). The correlation coefficient of that best-fit second-order polynomial curve is a respectable 0.96. It would seem that the greater eukaryotic linker lengths are indifferent to the reductive pressures correlated with increasing CDS in prokaryotes. Similar conclusions could be drawn from plots that use domain diversity instead of CDS (Fig. S7).
Discussion
The evolution of protein structure in general and linker sequences in particular is conditioned by the remarkably high ambient concentrations of cellular proteins, which typically correspond to 20% to 30% of cell mass (23–27). Such cellular densities are described in terms of “molecular crowding,” in which the space between macromolecules is much less than their diameters so diffusion of macromolecules is retarded. As a consequence, molecular crowding favors macromolecular associations into large complexes as well as networks of proteins and nucleic acids that can support kinetically efficient biological functions without requiring rapid diffusion of macromolecules (24, 25). High densities also enhance the diffusion rates of small molecules because the excluded volumes of the proteins reduce the effective volume through which small molecules diffuse. The sum of these effects is that the high macromolecular densities enhance the kinetic efficiencies of proteins through functional complex formation. Molecular crowding is similar in Archaea and Bacteria, but its effects are exaggerated in the much larger cells of Eukarya. Accordingly, subdivision of high densities of proteins into distinct compartments, each with its characteristic functionally interactive macromolecules, is a feature of Eukarya that promotes kinetically efficient use of proteins and nucleic acids (23). Our working hypothesis is that linker sequences evolve in support of functional protein complexes as well as to localize proteins in subcellular compartments (3, 11, 16–20).
Previous studies (1–4) have focused on the average lengths of pooled proteins from each of the superkingdoms. It was therefore natural to attribute these length variations to discrete differences between protein functions in the superkingdoms. For example, it was suggested that the presence of cellular compartments in eukaryotes necessitated the evolution of extra sequences to address proteins to specific compartments (3). Such addressing sequences were expected to be absent from Archaea as well as from Bacteria. In contrast, the data summarized in Fig. 2 show that the sequence lengths of linkers can be arranged in a continuous rank order. Thus, the expectation (3) that discrete blocks of addressing sequences would be present in eukaryote proteins and absent from prokaryote proteins is contradicted by this continuous variation of linker lengths (Fig. 2).
Our working hypothesis identifies a primary function of linker sequences with the provision of site-specific interactions with RNA, DNA, and proteins (3, 11, 16–20). Three factors may have an influence on the minimal lengths of binding sequences in the proteins of growth rate optimized cells (4, 5). First, the reductive selective pressure on protein lengths will decrease as the diversity of the proteome increases because the expression level of individual proteins decreases (4, 5). For this reason, protein linkers may vary with the coding capacity of prokaryote genomes (Fig. 3). Among Eukarya, the linker lengths exceed the expected minima in random ways (Fig. 3).
Second, minimum lengths of linker binding sites are required to ensure sequence specificity of binding either to proteins or to nucleic acids. The minimum length for this specificity would increase with the numbers and concentrations of competing nonspecific interactions with closely related ligands (20, 28, 29). As the expected numbers of competitors increase with the diversity of RNA, DNA, and protein targets, the minimum length of binding sequences is expected to increase with the complexity of CDSs in genomes. Here too, prokaryote linker lengths may be subject to reduction to the corresponding minimal lengths (Fig. 3).
Third, kinetics of association between correctly matched proteins and their targets limit complex formation rates progressively as the concentrations of proteins decreases as a result of increasing diversity. Increasing the lengths of binding sequences progressively as the diversity of the proteome increases in prokaryotes may compensate the loss in kinetic efficiency of association (30) (Fig. 3). Furthermore, the intrinsically disordered regions seem to be more frequent in the longer eukaryote linkers than in short prokaryote linkers. Accordingly, intrinsically disordered sequences may be selected to support rapid and efficient binding interactions that would be retarded by linker sequences that form transient secondary structures or tangled arrays.
In summary, all three of these factors may contribute to the increase in the linker lengths of the prokaryote linkers associated with increases in the CDS. As the eukaryotic linker lengths most often exceed these minimal values, we conclude that the growth rate maximization that is the basis of the reductive pressure exerted on Archaea and Bacteria (4–7, 23) may not be directly applicable to Eukarya.
We do not have to go far to discover reasons for the inapplicability of the growth rate maximization to Eukarya. Thus, the relatively small population sizes of Eukarya may reduce the intensity of selection pressure on their genomes (31–33). Indeed, another indicator of lower reductive pressure is the common finding that noncoding sequences can accumulate in great excess over CDSs in Eukarya, whereas noncoding sequences are commonly a minor fraction of genomes in Archaea and Bacteria (33). Second, the much greater coding capacities of most eukaryote genomes increases their vulnerability to random mutations that could otherwise facilitate the reductive pressure on CDS and noncoding sequence lengths (33). In effect, potential collateral damage caused by random deletions in very large CDSs of eukaryotes might preclude the magnitude of random deletion rates per nucleotide that, in microorganisms, can support the minimization of noncoding genome sequences (33). Nevertheless, it remains to be determined why, in general, eukaryote diploid sexual populations are unsuitable for the growth rate maximization that is apparently expressed as the reductive evolution of linker lengths in asexual haploid prokaryote populations.
The stable structures of folded domains are considered autogenic in the sense that the final fold is not dependent on heterologous molecular interactions. For this reason, the length minimization of the self assembled structures that form functional domains may be relatively insensitive to the cellular context as suggested by the near stability of domain sizes across the entire span of proteomes (Fig. 3). On the contrary, in a limited number of cases, there may be selection for functionally equivalent, smaller domains that are unstable in single domain proteins but that are stabilized by quaternary interactions in multidomain proteins, especially among Eukarya (Table 1).
It has been suggested that Archaea and Bacteria diverged from Eukarya in reductive trajectories that are reflected in the details of their modern proteomes (4, 7, 8, 23). The present data show that minimal linker lengths evolve normally in proportion to the relatively limited coding capacities of prokaryotic genomes.
Materials and Methods
A total of 2,983,835 protein sequences in 745 proteomes were downloaded from SUPERFAMILY (15), of which 64%, 34%, and 2% were of eukaryotic, bacterial, or archaeal origin, respectively. Lengths of domains and linkers for single domain and multidomain proteins, given in numbers of amino acids, were calculated and statistically compared for the three superkingdoms. Several statistical tests and experiments were carried out to detect the reliability and robustness of domain assignments with HMMs of structural recognition. We found no bias among different superkingdoms. We also estimated the probability of linker sequences matching cryptic domains to assess their potential influences on our conclusions. A detailed description of experimental procedures is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Otto Berg, David Penny, Linda Randall, and Irmgard Winkler for help with the manuscript. This work was supported by National Science Foundation Grants MCB-0343126 and MCB-074983607 (to G.C.-A.) and the International Atomic Energy Agency (G.C.-A.). Travel was supported by the Center for Advanced Study and Institute for Genomic Biology of the University of Illinois (G.C.-A.), the Royal Physiographic Society of Lund, (C.G.K.), and the Nobel Committee for Chemistry, Royal Swedish Academy of Sciences, Stockholm (C.G.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017361108/-/DCSupplemental.
References
- 1.Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. doi: 10.1016/s0168-9525(99)01922-8. [DOI] [PubMed] [Google Scholar]
- 2.Liang P, Riley M. A comparative genomics approach for studying ancestral proteins and evolution. Adv Appl Microbiol. 2001;50:39–72. doi: 10.1016/s0065-2164(01)50003-9. [DOI] [PubMed] [Google Scholar]
- 3.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33:3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurland CG, Canbäck B, Berg OG. The origins of modern proteomes. Biochimie. 2007;89:1454–1463. doi: 10.1016/j.biochi.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Ehrenberg M, Kurland CG. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984;17:45–82. doi: 10.1017/s0033583500005254. [DOI] [PubMed] [Google Scholar]
- 6.Bremer H, Dennis PP. In: Escherichia coli and Salmonella. Neidhardt FC, editor. Washington, DC: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 7.Wang M, Yafremava LS, Caetano-Anollés D, Mittenthal JE, Caetano-Anollés G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007;17:1572–1585. doi: 10.1101/gr.6454307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Caetano-Anollés G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure. 2009;17:66–78. doi: 10.1016/j.str.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Caetano-Anollés G, Wang M, Caetano-Anollés D, Mittenthal JE. The origin, evolution and structure of the protein world. Biochem J. 2009;417:621–637. doi: 10.1042/BJ20082063. [DOI] [PubMed] [Google Scholar]
- 10.Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem J. 2009;419:15–28. doi: 10.1042/BJ20090122. [DOI] [PubMed] [Google Scholar]
- 11.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 13.Radivojac P, et al. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: Families and functions. Nucleic Acids Res. 2007;35(database issue):D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton JM, Sibanda BL. Amino and carboxy-terminal regions in globular proteins. J Mol Biol. 1983;167:443–460. doi: 10.1016/s0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- 17.High S, Abell BM. Tail-anchored protein biosynthesis at the endoplasmic reticulum: the same but different. Biochem Soc Trans. 2004;32:659–662. doi: 10.1042/BST0320659. [DOI] [PubMed] [Google Scholar]
- 18.Dyson HJ, Wright PE. Elucidation of the protein folding landscape by NMR. Methods Enzymol. 2005;394:299–321. doi: 10.1016/S0076-6879(05)94011-1. [DOI] [PubMed] [Google Scholar]
- 19.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci USA. 2009;106:19819–19823. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 22.Apic G, Gough J, Teichmann SA. An insight into domain combinations. Bioinformatics. 2001;17(Suppl 1):S83–S89. doi: 10.1093/bioinformatics/17.suppl_1.s83. [DOI] [PubMed] [Google Scholar]
- 23.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 24.Ellis RJ. Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 25.Ellis RJ. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr Opin Struct Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 26.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: Volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 27.Pielak GJ. A model of intracellular organization. Proc Natl Acad Sci USA. 2005;102:5901–5902. doi: 10.1073/pnas.0501812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Hippel PH, Berg OG. On the specificity of DNA-protein interactions. Proc Natl Acad Sci USA. 1986;83:1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg OG. The evolutionary selection of DNA base pairs in gene-regulatory binding sites. Proc Natl Acad Sci USA. 1992;89:7501–7505. doi: 10.1073/pnas.89.16.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 33.Pettersson ME, Kurland CG, Berg OG. Deletion rate evolution and its effect on genome size and coding density. Mol Biol Evol. 2009;26:1421–1430. doi: 10.1093/molbev/msp054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.