Abstract
Bloom’s syndrome (BS) is a rare genetic disorder and the cells from BS patients show genomic instability and an increased level of sister chromatid exchange (SCE). We generated BLM–/– and BLM–/–/RAD54–/– DT40 cells from the chicken B-lymphocyte line DT40. The BLM–/– DT40 cells showed higher sensitivity to methyl methanesulfonate and elevated levels of SCE as expected. The targeted integration frequency was also increased remarkably in BLM–/– cells. The SCE frequency increase in BLM–/– cells was considerably reduced and the enhanced targeted integration observed in BLM–/– cells was almost completely abolished in BLM–/–/RAD54–/– cells, indicating that a large portion of the SCE in BLM–/– cells occurs via homologous recombination, and homologous recombination events increase with the defect of BLM function. The BLM–/–/RAD54–/– cells showed a slow growth phenotype and an increased incidence of chromosome-type breaks/gaps while each single mutant showed relatively small numbers of chromosome-type breaks/gaps.
Keywords: BLM/chromosome-type breaks/HR/RAD54/SCE
Introduction
Bloom’s syndrome (BS) is a rare genetic disorder characterized by small size, sunlight sensitivity, immunodeficiency and male infertility. The cells from BS patients show an abnormally high incidence of sister chromatid exchange (SCE) compared with cells from normal individuals (Chaganti et al., 1974; reviewed by German, 1993). The gene responsible for BS, BLM, encodes a protein belonging to the RecQ helicase family, which consists of 1417 amino acids with seven conserved helicase motifs (Ellis et al., 1995).
Recently, Werner’s syndrome (WS) responsible gene (WRN) has been cloned and revealed to encode a RecQ homolog (Yu et al., 1996). WS is a genetic disorder characterized by premature aging, and the cells from WS patients also show genomic instability (Gebhart et al., 1988; reviewed by Martin, 1982) but not increased SCEs.
In Saccharomyces cerevisiae, a sole gene encodes the RecQ homolog, SGS1 (slow growth suppressor 1), whose mutant allele was identified as a suppressor of the slow growth phenotype of top3 mutants (Gangloff et al., 1994). The sgs1 mutants show hyper-recombination and chromosome missegregation (Watt et al., 1995, 1996). It has been shown that Sgs1 interacts physically with DNA topoisomerase (topo) II and topo III (Gangloff et al., 1994; Watt et al., 1995) and genetically with topo I and III (Gangloff et al., 1994; Lu et al., 1996). Topoisomerases are essential for DNA replication, transcription, chromosome condensation and segregation (reviewed by Wang, 1996).
Biochemical analysis of BLM, WRN and Sgs1 showed that they were all ATP-dependent helicases with 3′→5′ directionality (Lu et al., 1996; Gray et al., 1997; Karow et al., 1997; Bennett et al., 1998). BLM, WRN and Sgs1 are similar in length and share limited sequence homology outside the helicase domain (Gangloff et al., 1994; Ellis et al., 1995; Yu et al., 1996). BLM and WRN partially suppressed the increased homologous and illegitimate recombination in the sgs1 mutants (Yamagata et al., 1998), suggesting that structures and functions are partially conserved among these proteins.
The most characteristic feature of BS cells is a high incidence of SCE. Although the phenomenon of SCE has long been recognized, the molecular mechanism behind it, especially the SCE in BS cells, remains unclear. There are two major models to explain SCE. First, SCE is mediated by homologous recombination (HR) (Painter, 1980; Cleaver, 1981; Sonoda et al., 1999). Secondly, topo II causes transient double-stranded DNA breaks (DSBs) during replication, and the proximity of DNA breaks on sister chromatids may result in incorrect rejoining, causing an SCE (Ishii and Bender, 1980; Pommier et al., 1985; Heartlein et al., 1987; Dillehay et al., 1989).
Studies on HR in S.cerevisiae defined the RAD52 epistasis group of genes, which are constituents of a pathway for the repair of DSBs by HR (reviewed by Game, 1993; Shinohara and Ogawa, 1995). The rad51, rad52 and rad54 mutants, which are involved in the RAD52 epistasis group, show similar phenotypes including high sensitivity to ionizing radiation and a reduced mitotic recombination frequency, and recent studies indicated that these genes are conserved in function from yeasts to vertebrate cells (Bezzubova et al., 1997; Essers et al., 1997; Sonoda et al., 1998; Yamaguchi-Iwai et al., 1998).
In this study, to elucidate the molecular mechanism to increase SCEs in BS cells and to examine whether the increased SCEs are mediated via HR, we generated BLM–/– and BLM–/–/RAD54–/– cells from the chicken B-lymphocyte line DT40 and characterized their phenotypes.
Results
BLM targeting constructs and generation of BLM–/– and BLM–/–/RAD54–/– clones
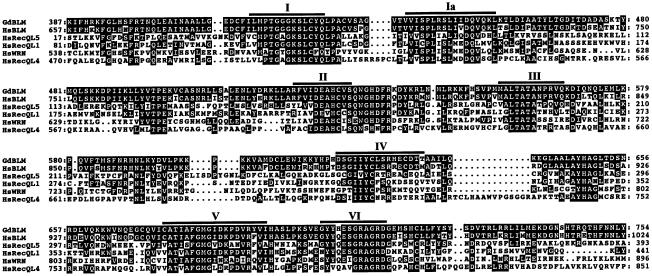
A chicken BLM cDNA fragment was amplified from chicken testis RNA by RT–PCR using primers based on mouse BLM sequences. The N-terminal region and the C-terminal region of chicken BLM were obtained by 5′-RACE and 3′-RACE, respectively. We obtained an ∼3.5 kb chicken BLM cDNA fragment containing the helicase domain. The helicase domain of chicken BLM shares 84.4% identity with that of human BLM at the amino acid level and 39.1, 47.5, 35.1 and 46.5% identity with those of human WRN, RecQL/DNA helicase Q1 (RecQL1), RecQL4 and RecQL5, respectively (Figure 1).
Fig. 1. Amino acid alignments in the helicase domain of chicken BLM (GdBLM) and human (Hs) BLM, WRN, RecQL1, RecQL4 and RecQL5. Thick lines indicate helicase motifs.
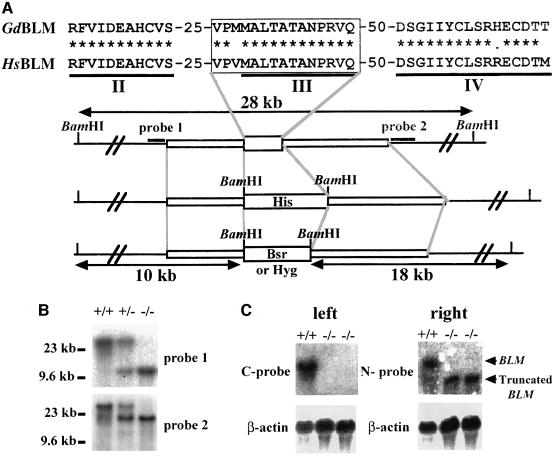
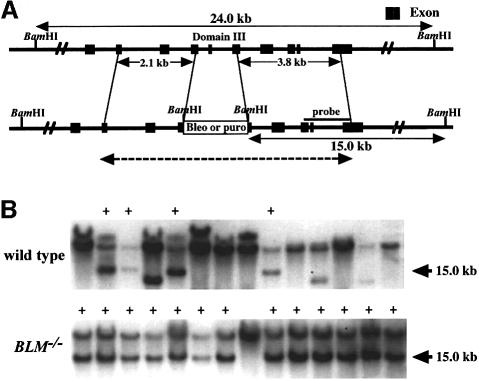
An ∼4 kb genomic DNA fragment was amplified by long-range PCR using primers synthesized according to the cDNA sequences within the helicase domain. To generate BLM targeting constructs, either histidinol (his), blasticidin (bsr) or hygromycin (hyg) resistance genes were inserted into the genomic sequence as shown in Figure 2A. Targeted integration of these constructs is expected to delete the helicase motif III, which is highly conserved among the RecQ helicase family. Two BLM targeting constructs were sequentially transfected into DT40 cells. Disruption of the BLM gene was verified by Southern blotting (Figure 2B). Northern blotting using N-probe indicated expression of truncated mRNA (Figure 2C).
Fig. 2. Generation of BLM–/– clones. (A) Schematic representation of disruption constructs. Motif III in chicken BLM genomic DNA cloned in a plasmid was replaced with a BamHI site by PCR, and his or bsr or hyg was inserted at this site. The regions of the probe used for Southern blot analysis are indicated. (B) Southern blot analysis. BamHI-digested genomic DNA prepared from wild-type, BLM+/– or BLM–/– cells was hybridized with the probes shown in (A). Lane 1, wild-type cells (+/+); lane 2, BLM+/– cells (+/–); lane 3, BLM–/– cells (–/–). (C) Northern blot analysis. Total RNA from wild-type or BLM–/– cells was hybridized with the chicken N- or C-terminal BLM cDNA probe synthesized by PCR as described in Materials and methods. The same filter was rehybridized with a chicken β-actin probe.
Since HR was reduced in RAD54–/– cells (Bezzubova et al., 1997), we generated BLM–/–/RAD54–/– clones from a BLM–/– clone. The two RAD54 targeting constructs containing selection markers of hygromycin or puromycin were sequentially transfected into a BLM–/– clone. The disruption of the RAD54–/–gene was also verified by Southern blotting (data not shown).
Proliferative properties of BLM–/–, RAD54–/– and BLM–/–/RAD54–/–cells
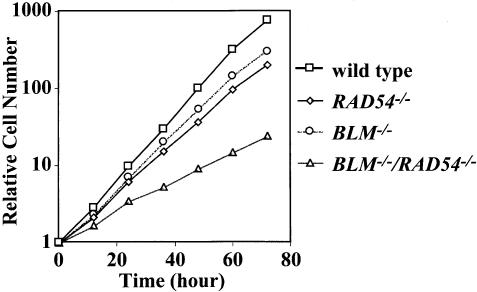
To examine the proliferative property, we monitored growth curves of wild-type, BLM–/–, RAD54–/– and BLM–/–/RAD54–/– cells (Figure 3). BLM–/–and RAD54–/– cells proliferated at a slightly lower rate than wild-type cells, while BLM–/–/RAD54–/– cells proliferated at a considerably lower rate than either single mutant. The doubling times of these cells were 7.4 ± 0.2, 8.2 ± 0.1, 8.9 ± 0.2 and 16.0 ± 0.5 h for wild-type, BLM–/–, RAD54–/– and BLM–/–/RAD54–/– cells, respectively.
Fig. 3. Growth curves of cells with various genotypes. Cells were inoculated into 3-cm dishes, and enumerated at the time indicated. The data show the average of the results from three independent experiments.
Sensitivity of BLM–/– cells to methyl methanesulfonate (MMS), VP16 and camptothecin (CAM)
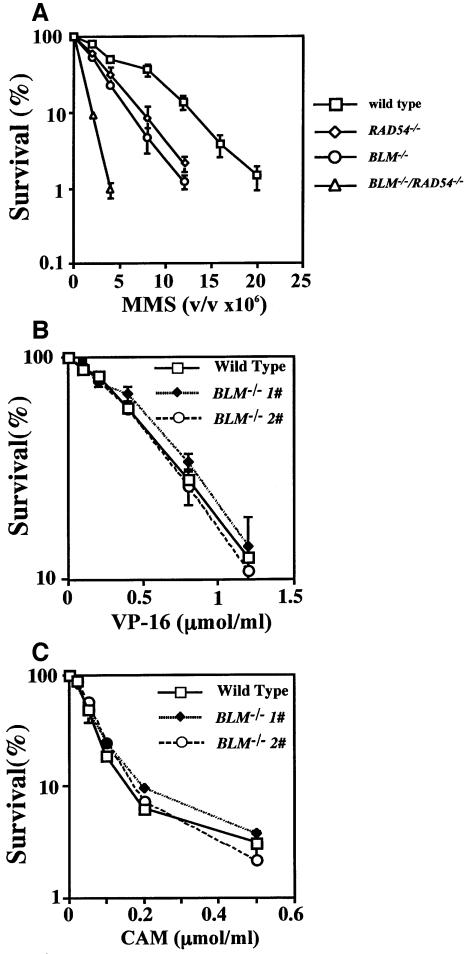
To examine the sensitivity of BLM–/– cells to MMS, they were grown in medium containing various concentrations of MMS. The BLM–/– cells were sensitive compared with wild-type cells (Figure 4A). Thus, we next examined the sensitivity of RAD54–/– and BLM–/–/RAD54–/– cells to MMS. RAD54–/–cells were moderately sensitive, like BLM–/– cells, and BLM–/–/RAD54–/– cells were highly sensitive to MMS.
Fig. 4. Sensitivity of cells with various genotypes to MMS, VP16 and CAM. Cells were treated with the indicated concentration of MMS (A), VP16 (B) or CAM (C) as described in Materials and methods. (A) The concentration of undiluted MMS (99%) is 1. The data show the average of the results from three independent experiments.
Since the yeast homolog for BLM, Sgs1, physically interacts with topo II and topo III and functional interaction between Sgs1 and topo I has been shown genetically, we examined the sensitivity of BLM–/– cells to the topo II inhibitor, VP16, which is known to induce DNA strand breaks and SCEs (Tominaga et al., 1986) and to the topo I inhibitor, CAM (Hsiang et al., 1989). As shown in Figure 4B and C, BLM–/– cells showed no significant difference in VP16 or CAM sensitivity compared with wild-type cells.
Increased SCE frequency in BLM–/– DT40 cells was reduced by disruption of the RAD54 gene
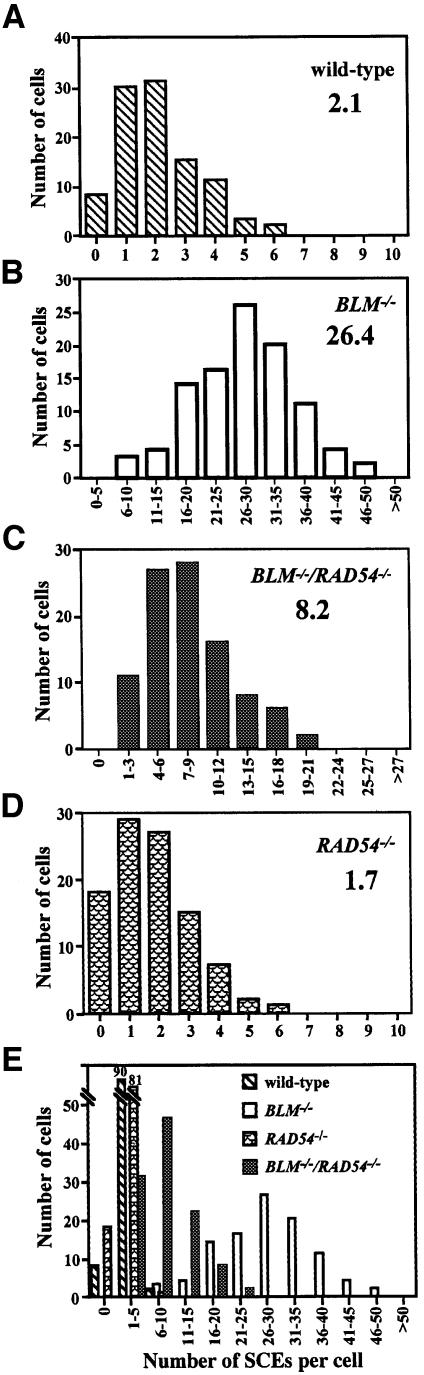
Lymphoblastoid cells from BS patients show a 10- to 15-fold higher number of SCEs than cells from normal individuals (Chaganti et al., 1974; reviewed by German, 1993). Thus, we examined the frequency of SCE in BLM–/– DT40 cells. Most of the large chromosomes of BLM–/– cells exhibited multiple SCEs as expected and wild-type cells showed a small number of SCEs. BLM–/– cells had 26.4 SCEs/cell on average (Figure 5B), while wild-type cells had only 2.1 SCEs/cell (Figure 5A).
Fig. 5. Histograms of SCE in wild-type, RAD54–/–, BLM–/– and BLM–/–/RAD54–/– cells. SCEs in the macrochromosomes were counted. Histograms show the frequency of cells with the indicated number of SCEs per cell. The mean number of SCEs per cell is shown in the upper right corner. (A) Wild-type cells; (B) BLM–/– cells; (C) BLM–/–/RAD54–/– cells; (D) RAD54–/– cells; (E) a superimposed figure of (A), (B), (C) and (D) expressed on the same scale.
Since it is not known whether the SCEs in BLM–/– cells are formed via HR, we measured the frequency of SCE in BLM–/–/RAD54–/– cells. BLM–/–/RAD54–/– cells showed 8.2 SCEs/cell (Figure 5C), and RAD54–/– cells showed 1.7 SCEs/cell (Figure 5D). Figure 5E shows Figure 5A–D expressed on the same scale. These results indicate that a considerable portion of the SCEs in BLM–/– cells are formed depending on Rad54 function, that is, they are formed via HR. However, BLM–/–/RAD54–/– cells still have a 4-fold higher frequency than the wild-type cells.
Targeted integration is increased in BLM–/– cells depending on RAD54
We next analyzed the targeted integration frequency using a RECQL/DNA helicase Q1 (RECQL1) targeting construct (Figure 6A), because the construct showed relatively low targeting efficiency and heterozygous cells displayed no defect in the recovery of clones that had undergone integration (our unpublished data). To analyze targeted integration events at the RECQL1 locus, genomic DNA of the transfectants was analyzed by Southern blotting (Figure 6B). The targeted integration frequency was considerably increased by disruption of BLM, from 32% (19/60) or 27% (16/60) for wild-type cells to 90% (54/60) or 82% (59/72) for BLM–/– cells (Table I). Disruption of the RAD54 gene of BLM–/– cells decreased the targeted integration frequency to 1.7% (1/60), indicating that the targeted integration in BLM–/– cells is Rad54-dependent HR.
Fig. 6. Southern blot analysis of drug-resistant clones. (A) Schematic representation of the wild-type allele and the targeted allele. The dotted line corresponds to the targeting construct. (B) The DNA samples were digested with BamHI and hybridized to the probe shown in (A). A typical Southern blot is shown. Arrowheads indicate the position of the fragment representing the targeted allele. +, targeted integration.
Table I. Targeted integration frequency.
| Targeting construct | Targeted integrated colonies/tested colonies |
||||
|---|---|---|---|---|---|
| Wild type | BLM–/– 1# | BLM–/– 2# | RAD54–/– | BLM–/–/RAD54–/– | |
| RECQL1-puro | |||||
| expt 1 | 7/24 | 21/24 | 22/24 | 0/48 | – |
| expt 2 | 12/36 | 33/36 | 32/36 | 0/48 | – |
| total | 19/60 (32%) | 54/60 (90%) | 54/60 (90%) | 0/96 (0) | – |
| transfection efficiency | 2.8 × 10–5 | 2.3 × 10–5 | 1.9 × 10–5 | 2.2 × 10–5 | |
| RECQL1-bleo | |||||
| expt 1 | 6/24 | 20/24 | – | 0/48 | 0/24 |
| expt 2 | 10/36 | 39/48 | – | 0/48 | 1/36 |
| total | 16/60 (27%) | 59/72 (82%) | – | 0/96 (0) | 1/60 (1.7%) |
| transfection efficiency | 2.2 × 10–5 | 1.6 × 10–5 | 2.1 × 10–5 | 1.3 × 10–5 | |
RECQL1 targeting constructs were transfected into cells of the genotypes indicated, and targeted integration events following selection were determined by Southern blot analysis and PCR.
Cell cycle analysis of BLM–/–/RAD54–/– cells
The growth curves shown in Figure 3 indicated that disruption of BLM and RAD54 had a synergistic effect on cell growth. Thus, we analyzed the cell-cycle phase distribution of asynchronous cells by flow cytometry. The cell-cycle phase distribution patterns for wild-type, RAD54–/– and BLM–/– cells were essentially the same, although the population in G2–M phase was increased slightly in RAD54–/–and BLM–/– cells compared with wild-type cells (Figure 7). In contrast, BLM–/–/RAD54–/– cells showed a remarkable increase in the G2–M phase population.
Fig. 7. Cell cycle analysis by flow cytometry. Distribution patterns of asynchronous culture of wild-type, BLM–/–, RAD54–/– and BLM–/–/RAD54–/– cells in the cell cycle.
Chromosome-type breaks/gaps are increased in BLM–/–/RAD54–/– cells
We next analyzed chromosomal breakage. DT40 cells display a stable karyotype with a modal chromosome number of 80 in total, which comprises 11 autosomal macrochromosomes, the ZW sex chromosomes and 67 microchromosomes (Sonoda et al., 1998). Since alterations in the minichromosomes are difficult to assess, analysis of chromosomal breakage was limited to the 11 autosomal macrochromosomes and the Z chromosome. BLM–/– and RAD54–/– cells showed slightly larger numbers of chromatid- and chromosome-type breaks/gaps than wild-type cells (Table II). BLM–/–/RAD54–/– cells exhibited many more chromosomal aberrations than either single gene mutant, that is, chromosome-type breaks/gaps were specifically increased in the double mutants.
Table II. Spontaneous chromosomes aberrations.
| Genotype | Chromatid breaks/gaps | Chromosome breaks/gaps | Chromatid exchanges | Total (per cell) |
|---|---|---|---|---|
| Wild type | 3 | 1 | 0 | 4 (0.013) |
| BLM–/– | 2 | 6 | 0 | 8 (0.027) |
| RAD54–/– | 10 | 4 | 0 | 14 (0.047) |
| BLM–/–RAD54–/– | 3 | 46 | 0 | 49 (0.163) |
Spontaneous chromosomal aberrations were analyzed in 300 cells of the genotypes indicated.
Discussion
BLM–/– DT40 cells show phenotypes characteristic of human BS cells
The BLM–/– DT40 cells showed the slow growth phenotype, a higher sensitivity to MMS and an elevated frequency of SCE, which are characteristic phenotypes of human BS cells (reviewed by German, 1993). The helicase domain of chicken BLM showed 84.4 and 83.8% identity to those of human and mouse BLM, respectively. These results suggest that the structure and function of BLM are conserved in vertebrates.
The results shown in Figure 5 indicate for the first time that a considerable portion of the SCEs in BLM–/– cells are formed via a process requiring Rad54 function, HR. The mechanism of SCE in BLM–/–/RAD54–/– cells is not clear at present. One possibility is that Rad54B, a homolog of Rad54, functions in place of Rad54 (Hiramoto et al., 1999) and another possibility is that the SCEs are formed by topo II (Ishii and Bender, 1980; Heartlein et al., 1987; Dillehay et al., 1989). These possibilities should be addressed in a future study.
BLM suppresses homologous recombination
Disruption of the BLM gene resulted in a remarkable increase in the targeted integration frequency (Table I), indicating that specific inhibitors for BLM, if any exist, would increase the targeted integration frequency of higher eukaryotic cells. As expected, the elevated targeted integration in BLM–/– cells was abolished by disruption of RAD54. In budding yeast sgs1 disruptants, the frequency of illegitimate recombination (Yamagata et al., 1998) as well as HR was elevated (Gangloff et al., 1994; Watt et al., 1996). In addition, an increase in illegitimate recombination was observed in Escherichia coli recQ mutants (Hanada et al., 1997). However, in the BLM disruptants, only targeted integration, i.e. HR, increased, indicating that BLM mainly suppresses HR.
Sgs1 and RecQ are the sole RecQ helicase in budding yeast and E.coli, respectively, and they carry out multiple functions. In contrast, Caenorhabditis elegans, Drosophila melanogaster and higher eukaryotes, including vertebrates, have multiple RecQ helicase genes (Kusano et al., 1999; Sekelsky et al., 1999). In human cells, five genes encoding RecQ homologs have been identified: RECQL1 (Puranam and Blackshear, 1994; Seki et al., 1994), BLM (Ellis et al., 1995), WRN (Yu et al., 1996), RECQL4 and RECQL5 (Kitao et al., 1998). Recently, RECQL4 has been shown to be a causative gene of Rothmund–Thomson’s syndrome (Kitao et al., 1999). Thus it seems likely that some of the five RecQ homologs share the functions carried out by Sgs1 in higher eukaryotic cells. In this context, it is interesting to determine which RecQ homolog suppresses illegitimate recombination.
Slow growth phenotype of BLM–/–/Rad54–/– cells is caused by DNA lesions to induce chromosome-type breaks/gaps
Flow cytometric analysis showed an accumulation of BLM–/–/RAD54–/– cells in G2–M phase, and a high incidence of chromosome-type breaks/gaps was observed in these cells. It is suggested that chromosome-type breaks are caused by DSBs generated prior to and during DNA replication, while chromatid-type breaks are caused by DSBs generated following DNA replication. Previous studies showed delayed DNA-chain maturation in BS cells (Hand and German, 1975; Giannelli et al., 1977). Considering both of the above, it seems likely that DSBs are generated in BLM–/–/RAD54–/– cells during DNA replication.
Chromosome analysis showed that ∼15% of BLM–/–/RAD54–/– cells contain chromosome-type breaks/gaps, and this value is probably underestimated because we can only analyze the chromosomes of the cells that passed through the the G2 phase. A single unrepaired DSB is sufficient to induce cell death in yeast (reviewed by Game, 1993), and RAD51–/– DT40 cells died rapidly showing chromosome-type breaks after inhibition of the expression of exogenous Rad51 (Sonoda et al., 1998). Therefore, a substantial fraction of BLM–/–/RAD54–/– cells are dying or dead.
A possible mechanism for the increase of SCE and targeted integration in BLM–/– cells
Zou and Rothstein (1997) showed that Holliday junctions accumulate spontaneously during DNA replication in mitotically growing yeast. Recently, a model for the occurrence of DSBs at arrested replication forks in E.coli was proposed (Seigneur et al., 1998). According to this model, Holliday junctions are formed by the annealing of two newly synthesized DNAs at arrested replication forks but are not formed via DSBs, and DSBs are formed by cleavage of the Holliday junctions. Indeed, annealed molecules consisting of two newly synthesized DNA were detected in the cells from BS patients (Waters et al., 1978). The BLM homolog of S.cerevisiae, Sgs1, has been shown to possess an activity to disrupt Holliday junctions (Bennett et al., 1999). If Holliday junctions are formed by the above mechanism, and the physiological function of BLM is to disrupt Holliday junctions, the defect of BLM will cause the formation of more Holliday junctions and more DSBs. In fact, induction of DSBs during DNA replication was indicated by the increase in chromosome-type DNA breaks/gaps in BLM–/–/RAD54–/– cells. It is therefore likely that the majority of DSBs formed during DNA replication due to the defect of BLM function are repaired by HR, resulting in an increase in SCE in Rad54-proficient cells.
The induction of a DSB in the genome by the expression of a restriction enzyme increased the allelic recombination frequency by a few hundred-fold, suggesting that induced DSBs stimulate allelic recombination in mammalian cells (Moynahan and Jasin, 1997). In this context, it is interesting that the allelic recombination frequency is increased in BS cells (Ellis et al., 1995). In addition, artificially introduced DSBs in the chromosomal locus of mammalian cells highly stimulate targeted integration (Smih et al., 1995). Thus, it is likely that DSBs transiently formed in the chromosomal locus in BLM–/– cells stimulate targeted integration.
Materials and methods
Construction of targeting vectors
A partial chicken BLM cDNA was obtained by RT–PCR of chicken testis RNA using primers synthesized based on the sequences in motifs I and VI of mouse BLM helicase domains (Seki et al., 1998). A partial N-terminal region and the C-terminal region of chicken BLM were obtained by 5′-RACE and 3′-RACE, respectively. We obtained an ∼3.5 kb chicken BLM cDNA fragment including the helicase domain. The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. AB040747. A partial genomic DNA segment corresponding to the helicase domain was amplified with the genomic DNA of DT40 cells as a template by long-range PCR. Chicken BLM targeting constructs, BLM-his and BLM-hyg and BLM-bsr, were made by replacing the helicase motif III with histidinol (his)-, hygromycin (hyg)-, or blasticidin (bsr)-selection marker cassettes. Chicken RAD54 targeting constructs were made as described previously using hyg and puromycin as selection markers (Bezzubova et al., 1997).
Cell culture and DNA transfection
Cells were cultured in RPMI 1640 supplemented with 100 µg/ml kanamycin, 10% fetal bovine serum and 1% chicken serum (Sigma, St Louis, MO) at 39.5°C. For gene targeting, 107 cells were electroporated with 30 µg of linearized BLM targeting constructs using a Gene Pulser apparatus (Bio-Rad, Hercules, CA) at 550 V and 25 µF. Drug-resistant colonies were selected in 96-well plates with medium containing 1 mg/ml his, 2 mg/ml hyg or 20 µg/ml bsr. Genomic DNA was isolated from drug-resistant clones. Gene disruption was confirmed by Southern and northern blot analysis.
Northern blotting
Northern blotting was performed according to the manual of Multiple Tissue Northern Blot (Clontech). Total RNA from wild-type or BLM–/– cells was hybridized with the chicken N- or C-terminal BLM cDNA probe synthesized by PCR. The primers used were forward primer 5′-ACCAGCGTGTGTCTCTGCTG-3′ and reverse primer 5′-GGATAACATAGCGTACGTCAG-3′ for the N-terminal probe, and forward primer 5′-CTATCATGCTGGCCTCACTG-3′ and reverse primer 5′-TGTCTACCATCATATTCAGTGTG-3′ for the C-terminal probe.
Measurements of MMS, VP16 and CAM sensitivity
To determine sensitivity to MMS, 3 × 102 cells were inoculated into dishes containing various concentrations of MMS in a medium supplemented with 1.5% (w/v) methylcellulose, 1.5% chicken serum and 15% fetal bovine serum. To determine sensitivity to VP16 and CAM, cells were treated with various concentrations of VP16 or CAM for 4 h, washed twice with PBS and then inoculated (3 × 102 cells) into dishes containing the growth medium supplemented with 1.5% methylcellulose. The colonies were enumerated 14 and 9 days after inoculation for BLM–/–/RAD54–/– cells and other cells, respectively. Survival was determined by comparing the number of colonies of untreated cells.
SCE analysis
Cells (5 × 105) were cultured for approximately two cycle periods with culture medium containing 10 µM bromodeoxyuridine (BrdU) and pulsed with 0.1 µg/ml colcemid for the last 2–3 h. The cells were harvested and treated with 75 mM KCl for 20 min at room temperature and then fixed with methanol–acetic acid (3:1) for 30 min. The cells were washed once with the fixative and suspended in a small volume of the fixative. The cell suspension was dropped onto ice-cold wet glass slides and air-dried. The cells on the slides were incubated with 10 µg/ml Hoechst 33258 in phosphate buffer pH 6.8 for 20 min and rinsed with MacIlvaine solution (164 mM Na2HPO4, 16 mM citric acid pH 7.0). The cells were exposed to black light (λ = 352 nm) at a distance of 1 cm for 30 min and incubated in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate) solution at 62°C for 30 min and then stained with 3% Giemsa solution at pH 6.8 for 10 min.
Chromosome aberration analysis
Cells were treated with 0.1 µg/ml colcemid for 3 h and harvested. The cells were treated with 75 mM KCl for 20 min at room temperature and fixed with methanol–acetic acid (3:1) for 30 min. The cell suspension was dropped onto ice-cold wet glass slides and air-dried. The cells on the slides were stained with 3% Giemsa solution at pH 6.8 for 10 min and examined with a light microscope.
Measurement of targeted integration frequency
To analyze targeted integration events at the chicken RECQL1 locus, targeting constructs (chicken RECQL1-puromycin or RECQL1-bleomycin) were transfected into cells, and cells were selected with 0.5 µg/ml puromycin or 1.5 mg/ml zeocin, a derivative of bleomycin. Genomic DNA of drug-resistant clones was isolated and targeted integration was confirmed by Southern blot analysis and PCR.
Analysis of cell-cycle phase distribution
Cells were prepared with CycleTESTTM PLUS DNA Reagent Kit (Becton Dickinson, CA, USA). Subsequent flow-cytometric analysis was performed with a FACScan (Becton Dickinson). Data were analyzed using the CellFIT software (Becton Dickinson).
Acknowledgments
Acknowledgements
We thank Dr O.Nakabayashi for providing the chicken testis RNA. This work was supported by Grants-in-Aid for Scientific Research and for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, Health Sciences Research Grants from the Ministry of Health and Welfare of Japan, and the Mitsubishi Foundation.
References
- Bennett R.J., Sharp,J.A. and Wang,J.C. (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem., 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a Rad54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg,S. and German,J. (1974) A many-fold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl Acad. Sci. USA, 71, 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J.E. (1981) Correlations between sister chromatid exchange frequencies and replicon sizes. A model for the mechanism of SCE production. Exp. Cell Res., 136, 27–30. [DOI] [PubMed] [Google Scholar]
- Dillehay L.E., Jacobson-Kram,D. and Williams,J.R. (1989) DNA topoisomerases and models of sister-chromatid exchange. Mutat. Res., 215, 15–23. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- Essers J., HenDriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse Rad54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Game J.C. (1993) DNA double-strand breaks and the RAD50–RAD57 genes in Saccharomyces. Semin. Cancer Biol., 4, 73–83. [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart E., Bauer,R., Raub,U., Schinzel,M., Ruprecht,K.W. and Jonas,J.B. (1988) Spontaneous and induced chromosomal instability in Werner syndrome. Hum. Genet., 80, 135–139. [DOI] [PubMed] [Google Scholar]
- German J. (1993) Bloom’s syndrome: a mendelian prototype of somatic mutational disease. Medicine, 72, 393–406. [PubMed] [Google Scholar]
- Giannelli F., Benson,P.F., Pawsey,S.A. and Polani,P.E. (1977) Ultraviolet light sensitivity and delayed DNA-chain maturation in Bloom’s syndrome fibroblasts. Nature, 265, 466–469. [DOI] [PubMed] [Google Scholar]
- Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nature Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- Hanada K., Ukita,T., Kohno,Y., Saito,K., Kato,J. and Ikeda,H. (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. and German,J. (1975) A retarded rate of DNA chain growth in Bloom’s syndrome. Proc. Natl Acad. Sci. USA, 72, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heartlein M.W., Tsuji,H. and Latt,S.A. (1987) 5-Bromodeoxyuridine-dependent increase in sister chromatid exchange formation in Bloom’s syndrome is associated with reduction in topoisomerase II activity. Exp. Cell Res., 169, 245–254. [DOI] [PubMed] [Google Scholar]
- Hiramoto T. et al. (1999) Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene, 18, 3422–3426. [DOI] [PubMed] [Google Scholar]
- Hsiang H., Liu,L.F., Wall,M.E., Wani,M.C., Nicholas,A.W., Manikumar,G., Kirschenbaum,S., Silber,R. and Potmesil,M. (1989) DNA topoisomerase I-mediated DNA cleavage and cytotoxicity of camptothecin analogues. Cancer Res., 49, 4385–4389. [PubMed] [Google Scholar]
- Ishii Y. and Bender,A. (1980) Effects of inhibitors of DNA synthesis on spontaneous and ultraviolet light-induced sister-chromatid exchanges in Chinese hamster cells. Mutat. Res., 79, 19–32. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′ to 5′ DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- Kitao S., Ohsugi,I., Ichikawa,K., Goto,M., Furuichi,Y. and Shimamoto,A. (1998) Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics, 54, 443–452. [DOI] [PubMed] [Google Scholar]
- Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- Kusano K., Berres,M.E. and Engels,W.R. (1999) Evolution of the RECQ family of helicases: A Drosophila homolog, Dmblm, is similar to the human Bloom syndrome gene. Genetics, 151, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Mullen,J.R., Brill,S.J., Kleff,S., Romeo,A.M. and Sternglanz,R. (1996) Human homologues of yeast helicase. Nature, 383, 678–679. [DOI] [PubMed] [Google Scholar]
- Martin G.M. (1982) Syndromes of accelerated aging. Natl Cancer Inst. Monogr., 60, 241–247. [PubMed] [Google Scholar]
- Moynahan M.E. and Jasin,M. (1997) Loss of heterozygosity induced by a chromosomal double-strand break. Proc. Natl Acad. Sci. USA, 94, 8988–8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter R.B. (1980) A replication model for sister-chromatid exchange. Mutat. Res., 70, 337–341. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Zwelling,L.A., Kao-Shan,C.S., Whang-Peng,J. and Bradley,M.O. (1985) Correlation between intercalator-induced DNA strand breaks and sister chromatid exchanges, mutations and cytotoxicity in Chinese hamster cells. Cancer Res., 45, 3143–3149. [PubMed] [Google Scholar]
- Puranam K.L. and Blackshear,P.J. (1994) Cloning and characterization of RecQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem., 269, 29838–29845. [PubMed] [Google Scholar]
- Seigneur M., Bidnenko,V., Ehrlich,S.D. and Michel,B. (1998) RuvAB acts at arrested replication forks. Cell, 95, 419–430. [DOI] [PubMed] [Google Scholar]
- Sekelsky J.J., Brodsky,M.H., Rubin,G.M. and Hawley,R.S. (1999) Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res., 27, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M. et al. (1994) Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli RecQ helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res., 22, 4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T., Wang,W.S., Okumura,N., Seki,M., Katada,T. and Enomoto,T. (1998) cDNA cloning of mouse BLM gene, the homologue to human Bloom’s syndrome gene, which is highly expressed in the testis at the mRNA level. Biochim. Biophys. Acta, 1398, 377–381. [DOI] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa,T. (1995) Homologous recombination and the roles of double-strand breaks. Trends Biochem. Sci., 20, 387–391. [DOI] [PubMed] [Google Scholar]
- Smih F., Rouet,P., Romanienko,P.J. and Jasin,M. (1995) Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res., 23, 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K., Shinkai,T., Saijo,N., Nakajima,T., Ochi,H. and Suemasu,K. (1986) Cytogenetic effects of etoposide (VP-16) on human lymphocytes; with special reference to the relation between sister chromatid exchange and chromatid breakage. Jpn. J. Cancer Res., 77, 385–391. [PubMed] [Google Scholar]
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- Waters R., Regan,J.D. and German,J. (1978) Increased amounts of hybrid (heavy/heavy) DNA in Bloom’s syndrome fibroblasts. Biochem. Biophys. Res. Commun., 83, 536–541. [DOI] [PubMed] [Google Scholar]
- Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E.coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- Watt P.M., Hickson,I.D., Borts,R.H. and Louis,E.J. (1996) Sgs1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics, 144, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K., Kato,J.I., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: Implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Sonoda,E., Buerstedde,J.M., Bezzubova,O., Morrison,C., Takeda,M., Shinohara,A. and Takeda,S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in Rad52. Mol. Cell. Biol., 18, 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein,R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]