Abstract
Huntington disease (HD) is a neurodegenerative disorder caused by an expansion of polyglutamines in the first exon of huntingtin (HTT), which confers aggregation-promoting properties to amino-terminal fragments of the protein (N-HTT). Mutant N-HTT aggregates are enriched for ubiquitin and contain ubiquitin E3 ligases, thus suggesting a role for ubiquitination in aggregate formation. Here, we report that tumor necrosis factor receptor-associated factor 6 (TRAF6) binds to WT and polyQ-expanded N-HTT in vitro as well as to endogenous full-length proteins in mouse and human brain in vivo. Endogenous TRAF6 is recruited to cellular inclusions formed by mutant N-HTT. Transient overexpression of TRAF6 promotes WT and mutant N-HTT atypical ubiquitination with Lys6, Lys27, and Lys29 linkage formation. Both interaction and ubiquitination seem to be independent from polyQ length. In cultured cells, TRAF6 enhances mutant N-HTT aggregate formation, whereas it has no effect on WT N-HTT protein localization. Mutant N-HTT inclusions are enriched for ubiquitin staining only when TRAF6 and Lys6, Lys27, and Lys29 ubiquitin mutants are expressed. Finally, we show that TRAF6 is up-regulated in post-mortem brains from HD patients where it is found in the insoluble fraction. These results suggest that TRAF6 atypical ubiquitination warrants investigation in HD pathogenesis.
Keywords: E3 Ubiquitin Ligase, Huntington Disease, Polyglutamine Disease, TRAF, Ubiquitin
Introduction
Huntington disease (HD)2 is a dominantly inherited neurodegenerative disorder caused by the expansion of a polymorphic CAG sequence in the first exon of the gene encoding for huntingtin (HTT) protein (1). The onset of HD symptoms, including motor dysfunction and cognitive decline, occurs when the length of CAG exceeds 36 repeats (2). CAG triplets are translated into a series of uninterrupted glutamine residues (polyQ) at the N terminus of HTT, a ubiquitously expressed protein of ∼350 kDa. HTT is found both in the nucleus and in the cytoplasm where it associates with a variety of organelles, including endoplasmic reticulum, Golgi apparatus, and mitochondria (3–5). This widespread subcellular localization, associated with the ability to interact with a large number of proteins, implicates HTT in diverse cellular activities ranging from transcription and energy metabolism to vesicle trafficking (6).
The elongation of the polyQ sequence in HTT protein induces HD pathogenesis largely through a gain-of-function mechanism. One hallmark of HD is the presence of neuronal aggregates that contain N-terminal fragments of polyQ HTT (N-HTT). Aggregates or inclusions are found primarily in the striatum and in the cortex of HD patients and are located mainly in nuclear, perinuclear, and cytoplasmic regions of affected neurons (7). Although it is still unclear whether inclusions are protective or detrimental for cell viability, their formation seems to be strikingly connected to the propensity of polyQ N-HTT to misfold. In addition to mutant N-HTT, inclusions are enriched for components of the protein quality control machinery, such as ubiquitin, proteasome subunits, and chaperones (7, 8).
An extensive proteomic study has demonstrated that global changes to the ubiquitin system are associated with HD pathology (9). Although the E3 ligase Hdr1 and the ubiquitin-conjugating enzymes E2–25K interact and ubiquitinate both WT and mutant HTT (10, 11), parkin and CHIP (C terminus of Hsc70-interacting protein) selectively act on the mutant protein (12, 13). All of these are localized in the aggregates of HD post-mortem brains and are able to decrease aggregate formation in vitro.
Ubiquitin contains seven lysine residues, and it is assumed that all of these can be involved in the formation of polyubiquitin chains on target proteins. Canonical Lys48- and Lys63-linked polyubiquitination has been related respectively to proteasome degradation of the substrate and to cell signaling or aggresome formation (14). The role of nonconventional chains via Lys6, Lys11, Lys27, Lys29, and Lys33 is much less clear (15). It has been shown that Lys6-specific modification can protect substrates from proteolysis (16, 17) or regulate their enzymatic activity (18). Similarly, nondegradative functions have been associated with Lys27 and Lys29 chains, which have been shown to control subcellular localization and activity of targeted proteins (19–21).
Tumor necrosis factor receptor-associated factor 6 (TRAF6) is an E3 ubiquitin ligase that promotes Lys63-specific chain assembly in the signal transduction pathway that leads ultimately to NFκB activation (22). In the brain, TRAF6 activity has been associated with the transduction cascade of the neurotrophin receptors p75 and TrkA (23, 24). Interestingly, TRAF6 localizes to Tau inclusions and Lewy bodies in brains of Alzheimer disease and Parkinson disease (PD) patients, respectively (25, 26), supporting the idea that E3 ligases tend to accumulate at the site of protein inclusions. Furthermore, TRAF6 was found to promote atypical Lys6-, Lys27-, and Lys29-mediated poly-ubiquitination of mutant DJ-1 and α-synuclein, proteins relevant for PD pathogenesis. Atypical ubiquitin ligase activity of TRAF6 was shown to induce mutant DJ-1 aggregation (26).
In the current study, we provide evidence that TRAF6 interacts with and ubiquitinates WT and mutant HTT. In analogy to PD targets, TRAF6 promotes an atypical mode of polyubiquitin chain formation onto N-HTT proteins with selective choice of Lys6, Lys27, and Lys29 linkages. We also show that both overexpressed and endogenous TRAF6 is recruited at the site of mutant N-HTT aggregates. Furthermore, TRAF6-mediated ubiquitination stimulates the formation of mutant N-HTT inclusions. Only when atypical Lys6, Lys27, and Lys29 ubiquitins are expressed, N-HTT aggregates are strongly positive for ubiquitin. Finally, in post-mortem brains of HD patients, TRAF6 expression is induced, and the protein preferentially accumulates in the insoluble fraction. Altogether, our data imply a novel role for TRAF6 and atypical ubiquitination in HD pathogenesis.
EXPERIMENTAL PROCEDURES
Plasmids and Cells
N-HTT-GFP Gln21 and Gln150 constructs were described previously (27). N-HTT-GFP Gln60 was a serendipitous construct obtained during Gln150 plasmid preparation. It was sequence-verified and controlled during each preparation. FLAG-TRAF6 (WT and DN) and HA-ubiquitin (WT and mutants) were described elsewhere (26).
HEK 293 cells (Sigma) were maintained in culture in DMEM (Invitrogen) with 10% FBS (Sigma). Transfections were performed with the standard calcium phosphate method or with Lipofectamine 2000 (Invitrogen), as required.
Immunoprecipitation and Western Blot
For co-immunoprecipitation experiments, cells were lysed in TRAF6 buffer (200 mm NaCl, 50 mm Tris, pH 7.5, 0.5% Nonidet P-40, 10% glycerol) supplemented with anti-protease mixture (Roche Applied Science) and 5 mm N-ethylmaleimide. Cell lysates were incubated with anti-FLAG-agarose beads (Sigma) or with anti-GFP antibody (Invitrogen). For co-immunoprecipitation of endogenous proteins anti-HTT mouse monoclonal (Chemicon, catalog nos. MAB 2166 and MAB 5490) and rabbit polyclonal HP1 (27) antibodies were used. Cortex was dissected from homozygous HdhQ111 knock-in mice (28) or control littermates, snap-frozen on liquid nitrogen, and stored at −80 °C. Parietal cortex from HD patients was used for co-immunoprecipitation of endogenous proteins in the human brain sample. Lysates were prepared homogenizing tissue in modified TRAF6 buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 0.5% Nonidet P-40, 10% glycerol) supplemented with anti-protease mixture. Total protein content was measured, and an equivalent amount was used for co-immunoprecipitation. After washing, immunoprecipitated proteins were eluted with 2× SDS sample buffer, boiled, and analyzed by Western blot. The following antibodies were used: anti-FLAG (1:2000; Sigma), anti-GFP (1:1000; Invitrogen), anti-HA (1:1000; from hybridoma supernatant, kindly provided by Dr. Licio Collavin), anti-HTT (Chemicon, catalog no. MAB 2166), anti-TRAF6 (Santa Cruz Biotechnology, catalog no. sc-7221), and anti-β-actin (1:5000; Sigma). For detection, anti-mouse-HRP and anti-rabbit-HRP (Dako) or protein A-HRP (Upstate) in combination with ECL (GE Healthcare) were used. For total protein extracts, cells were lysed directly in 2× SDS sample buffer and analyzed by Western blot.
Cell-based Ubiquitination Assay
For cell-based ubiquitination assays, HEK cells were transfected with HA-ubiquitin, FLAG-TRAF6 (WT or DN), and N-HTT-GFP constructs. After transfection, cells were lysed with radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 1% Triton X-100, 1% deoxycholic acid, 0.1% SDS), and samples were briefly sonicated. After centrifugation, clear lysates were immunoprecipitated with anti-GFP or anti-FLAG antibodies. Immunocomplexes were analyzed by Western blot using anti-HA antibody to detect ubiquitin conjugates. For development, protein A-HRP was used.
Immunocytochemistry
Indirect immunofluorescence was carried out following standard methods (29). Anti-FLAG (1:1000; SIGMA), anti-ubiquitin (1:50; Dako) and anti-TRAF6 (1:100; catalog no. sc-8409, Santa Cruz Biotechnology) antibodies were used. For detection, Alexa Fluor 405 (blue) or 594 (red) (Invitrogen) labeled anti-mouse or anti-rabbit antibodies were used. All images were collected using a confocal microscope (Leica TCS SP2). The analysis of aggregates was performed on high resolution images using ImageJ software. At least 200 cells from two independent experiments were counted and scored for percentage of cells with aggregates. For the analysis of aggregate size, only cells (n = 200) with comparable levels of TRAF6 expression, as determined by fluorescence intensity normalized to cell area, were used. N-HTT cytoplasmic staining was quantified on the same cells on a randomly selected area. Background fluorescence was quantified from an area placed outside the cells and was subtracted to each signal.
Human Post-mortem Brain Samples
Control and HD post-mortem brain tissues used in this study were collected by the Harvard Brain Tissue Resource Center, McLean Hospital (Belmont, MA) and described previously (30). HD brains were assigned Vonsattel grade 3 pathology (31). Post-mortem intervals were from 12 to 32 h for controls and from 8 to 30 h for HD brains. Total proteins were extracted from parietal cortex in 10% SDS. Fractionation in soluble and insoluble protein extracts was performed from the cortex as follows; soluble fractions were extracted in 150 mm sucrose, 15 mm Hepes, pH 7.9, 60 mm KCl, 15 mm NaCl, 5 mm EDTA, 1 mm EGTA, 1% Triton X-100), supplemented with protease inhibitors (Roche Applied Science). After centrifugation, the soluble fraction was removed, and the insoluble fractions were extracted from pellets in 10% SDS. For Western blot analysis, anti-TRAF6 polyclonal (Santa Cruz Biotechnology, catalog no. sc-7221) and monoclonal (Santa Cruz Biotechnology, catalog no. sc-8409) antibodies were used.
Quantitative Real-time PCR
Total RNA from cortex of pathologically confirmed HD and control subjects was isolated using the TRIzol reagent (Invitrogen). cDNA was obtained from 1 μg of purified RNA using the iSCRIPTTM cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR was performed using SYBR-Green PCR Master Mix (Applied Biosystem). Expression of TRAF6 was analyzed as already described (26).
Statistical Analysis
All experiments were repeated in duplicate or more. Confocal images were analyzed with ImageJ software. Western blot scans were quantified with Adobe Photoshop CS3. Data represent the mean with standard deviation. When necessary, each group was compared individually with reference to the control group using Student's t test (Microsoft Excel software).
RESULTS
E3 Ubiquitin Ligase TRAF6 Interacts with WT and Mutant HTT and Associates with Polyglutamine Aggregates
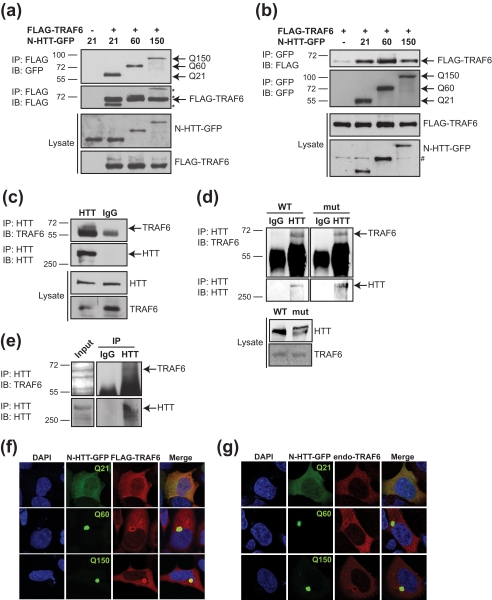
To evaluate whether TRAF6 might have a role in HD, we studied its ability to bind huntingtin amino-terminal fragments (residues 1–171) fused to green fluorescent protein (N-HTT-GFP), with either a physiological (Gln21), or short (Gln60) or long (Gln150) pathological polyQ stretch. N-HTT-GFP constructs were transfected in HEK 293 cells with FLAG-TRAF6 or FLAG-empty vector, as control. Immunoprecipitation of transfected cell lysates revealed a specific binding between TRAF6 and N-HTT with no evident selectivity for polyQ length (Fig. 1a). Reverse co-immunoprecipitation of transfected proteins confirmed that TRAF6 physically associated with N-HTT with a physiological and pathological polyQ stretch (Fig. 1b).
FIGURE 1.
TRAF6 interacts with WT and mutant N-HTT in vitro and full-length proteins in vivo and accumulates in mutant N-HTT aggregates. a, HEK 293 cells were transfected with FLAG-TRAF6, and the huntingtin N-terminal fragment fused to GFP (N-HTT-GFP) with WT (Gln21, Q21) or mutated (Gln60, Q60 and Gln150, Q150) polyglutamine expansion. Lysates were immunoprecipitated (IP) with anti-FLAG beads, and bound proteins were revealed by immunoblot (IB) with anti-GFP, Q150 and anti-FLAG antibodies. Lysates were tested for the expression of TRAF6 and N-HTT proteins. Molecular mass markers (kDa) are indicated on the left. b, cells were transfected with FLAG-TRAF6 and N-HTT-GFP constructs as indicated. Lysates were immunoprecipitated with anti-GFP. Bound proteins and lysates were analyzed with anti-FLAG and anti-GFP antibodies. An asterisk represents N-HTT bands from previous development of the same gel. # represents an unspecific band. c, HEK 293 cell lysates were immunoprecipitated with anti-HTT or control IgG (as indicated), and bound endogenous proteins were revealed with anti-TRAF6 and anti-HTT antibodies. d, the cortex from WT and homozygous HdhQ111 (mut) mice was dissected, lysed, and used for immunoprecipitation of endogenous TRAF6 and full-length HTT proteins. e, the parietal cortex from HD post-mortem brain was lysed and processed for co-immunoprecipitation of endogenous proteins as in d. f, HEK 293 cells were transfected as in a. TRAF6 was visualized by indirect immunofluorescence with anti-FLAG antibody (red). N-HTT was visible by GFP autofluorescence (green). Nuclei were visualized with DAPI (blue). g, cells were transfected with N-HTT-GFP constructs. Endogenous TRAF6 was stained with anti-TRAF6 antibody (red).
We then performed co-immunoprecipitation experiments in HEK 293 cells on endogenous proteins proving TRAF6 interaction with full-length WT HTT in vitro (Fig. 1c). The cortex from control WT and HdhQ111 knock-in mice were also analyzed, showing that TRAF6 specifically binds full-length HTT in vivo with no preference for WT or expanded polyQ-containing protein (Fig. 1d). This interaction was confirmed in vivo in the parietal cortex from HD post-mortem brains (Fig. 1e).
We then analyzed TRAF6 localization in cells that express WT or mutant N-HTT. WT N-HTT was located both in the cytoplasm and the nucleus, with a diffused staining throughout the cell. TRAF6 was mainly present in the cytoplasm and partially in the nucleus, where it co-localized with WT N-HTT (Fig. 1, f and g). Similar data were obtained with overexpressed and endogenous TRAF6. Overexpression of WT N-HTT did not alter TRAF6 cellular localization (data not shown). When transfected in cells, mutant N-HTT-GFP (Gln60 and Gln150) developed fluorescent aggregates, as expected, located in the cytoplasm and concentrated in perinuclear regions. The presence of mutant N-HTT aggregates induced the recruitment of TRAF6 at the site of N-HTT inclusions, with a more prominent accumulation at the border (Fig. 1, f and g).
Altogether, these results demonstrate that TRAF6 binds huntingtin in vitro and in vivo in a polyQ length-independent manner. Furthermore, mutant protein can sequester TRAF6 into cellular aggregates.
TRAF6 Facilitates Atypical Polyubiquitination of WT and Mutant N-HTT
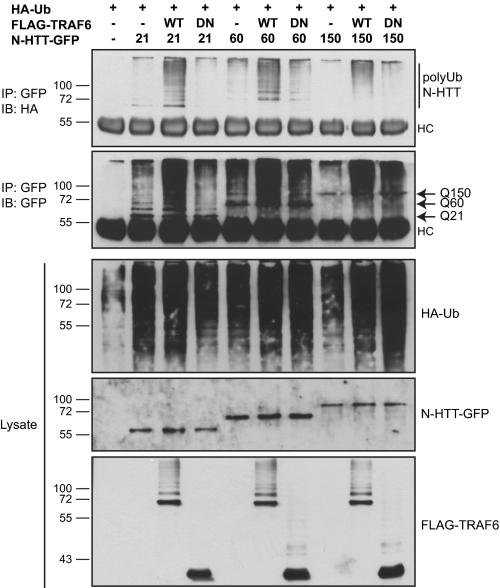
The interaction between TRAF6 and N-HTT and their co-localization in polyglutamine aggregates raised the possibility that N-HTT might be target of TRAF6 E3 ubiquitin ligase activity. To test this hypothesis, we performed ubiquitination assay in HEK 293 cells. Cells were transfected with HA-ubiquitin, N-HTT-GFP with 21, 60, or 150 polyQ segment and with or without FLAG-TRAF6. A TRAF6 mutant deleted of the N-terminal E3 ligase domain (DN) also was included as control. We found that overexpression of WT TRAF6, but not of DN mutant, facilitated the formation of high molecular weight polyubiquitinated forms of N-HTT (Fig. 2). TRAF6 activity was evident toward N-HTT with physiological length of polyQ stretch (Gln21) and for pathological mutants (Gln60 and Gln150). Therefore, TRAF6 ubiquitin-ligase activity did not seem to be polyQ-dependent, as already seen for its binding capability.
FIGURE 2.
TRAF6 enhances ubiquitination of WT and mutant N-HTT. For ubiquitination assay, HEK 293 cells were transfected with HA-ubiquitin and N-HTT-GFP Gln21 (Q21), Gln60 (Q60), and Gln150 (Q150) constructs with WT or DN FLAG-TRAF6. Lysates were immunoprecipitated (IP) with anti-GFP antibody, and ubiquitinated N-HTT protein was revealed with anti-HA. Input lysates were analyzed with anti-HA, anti-GFP, and anti-FLAG antibodies. Molecular mass markers (kDa) are indicated on the left. HC, heavy chain; IB, immunoblot; Ub, ubiquitin.
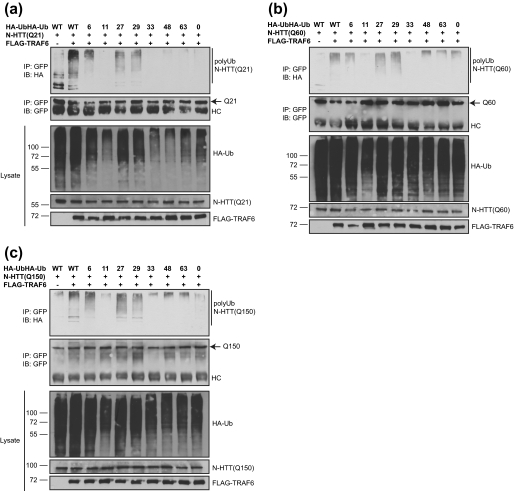
Because we have shown recently that TRAF6 can promote unconventional ubiquitin linkages on target proteins (26), we investigated the mode of polyubiquitination mediated by TRAF6 onto N-HTT. We thus took advantage of a set of ubiquitin mutants that we have generated with only one lysine available for chain formation (remaining lysine residues were substituted with arginine). An ubiquitin mutant with all lysines substituted with arginine was also included as negative control (Lys0). Cell-based ubiquitination assay was performed with all N-HTT constructs with or without TRAF6 and in combination with ubiquitin mutants. TRAF6 promoted a robust polyubiquitination of WT N-HTT in the presence of Lys6, Lys27, and Lys29 mutants. Background signals were observed with Lys0 mutant (Fig. 3a). Similar results were obtained with mutant N-HTT with short (Gln60) or long (Gln150) polyQ stretch (Fig. 3, b and c).
FIGURE 3.
TRAF6 promotes atypical ubiquitination of WT and mutant N-HTT. HA-ubiquitin (Ub) mutants were used in which only the indicated lysine is available for chain formation. WT and Lys0 ubiquitin were included as controls. HEK 293 cells were transfected with HA-ubiquitin mutant (as indicated), FLAG-TRAF6, and N-HTT-GFP Gln21 (Q21) (a), Gln60 (Q60) (b), and Gln150 (Q150) (c). Ubiquitination was monitored with anti-HA, anti-GFP, and anti-FLAG antibodies. IB, immunoblot; IP, immunoprecipitation; HC, heavy chain.
Therefore, we conclude that TRAF6 promotes an atypical polyubiquitination of WT and mutant N-HTT that is independent from polyQ length. Lys6, Lys27, and Lys29 are the preferred lysine residues for linkage formation.
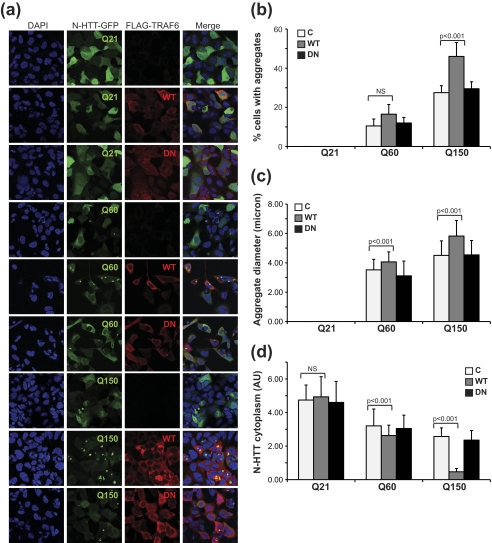
TRAF6-mediated Ubiquitination Increases Mutant N-HTT Aggregates
When expressed in cells, Gln60 and Gln150 mutant, but not Gln21 N-HTT, generated aggregates, with an average size and number that correlate with polyQ length. We examined the influence of TRAF6-mediated ubiquitination on mutant N-HTT aggregation by transient transfection in HEK 293 cells. We included WT N-HTT as an internal reference in all our experiments. Only cells with comparable levels of TRAF6 expression (WT and DN), as determined by fluorescence intensity normalized to cell area, were used for analysis of aggregate number and size. The residual diffuse cytoplasmic staining was also quantified as additional indication of protein segregation into aggregates. TRAF6 overexpression had no influence on WT N-HTT cellular localization, with a diffuse cytoplasmic and nuclear staining as expected (Fig. 4a). Mutant N-HTT formed cytoplasmic and perinuclear aggregates, whose number and size correlated with polyQ length (Fig. 4a). Overexpression of TRAF6 increased the percentage of cells with aggregates and promoted the formation of inclusions with an average larger size (Fig. 4, b and c). This is likely due to a redistribution of the mutant protein to inclusions, as total protein levels were not affected by TRAF6 (supplemental Fig. S1) (compatible with a nondegradative role of atypical ubiquitination) and diffuse cytoplasmic staining decreased when aggregate number and size increased (Fig. 4d). The expression of TRAF6 DN mutant had no effect on percentage, size, and distribution of N-HTT aggregates, thus indicating that TRAF6 E3 ligase activity is required for its function on mutant N-HTT inclusion formation.
FIGURE 4.
TRAF6 enhances aggregate formation. a, HEK 293 cells were transfected with N-HTT-GFP alone (C, control), or with FLAG-TRAF6 (WT or DN), as indicated. Immunofluorescence was performed with anti-FLAG (red) antibody. Nuclei were visualized with DAPI (blue). GFP was followed by autoflorescence. Confocal images from two independent experiments were scored for percentage of cells with aggregates (b), aggregate average size (c), and N-HTT cytoplasmic diffuse staining (d). At least 200 cells per experimental condition were counted. Data were analyzed with ImageJ software. Statistical analysis was performed with a t test. NS, not significant. AU, arbitrary units.
Because TRAF6 is recruited at the site of N-HTT aggregates and affects inclusion formation, we asked whether mutant protein might affect TRAF6 E3 ligase activity. To do so, we followed TRAF6 autoubiquitination by immunoprecipitation assay. We included WT N-HTT as an internal control. We found no differences in the levels of ubiquitinated TRAF6 when Gln21, Gln60, or Gln150 N-HTT was expressed (supplemental Fig. S2). Altogether, these data suggest a scenario in which TRA6-mediated ubiquitination may control N-HTT recruitment into aggregate structures without affecting its steady-state levels.
TRAF6-mediated Atypical Ubiquitination Is Involved in Mutant N-HTT Aggregates
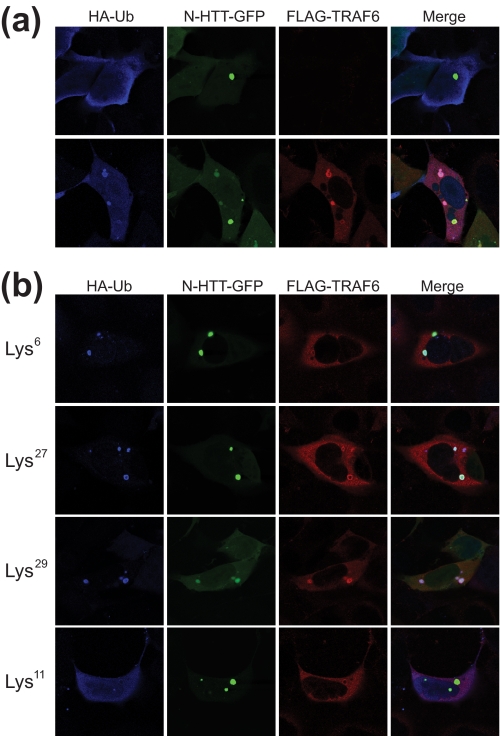
To functionally link TRAF6-mediated atypical ubiquitination with mutant N-HTT aggregates, HEK 293 cells were transfected with HA-ubiquitin, FLAG-TRAF6, and N-HTT-GFP Gln150. Aggregates were analyzed by double immunofluorescence coupled with GFP autofluorescence. First, we set the system using WT ubiquitin (Fig. 5a). Overexpression of ubiquitin per se enhanced N-HTT inclusion formation, as expected for the well established role of the UPS in mutant N-HTT aggregation. Nonetheless, under these conditions, ubiquitin staining was diffuse throughout the cell and did not accumulate at the site of N-HTT inclusions (Fig. 5a, upper panels). When TRAF6 was added to the system, all of the components coalesced into the aggregates, which appeared strongly stained with both ubiquitin and TRAF6 (Fig. 5a, lower panels). To prove that TRAF6 action on mutant N-HTT aggregates involved atypical ubiquitination, we carried out analogous experiments with ubiquitin mutants that were shown to be involved in N-HTT ubiquitination (Lys6, Lys27, and Lys29). As control, we used a Lys11 mutant that mediates polychain formation but is not used by TRAF6. Lys6, Lys27, and Lys29 were able to recapitulate the phenotype observed with WT ubiquitin (Fig. 5b). On the contrary, the Lys11 mutant did not accumulate into N-HTT inclusions. Together, these data provide evidence that TRAF6-mediated atypical activity is involved in controlling the composition of mutant N-HTT aggregates.
FIGURE 5.
Atypical ubiquitination by TRAF6 is localized at the aggregates. a, HEK 293 cells were transfected with Gln150 N-HTT-GFP with FLAG-TRAF6 and HA-ubiquitin (Ub) WT. Localization of ubiquitin (blue) and TRAF6 (red) at N-HTT aggregates were analyzed by double immunofluorescence coupled with GFP autofluorescence. b, cells were transfected with Gln150 N-HTT-GFP, FLAG-TRAF6, and HA-ubiquitin mutants Lys6, Lys27, Lys29, and Lys11 as indicated. Aggregates were analyzed by immunofluorescence as in a.
TRAF6 Is Up-regulated in Post-mortem Brains from HD Patients and Accumulates in Insoluble Protein Fraction
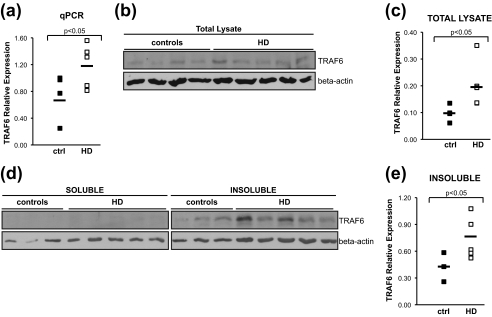
To obtain further evidence supporting the role of TRAF6 in HD pathogenesis, we assessed the expression of TRAF6 in human post-mortem brains from patients and age-matched healthy controls. First, TRAF6 mRNA expression was monitored by quantitative real-time PCR in RNA extracted from parietal cortex of controls and HD (Vonsattel grade 3 neuropathology) brains. TRAF6 mRNA levels were significantly up-regulated in HD compared with controls, with an average increase of 1.6-fold (Fig. 6a). We then tested whether TRAF6 up-regulation also was present at the protein level. Samples dissected from the parietal cortex of HD and control patients were used to prepare total protein extracts. Lysates were analyzed by Western blotting. To avoid artifacts due to unspecific signals, experiments were carried out using two different, commercially available anti-TRAF6 antibodies. Endogenous TRAF6 protein was slightly up-regulated in HD versus control samples (Fig. 6b). Quantification of TRAF6 signal demonstrated that the increase was statistically significant (Fig. 6c).
FIGURE 6.
In HD post-mortem brain, TRAF6 expression is increased, and TRAF6 protein accumulates in the insoluble fraction. a, total RNA was extracted from parietal cortex of HD and control brains. TRAF6 mRNA was measured by quantitative real-time PCR relative to β-actin. b, total protein lysates from HD and controls were prepared, and endogenous TRAF6 expression was monitored with anti-TRAF6 antibody. Loading was normalized by protein quantification and verified with anti-β-actin antibody. Representative images from three independent experiments are shown. c, densitometric analysis of protein bands was performed using Adobe Photoshop CS3. Expression of endogenous TRAF6 in total lysates was normalized relative to β-actin. Statistical analysis was performed with a Student's t test. d, soluble and insoluble fractions from HD and control brains were prepared and analyzed for TRAF6 distribution. Lysates were normalized for total protein content. Anti-TRAF6 and actin antibodies were used. Images are representative of three independent experiments. e, quantification of TRAF6 expression in insoluble fraction was done as described in c.
Because we found that mutant N-HTT promotes TRAF6 accumulation at the aggregates in cell culture system, TRAF6 distribution was studied in soluble and insoluble protein fractions prepared from human brain samples. Loading was normalized for total protein quantity in each of the two fractions. In HD and controls, TRAF6 was present mainly in the insoluble fraction. In this compartment, there was a statistically significant accumulation of TRAF6 in HD versus controls. Altogether, these data suggest an effect of mutant huntingtin on TRAF6 expression and solubility.
DISCUSSION
The ubiquitin proteasome system is under intense scrutiny in the study of neurodegenerative diseases. Different E2 conjugating enzymes and E3 ligases have been shown to ubiquitinate aggregate-prone mutant proteins via Lys48 and Lys63 moieties.
In the present study, we have found that the E3 ubiquitin ligase TRAF6 interacts with WT and mutant N-HTT in vitro as well as with endogenous full-length proteins in vivo and ubiquitinates both WT and mutant N-HTT through conjugation of atypical ubiquitin chains linked at Lys6, Lys27, and Lys29.
Unconventional ubiquitination recently has provided new moieties to regulate cell signaling, intracellular trafficking, and biochemical activities (15). A variety of cellular functions have been associated to atypical ubiquitination with Lys6, Lys27, and Lys29 linkages that include protein degradation, regulation of proteasome activity as well as control of target protein localization and function. The E3 ubiquitin ligase parkin, found mutated in genetic cases of PD, is an HTT interactor that has been shown to act with atypical Lys27 linkage on mitochondrial target VDAC1 to control mitophagy (32). Transcriptional activity of androgen receptor, another polyQ containing protein, is regulated by Lys6 and Lys27 ubiquitination through modulation of cofactor recruitment to a subset of androgen receptor target genes (21). In both cases, atypical ubiquitination regulates protein function without affecting its degradation rate.
Interestingly, we found that both WT and mutant N-HTT are ubiquitinated with no evident selectivity for polyQ length. It is conceivable to hypothesize that atypical ubiquitination of N-HTT might exert diverse functions when associated to WT or mutant polyQ stretch even if the mode of ubiquitination is maintained. Although the ubiquitination of mutant N-HTT by TRAF6 was found to induce aggregate formation, the role of this post-translational modification in WT N-HTT is unclear and deserves further investigation.
Recent data have proven the accumulation of ubiquitinated proteins with unconventional ubiquitin linkages in the brain of HD mouse models (9). In HD, it is well established that neuronal inclusions that contain mutant HTT in human post-mortem brains and in mouse models are stained heavily for ubiquitin (7, 33). In vitro, our data demonstrated that atypical ubiquitin, N-HTT, and TRAF6 were all localized to the aggregates. Differently from all of the ubiquitin ligases studied so far in HD, TRAF6 promoted aggregate formation and segregated ubiquitin at the site of N-HTT inclusion. This is in agreement with previous studies showing that TRAF6 elicited aggresome formation of the PD-associated mutant DJ-1 L166P (26).
An important debate is ongoing on the effects of aggregate formation on cell viability in vivo. Among the various functional consequences, it has been hypothesized that inclusions can act as a sink for prosurvival proteins, thus depleting the neuronal cell of soluble, active molecules. In HD postmortem brains, we found that TRAF6 was up-regulated and accumulated in the insoluble protein fraction. For the majority of the ubiquitin ligases found trapped in cellular inclusions, a concomitant increase in expression has been measured in post-mortem brains, suggesting a homeostasis response aimed to preserve a physiological concentration of the molecule. Under this model, we speculate that TRAF6 sequestration in vivo may deplete soluble TRAF6 in neurons impairing some of its physiological functions.
In vitro TRAF6 was able to increase both the number and size of intracellular aggregates via an atypical mode of ubiquitination. In general, larger aggregates tend to be disposed by the autophagy/lysosomal pathway. TRAF6 recently has been shown to be involved in autophagy via its Lys63-specific activity on Beclin-1 in the context of innate immunity (34). Furthermore, TRAF6 was able to bind p62/SQSTM1 (25), which anchors aggregated structures to LC3 and autophagosomes. Importantly, p62 is a component of the ubiquitinated proteins found in the inclusions of exon 1 mice and in cells overexpressing the N-terminal fragment of mutant HTT (35, 36). Because Lys27 ubiquitination of Jun and Lys29 of Deltex have been recently involved in their recruitment to autophagic and/or storage vesicles (20, 37), it will be interesting to study whether TRAF6 atypical ubiquitin ligase activity has an impact on autophagy in neurodegenerative disorders. It recently has been demonstrated that, similarly to Lys63, Lys27-linked ubiquitin chains are recognized by p62/SQSTM1 to label target proteins for autophagy (32). Our preliminary results (data not shown) indicate that TRAF6 is able to bind LC3 in transfected cells, further supporting our hypothesis that TRAF6 and atypical ubiquitination may favor alternative fates of misfolded aggregated proteins.
In the future, the generation of reagents to discriminate among Lys6, Lys27, and Lys29 ubiquitin chains and the improvement of proteomic techniques to be used on small amount of human autoptic samples will definitely help the study of atypical ubiquitination in various cellular processes in vivo and from diseased brains.
Supplementary Material
Acknowledgments
We are indebted to all members of the Persichetti and Gustincich laboratories for thought-provoking discussions and to Cristina Leonesi for technical support. We thank Professors Marcy MacDonald (Boston, USA) and Giannino Del Sal (Trieste, Italy) for helpful comments.
This work was supported by the Telethon Foundation of Italy (Grant GGP07185) and the Italian Institute of Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HD
- Huntington disease
- HTT
- huntingtin
- polyQ
- polyglutamine
- N-HTT
- amino-terminal fragment of huntingtin
- TRAF6
- TNF receptor-associated factor 6
- PD
- Parkinson disease
- DN
- deleted of the N terminus.
REFERENCES
- 1. Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Hendricks A. E., Latourelle J. C., Lunetta K. L., Cupples L. A., Wheeler V., MacDonald M. E., Gusella J. F., Myers R. H. (2009) Am. J. Med. Genet. A 149A, 1375–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hilditch-Maguire P., Trettel F., Passani L. A., Auerbach A., Persichetti F., MacDonald M. E. (2000) Hum. Mol. Genet. 9, 2789–2797 [DOI] [PubMed] [Google Scholar]
- 4. Panov A. V., Gutekunst C. A., Leavitt B. R., Hayden M. R., Burke J. R., Strittmatter W. J., Greenamyre J. T. (2002) Nat. Neurosci. 5, 731–736 [DOI] [PubMed] [Google Scholar]
- 5. Velier J., Kim M., Schwarz C., Kim T. W., Sapp E., Chase K., Aronin N., DiFiglia M. (1998) Exp. Neurol. 152, 34–40 [DOI] [PubMed] [Google Scholar]
- 6. Zuccato C., Valenza M., Cattaneo E. (2010) Physiol. Rev. 90, 905–981 [DOI] [PubMed] [Google Scholar]
- 7. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 8. Wyttenbach A., Carmichael J., Swartz J., Furlong R. A., Narain Y., Rankin J., Rubinsztein D. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 10. de Pril R., Fischer D. F., Roos R. A., van Leeuwen F. W. (2007) Mol. Cell. Neurosci. 34, 10–19 [DOI] [PubMed] [Google Scholar]
- 11. Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D. C., Monteiro M. J., Fang S. (2007) Exp. Cell Res. 313, 538–550 [DOI] [PubMed] [Google Scholar]
- 12. Jana N. R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K., Nukina N. (2005) J. Biol. Chem. 280, 11635–11640 [DOI] [PubMed] [Google Scholar]
- 13. Tsai Y. C., Fishman P. S., Thakor N. V., Oyler G. A. (2003) J. Biol. Chem. 278, 22044–22055 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z. J., Sun L. J. (2009) Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 15. Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cripps D., Thomas S. N., Jeng Y., Yang F., Davies P., Yang A. J. (2006) J. Biol. Chem. 281, 10825–10838 [DOI] [PubMed] [Google Scholar]
- 17. Shang F., Deng G., Liu Q., Guo W., Haas A. L., Crosas B., Finley D., Taylor A. (2005) J. Biol. Chem. 280, 20365–20374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 19. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. (2008) Biochem. J. 411, 249–260 [DOI] [PubMed] [Google Scholar]
- 20. Ikeda H., Kerppola T. K. (2008) Mol. Biol. Cell 19, 4588–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu K., Shimelis H., Linn D. E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Chen H., Kong X., Melamed J., Fang S., Xiao Z., Veenstra T. D., Qiu Y. (2009) Cancer Cell 15, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Z. J. (2005) Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geetha T., Jiang J., Wooten M. W. (2005) Mol. Cell 20, 301–312 [DOI] [PubMed] [Google Scholar]
- 24. Khursigara G., Orlinick J. R., Chao M. V. (1999) J. Biol. Chem. 274, 2597–2600 [DOI] [PubMed] [Google Scholar]
- 25. Babu J. R., Geetha T., Wooten M. W. (2005) J. Neurochem. 94, 192–203 [DOI] [PubMed] [Google Scholar]
- 26. Zucchelli S., Codrich M., Marcuzzi F., Pinto M., Vilotti S., Biagioli M., Ferrer I., Gustincich S. (2010) Hum. Mol. Genet. 19, 3759–3770 [DOI] [PubMed] [Google Scholar]
- 27. Persichetti F., Trettel F., Huang C. C., Fraefel C., Timmers H. T., Gusella J. F., MacDonald M. E. (1999) Neurobiol. Dis. 6, 364–375 [DOI] [PubMed] [Google Scholar]
- 28. White J. K., Auerbach W., Duyao M. P., Vonsattel J. P., Gusella J. F., Joyner A. L., MacDonald M. E. (1997) Nat. Genet. 17, 404–410 [DOI] [PubMed] [Google Scholar]
- 29. Zucchelli S., Vilotti S., Calligaris R., Lavina Z. S., Biagioli M., Foti R., De Maso L., Pinto M., Gorza M., Speretta E., Casseler C., Tell G., Del Sal G., Gustincich S. (2009) Cell. Death Differ. 16, 428–438 [DOI] [PubMed] [Google Scholar]
- 30. Fossale E., Wheeler V. C., Vrbanac V., Lebel L. A., Teed A., Mysore J. S., Gusella J. F., MacDonald M. E., Persichetti F. (2002) Hum. Mol. Genet. 11, 2233–2241 [DOI] [PubMed] [Google Scholar]
- 31. Vonsattel J. P., Myers R. H., Stevens T. J., Ferrante R. J., Bird E. D., Richardson E. P., Jr. (1985) J. Neuropathol. Exp. Neurol. 44, 559–577 [DOI] [PubMed] [Google Scholar]
- 32. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell. Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 33. Sieradzan K. A., Mechan A. O., Jones L., Wanker E. E., Nukina N., Mann D. M. (1999) Exp. Neurol. 156, 92–99 [DOI] [PubMed] [Google Scholar]
- 34. Shi C. S., Kehrl J. H. (2010) Sci. Signal. 3, ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagaoka U., Kim K., Jana N. R., Doi H., Maruyama M., Mitsui K., Oyama F., Nukina N. (2004) J. Neurochem. 91, 57–68 [DOI] [PubMed] [Google Scholar]
- 37. Chastagner P., Israël A., Brou C. (2006) EMBO Rep. 7, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.