Abstract
Camptothecin (CPT) and related chemotherapeutic drugs induce formation of DNA topoisomerase I (Top1) covalent or cleavage complexes (Top1ccs) that block leading-strand DNA synthesis and elicit DNA Double Stranded Breaks (DSB) during S phase. The Fanconi Anemia (FA) pathway is implicated in tolerance of CPT-induced DNA damage yet the mechanism of FA pathway activation by Top1 poisons has not been studied. We show here that the FA core complex protein FANCA and monoubiquitinated FANCD2 (an effector of the FA pathway) are rapidly mobilized to chromatin in response to CPT treatment in several human cancer cell lines and untransformed primary human dermal fibroblasts. FANCD2 depletion using siRNA leads to impaired recovery from CPT-induced inhibition or DNA synthesis, persistence of γH2AX (a DSB marker) and reduced cell survival following CPT treatment. The E3 ubiquitin ligase Rad18 is necessary for CPT-induced recruitment of FANCA and FANCD2 to chromatin. Moreover, Rad18-depletion recapitulates the DNA synthesis and survival defects of FANCD2-deficiency in CPT-treated cells. It is well-established that Rad18 promotes FA pathway activation and DNA damage tolerance in response to bulky DNA lesions via a mechanism involving PCNA monoubiquitination. In contrast, PCNA monoubiquitination is not involved in Rad18-mediated FA pathway activation or cell survival following acquisition of CPT-induced DSB. Moreover, while Rad18 is implicated in recombinational repair of DSB via an E3 ligase-independent mechanism, we demonstrate that Rad18 E3 ligase activity is essential for appropriate FA pathway activation and DNA damage tolerance after CPT treatment. Taken together, our results define a novel pathway of Rad18-dependent DSB repair that is dissociable from known Rad18-mediated DNA repair mechanisms based on its independence from PCNA ubiquitination and requirement for E3 ligase activity.
Key words: camptothecin, Rad18, topoisomerase I, double strand breaks, Fanconi anemia
Introduction
DNA Topoisomerase I (Top1) plays a critical role in maintenance of genome integrity by resolving topological strain during vital cellular processes such as DNA replication, transcription and chromatin remodeling.1–5 Top1 relaxes DNA supercoils by generating and quickly resealing a single-stranded break in the duplex DNA.6 Top1 is also a molecular target of the anti-cancer drug camptothecin (CPT) and related chemotherapeutics (including topotecan, irenotecan). CPT and related drugs reversibly trap Top1-DNA covalent or cleavage complexes (Top1ccs) by intercalating into the Top1-DNA nick and prevent the scissile strand's religation.7–10 Stabilization of Top1-DNA complexes by CPT induces aberrant DNA structures and accumulation of positive supercoils, which, following encounters with the DNA replication and transcription machinery, may lead to lethal DNA double-strand breaks (DSB).9–11 However, the mechanisms underlying the repair of CPT-induced lesions are not well understood. Studies in yeast and mammalian cells have identified multiple DNA damage signaling and repair pathways that respond to CPT, including those involved in DNA replication and cellular responses to single and double-stranded DNA breaks.12–16 Nevertheless, the molecular networks that integrate DNA replication, DNA repair in CPT-treated cells are poorly defined.
Rad18 is an E3 ubiquitin ligase whose role in DNA damage tolerance is best understood for a post-replication repair mechanism termed “Trans-lesion Synthesis” (TLS).17–19 In response to replication fork-stalling, bulky DNA lesions such as cyclobutane pyrimidine dimers (CPD) or Benzo[a]pyrene di-hydro-diol epoxide (BPDE) adducts, Rad18 monoubiquitinates PCNA. Monoubiquitinated PCNA, in turn, promotes the recruitment of specialized “Y-family” DNA polymerases Polη, Polκ, Polι and REV1 to sites of DNA damage, thereby facilitating replication of damaged DNA templates.17–21 Recent work from several laboratories indicates that recruitment of TLS polymerases to damaged DNA is not restricted to S phase.22 Therefore, Y-family polymerase activities may be involved in replication fork-independent repair and processing of adducted DNA. In many instances, Rad18-deficiency confers hypersensitivity to bulky DNA lesions, presumably due to its role in promoting lesion bypass by the Y-family DNA polymerases.
Rad18-deficient cells are also sensitive to DSB-inducing agents, such as ionizing radiation (IR) and CPT.23–25 Work from the Chen laboratory defined a novel role for Rad18 in recruiting Rad51 and its paralogs to DSB, thereby promoting DNA repair via homologous recombination (HR).23,26 Interestingly, Rad18 E3 ubiquitin ligase activity (which is required for PCNA monoubiquitination and efficient TLS) is dispensable for Rad18-mediated Rad51 recruitment and resistance to DSB-inducing agents.23 Thus, Rad18-mediated TLS and HR mechanisms are dissociable via their dependence on E3 ubiquitin ligase activity.23
Our recent studies have shown that Rad18 E3 ubiquitin ligase activity and PCNA ubiquitination are required for appropriate activation of the Fanconi Anemia (FA) pathway in response to bulky adducts induced by BPDE and UV irradiation.27 Thus, Rad18 provides a mechanism for coupling FA pathway activation with TLS, ensuring appropriate processing of bulky DNA lesions. FA is a cancer predisposition syndrome in humans. To date, at least 13 complementation groups of FA have been identified (designated ‘FANC’ A–M). The FA proteins (FANCs) are thought to work cooperatively in a common pathway to repair interstrand crosslinks.28 The eight-protein core complex (complementation groups, A–C, E–G, L and M) acts as an E3 ligase to monoubiquitinate FANCD2 and FANCI in response to acquisition of DNA damage and replication stress during S phase. Upon monoubiquitination, the FANCD2/I complex is recruited to chromatin and promotes DNA repair.29–31 The remaining three FA proteins are breast cancer susceptibility related genes (BRCA) FANCD1/ BRCA2, FANCJ, FANCN, which are thought to function downstream of FANCD2/I.28,32–35 Although the FA pathway is widely hypothesized to participate in DNA repair and maintenance of genomic stability, its precise biochemical role(s) are unknown. The FA pathway is implicated in the repair of DNA interstrand crosslinks and fork-stalling lesions.31,32 However, very little is known regarding roles of the FA pathway in processing and repair of Top1 inhibitor-induced DNA lesions. Since Top1 inhibitors induce DNA replication-dependent lesions, it is likely that the FA pathway participates in processing and repair of Top1-inhibitor-induced DNA damage. Recently, Elledge and colleagues reported that FANCD2, FANCI and FANCJ-deficient cells are hypersensitive to CPT yet display only moderate IR-sensitivity.36
Covalent trapping of Top1 on DNA by CPT (Top1ccs) induces aberrant DNA structures and lethal DNA lesions, such as accumulation of positive supercoils and DSB.8,10,37 Several DNA repair proteins that interact either directly or indirectly with FANCs are required for tolerance of Top1-mediated DNA damage. Indeed, deficiency or mutations in multiple components of the FA pathway (FANCB, FANCM, FANCJ, FANCD1, FANCD2 and FANCI) leads to CPT sensitivity.36,38–40 However, little is known regarding activation mechanisms of the FA pathway and its role in response to Top1-inhibitors-induced DNA damage.
Because we and others recently showed that Rad18 contributes to FA pathway activation in response to bulky DNA adducts,27 it was of interest to test the potential role (if any) of Rad18 in CPT-induced FA pathway activation. In response to bulky DNA lesions, Rad18-dependent FA pathway activation is mediated via PCNA ubiquitylation and requires Polη.27 We show here that in contrast with bulky DNA lesions, CPT does not induce PCNA ubiquitination. Surprisingly, however, we demonstrate that Rad18-mediated FA pathway activation by CPT and tolerance of CPT-induced damage are dependent on Rad18 E3 ubiquitin ligase activity. This work defines a new role for Rad18 in DNA repair, which is dissociable from its known functions in TLS and HR (which require PCNA ubiquitination and are E3 ligase-independent respectively). Moreover, this study indicates the existence of Rad18 substrates other than PCNA that are involved in FA pathway activation and tolerance of CPT-induced DNA lesions.
Results
CPT activates the FA pathway.
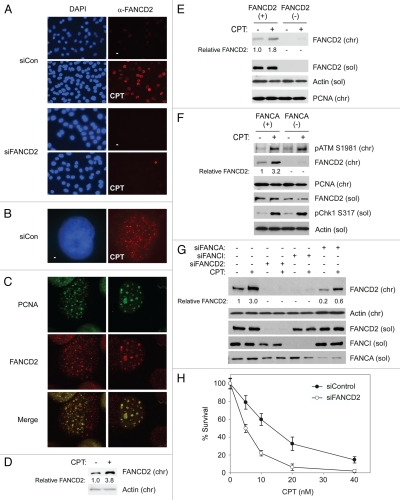
Several reports implicate the FA pathway in tolerance and repair of DNA damage due to CPT and other Top1 inhibitors.39,41 However, mechanisms of FA pathway activation in response to CPT have not been studied. To determine whether the FA pathway is activated in response to CPT, we treated H1299 lung carcinoma cells with 100 nM CPT for 1 hour and then examined the sub-cellular distribution and chromatin-binding of FANCD2 using immunofluorescence (IF) microscopy and immunoblotting (IB) respectively. As shown in Figure 1A (40x magnification) and 1B (100x magnification), CPT treatment induced an increase in the number of FANCD2 nuclear foci (likely corresponding to sites of DNA repair) in H1299 cells. Camptothecins are well established S phase-specific chemotherapeutic agents, and most of their cytotoxicity at sub-micromolar concentrations is dependent on DNA replication.9 Since we have used relatively low doses of CPT in this study (100 nM), the FANCD2 nuclear foci we observed in CPT-treated cells likely correspond to stalled/collapsed replication forks. Indeed, the FANCD2 foci co-localized with PCNA (Fig. 1C) and therefore represent sites of DNA replication. Since CPT induces DSB at sites of DNA replication, we conclude that the FANCD2 foci represent replication-induced DSB. CPT treatment also induced an increase in the levels of chromatin-bound FANCD2, the monoubiquitinated species of FANCD242 in H1299 cells (Fig. 1D) and in FANCD2-complemented PD-20 cells (Fig. 1E). Supplemental Figure 1 also demonstrates that the CPT-induced chromatin-associated FANCD2 corresponds to the electrophoretically retarded (monoubiquitinated) species.
Figure 1.
CPT activates the FA pathway. (A) Exponentially growing H1299 cells in chamber slides were treated with 100 nM CPT (or DMSO for controls). After 1 hr, cells were fixed with 4% formaldehyde, permeabilized to remove soluble proteins, stained with α-FANCD2 antibodies and visualized by using immunofluorescence microscopy at 40x magnification, as described under Materials and Methods. (B) A representative cell from a CPT-treated culture showing FANCD2 nuclear foci (shown at 100x magnification). (C) H1299 Cells were treated with CPT and fixed as described for (A) above. Fixed cells were co-stained with α-FANCD2 and α-PCNA antibodies and visualized by using immunofluorescence microscopy at 100x magnification. The representative cells shown have patterns of PCNA and FANCD2 foci suggestive of early (left) and late (right) DNA replication. (D) In parallel with the experiment described in (A), exponentially growing H1299 cells were treated with 100 nM CPT (or DMSO for controls). After 1 hr, chromatin was extracted from the cells and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (E) Exponentially growing PD20 FANCD2 (−) cells and isogenic FANCD2 (+) cells expressing stably reconstituted wild-type FANCD2 were treated with 100 nM CPT for 1 hr. Chromatin and soluble extracts from the cells were resolved by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (F) Exponentially growing GM6914 FANCA (−) fibroblasts or isogenic FANCA (+) cells expressing stably reconstituted wild-type FANCA were treated with CPT and analyzed as described for (E) above. (G) Replicate cultures of exponentially growing H1299 cells were transfected with siRNAs targeting FANCD2, FANCI and FANCA (or with non-targeting siRNA for controls) as described under Experimental Procedures. The resulting cells were treated with CPT and analyzed for expression of various FANCs as described under Materials and Methods. (H) H1299 cells were transfected with siFANCD2 or non-targeting siCon RNAs as described under Materials and Methods. Forty-eight hr post-transfection, siFANCD2 and siCon cells were seeded in 6-well plates and treated with the indicated concentrations of CPT in triplicate for 16 hours. After washout, cells were given fresh medium and allowed to form colonies. After 7–10 days, colonies containing >50 cells were enumerated. The graph shows the result of a representative experiment. Each data point represents the mean of triplicate determinations and the error bars represent standard deviations.
To determine the role of the FA core complex in FA pathway activation by CPT, we compared CPT-induced FANCD2 ubiquitination in FANCA (−) GM6914 cells and an isogenic FANCA (+) cell line complemented with a functional FANCA cDNA. As shown in Figure 1F, CPT treatment induced chromatin-binding of monoubiquitinated FANCD2 in FANCA (+) cells. However, even though levels of soluble FANCD2 were comparable in FANCA (+) and FANCA (−) cells, CPT did not induce chromatin association of FANCD2 in the absence of FANCA. In siRNA experiments using H1299 cells, we achieved partial FANCA-depletion, which resulted in attenuation of CPT-induced FANCD2 ubiquitination (Fig. 1G). We infer that CPT-induced FANCD2 ubiquitination is FA core complex-dependent. Consistent with previous reports using other cell lines,39 FANCD2-depleted H1299 cells exhibited increased sensitivity to CPT when compared with control siRNA-transfected cells (Fig. 1H). Therefore, CPT induces FA core complex-mediated FANCD2 monoubiquitination. Moreover, FANCD2 is necessary for tolerance of CPT-induced DNA damage.
Rad18 mediates CPT-induced FA pathway activation.
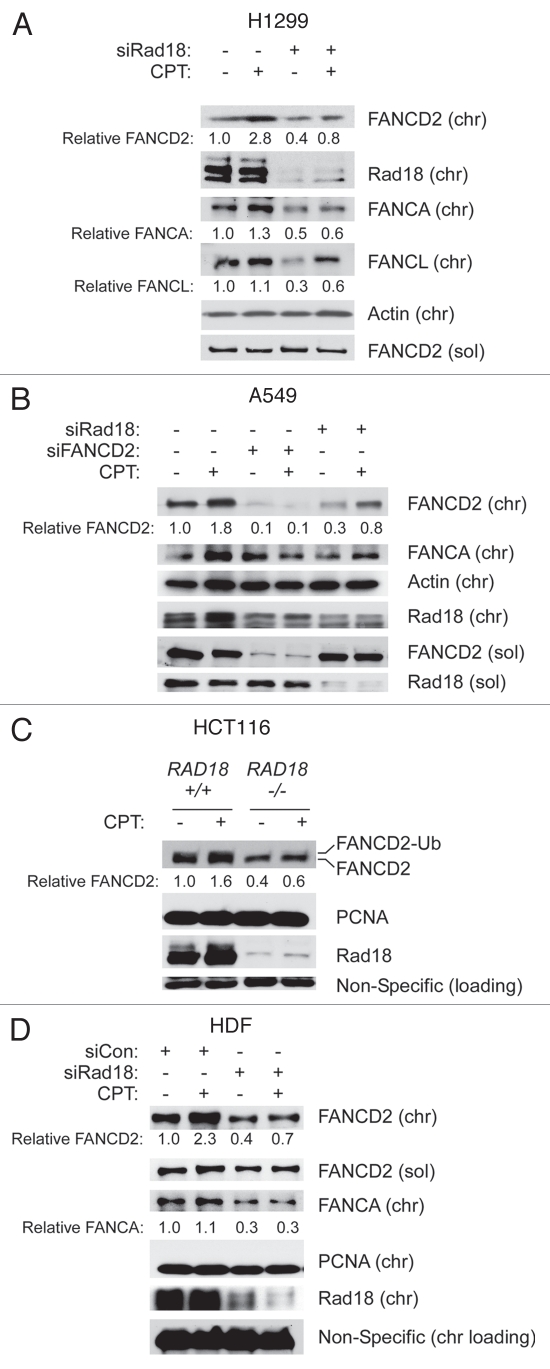
Rad18 contributes to FA pathway activation by various bulky DNA lesions, including BPDE, CPD and cis-Pt adducts.27,43 In those studies, the Rad18-dependent component of FANCD2 ubiquitination was mediated via PCNA monoubiquitination. Because CPT does not induce detectable PCNA monoubiquitination,44 we predicted that CPT-induced FANCD2 monoubiquitination would be Rad18-independent. Therefore we tested the effect of Rad18-deficiency on CPT-induced FA pathway activation in several cell lines, including H1299 (p53-deficient lung carcinoma), A549 (p53-expressing lung carcinoma), HCT 116 (colon carcinoma) and non-transformed Human Dermal Fibroblasts (HDF). Surprisingly, FANCD2 monoubiquitination was compromised as a result of Rad18-depletion in several cell lines, including H1299, A549 and Human Dermal Fibroblasts (Fig. 2A–C). CPT-induced FANCD2 ubiquitination was also attenuated in RAD18−/− HCT116 cells compared with isogenic RAD18+/+ parental HCT116 cells (Fig. 2D). Therefore, FA pathway activation in response to CPT is Rad18-dependent. Note that we typically observed slight decrease in Rad18 levels following FANCD2-depletion (see for example Figs. 2B and 3B). In a recent study, we reported that FANCD2 and Rad18 may be co-immunoprecipitated as a complex.45 Whether Rad18 and FANCD2 associate directly or via other proteins is not known and is beyond the focus of this study. However, it is very common for DNA repair proteins to stabilize their binding partners, and this provides a plausible explanation for the effect of FANCD2-depletion on Rad18 levels.
Figure 2.
CPT-induced FANCD2 ubiquitination is Rad18-mediated. Replicate plates of H1299 cells (A), A549 cells (B), HCT116 cells (C) and HDF (D) differing with respect to Rad18 status were treated with CPT for 1 hr, separated to give soluble and chromatin fractions and analyzed using SDS-PAGE and immunoblotting with antibodies against the indicated proteins. Note that the commercially available Rad18 antibody detects a minor non-specific band of approximately the same size as human Rad18 (evident in the Rad18−/− cell extracts).
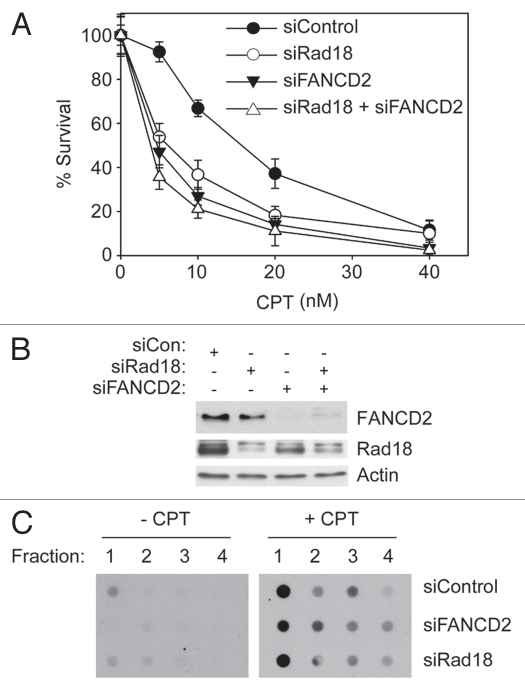
Figure 3.
Rad18-depletion confers CPT-sensitivity. (A) H1299 cells were transfected individually or in combination with siRNAs against Rad18 and FANCD2 (or with non-targetting siCon RNA). The resulting cultures were seeded in 6-well plates and treated with the indicated concentrations of CPT in triplicate for 16 hours. After washout, cells were given fresh medium and allowed to form colonies. After 7–10 days, colonies containing >50 cells were enumerated. The graph shows the result of a representative experiment. Each data point represents the mean of triplicate determinations and the error bars represent standard deviations. (B) In parallel with the experiment described in (A) above, siRNA-transfected cells were analyzed by SDS-PAGE and immunoblotting of whole cell extracts to confirm changes in FANCD2 and Rad18 expression. (C) In parallel with experiments described in (A and B) above, siCon-, siFANCD2- and siRad18-transfected cells were lysed in 1% sarkosyl following CPT treatment. Bottom four fractions of the CsCl gradient fractionated DNA were immobilized on membranes and Top1-DNA covalent or cleavage complexes were detected with α-Top1 antibody as described under Materials and Methods.
In clonogenic survival assays Rad18-depleted H1299 cells were sensitized to CPT-induced killing relative to control Rad18-replete cultures (Fig. 3A). CPT-sensitivity following Rad18 ablation was also observed in experiments using A549 cells (data not shown). Co-depletion of Rad18 and FANCD2 did not have a significant additive effect on sensitivity to killing by CPT (Fig. 3A). Nevertheless, our partial co-depletion experiments (Fig. 3A) suggest that Rad18 and FANCD2 function in a common pathway, or at least a partially overlapping pathway to confer tolerance of CPT-induced lesions.
We considered the formal possibility that Rad18 or FANCD2 status affects acquisition of Top1ccs following CPT treatment. Therefore, we used in vivo complex Enzyme (ICE) bioassays to determine the effect of Rad18 and FANCD2 status on levels of CPT-induced Top1ccs. Lysates from control and CPT-treated cells were fractionated by centrifugation on CsCl gradients. The individual fractions were dot-blotted onto nitrocellulose filters and probed with a Top1 antibody. For each experimental sample from CPT-treated cells (but not control cultures), Fraction 1 (enriched in genomic DNA) showed increased reactivity with anti-Top1 antibodies (Fig. 3C), indicating the presence of CPT-induced Top1ccs. We did not observe significant changes in spontaneous or CPT-induced Top1ccs formation as a result of Rad18- or FANCD2 depletion. Therefore, Rad18 and FANCD2 do not influence formation of CPT-induced Top1ccs, but affect distal events in the response to CPT-induced DNA damage. Taken together, these data suggest that Rad18 contributes to CPT-induced FA pathway activation and tolerance of CPT-induced DNA damage.
Rad18-deficient cells are defective in repair and recovery of CPT induced DNA damage.
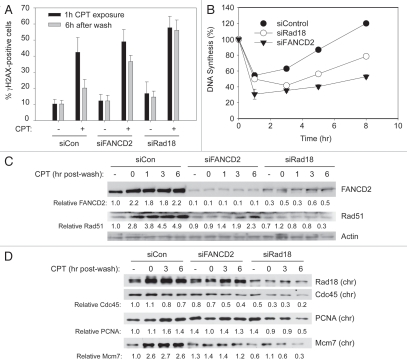
CPT-stabilized Top1ccs are known to interfere with ongoing DNA synthesis, potentially leading to DSB and replication fork collapse.9 We determined the effect of Rad18 and FANCD2 status on accumulation of γH2AX foci (a marker of ATM and ATR signaling) at different times after CPT treatment. As shown in Figure 4A, 40.3% (±9.2) of the control (siCon-transfected) cells contained γH2AX foci one hour after CPT treatment. At this time point, FANCD2- and Rad18-depleted cells accumulated slightly higher levels of γH2AX foci when compared to siCon-transfected cultures (48.9 ± 7.5% and 57.6 ± 7.1 cells contained nuclear γH2AX foci in siFANCD2 and siRad18-transfected cultures, respectively). Six hours after CPT washout, the number of cells containing γH2AX foci declined to 20.1% (±5.3) in the siCon-transfected cultures, indicating that approximately half of the CPT-induced DNA damage was repaired (Fig. 4A). Interestingly, however, levels of γH2AX foci remained relatively high 6 hr after CPT washout in the siFANCD2 and siRad18-transfected cells (36.7% ± 3.8 and 56.1% ± 6.4 cells retained γH2AX foci in FANCD2- and Rad18-depleted cultures, respectively). The persistence of γH2AX foci in Rad18- and FANCD2-depleted cells is an indication of a residual DNA damage signaling and suggests a role for Rad18 and the FA pathway in processing and/or repair of CPT-induced DSBs.
Figure 4.
Recovery from CPT-induced DNA damage is compromised in Rad18- and FANCD2-deficient cells. (A) Duplicate cultures of A549 cells growing in chamber slides were transfected with siRNAs targeting Rad18 and FANCD2 (or non-targeting RNA for control). For each replicate culture, one well was treated with 1 µM CPT for 1 hr and fixed immediately. Cells in the other duplicate well were washed extensively to remove CPT and were then returned to the incubator and allowed to recover in drug-free medium for 6 hr before fixing. Fixed cells were stained with γH2AX antibodies and mounted in DAPI. In a blinded analysis, ten fields (each containing 20–40 cells) were examined. Cells containing >5 γH2AX foci were scored “positive.” For each field the percentage of γH2AX-positive cells was calculated. Each data point represents the mean percentage of γH2AX-positive cells from ten fields and the error bars represent standard deviations. (B) In parallel with the experiment described in (A), A549 cells transfected with siRad18, siFANCD2 or siCon were plated in 24-well culture dishes. The resulting cultures were treated with 500 nM CPT (or were left untreated for controls). Rates of DNA synthesis were determined by measuring [3H]-thymidine incorporation for 1 hr immediately prior to CPT addition or at various times after CPT-washout in triplicate as described under Materials and Methods. For each siRNA transfection rates of DNA synthesis were normalized and expressed as a percent of [3H]-thymidine incorporation rates immediately prior to CPT treatment. Each data point represents a mean of triplicate determinations. (C and D) In parallel with the experiment described in (B) above A549 cells transfected with siRad18, siFANCD2 or siCon were plated in 10 cm culture dishes in replicate. The resulting cultures were treated with 500 nM CPT for 1 hr (or were left untreated for controls). Cells were harvested immediately after 1 hr of CPT addition or at various times after CPT washout. Extracts were separated to give chromatin and soluble fractions and these were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Note that (C and D) show the results of immunoblots using extracts from the same experiment. In (D) extracts from the 1 hr CPT washout time point were omitted.
Failure to repair replication-associated DNA damage often results in more persistent inhibition of DNA synthesis following genotoxin exposure.20,46 Therefore, we determined the effects of CPT treatment on rates of DNA synthesis in control and Rad18- and FANCD2-depleted cells. As shown in Figure 4B, in control (siCon-transfected) cultures, DNA synthesis was transiently inhibited by 54.4% 1 hr after CPT treatment. However, DNA synthesis recovered and normal rates of replication were resumed 8 hr after CPT treatment in the control cultures. In Rad18- and FANCD2-depleted cells, DNA synthesis was more sensitive to inhibition by CPT (49.8 and 30.5% inhibition 1 hr after CPT). Moreover, rates of DNA synthesis were inhibited more persistently following CPT treatment in Rad18- and FANCD2-depleted cells and failed to resume control levels within the time frame of this experiment (Fig. 4B). Whether resumption of DNA synthesis occurs due to repriming, dormant origin firing or restart via other mechanisms cannot be inferred from these experiments. Nevertheless, our results demonstrate that FANCD2 and Rad18 are required for maintenance of DNA replication in CPT-treated cells.
In parallel we also assessed the kinetics of FANCD2 and Rad51 chromatin loading in CPT-treated cells. As shown in Figure 4C, CPT treatment induced a ∼2-fold increase in the chromatin-association of FANCD2 and ∼2–5-fold increases in chromatin-binding of Rad51 in control cells. The increased chromatin association of FANCD2 and Rad51 persisted for at least 6 hours after CPT washout. In Rad18-depleted cells chromatin-loading of FANCD2 and Rad51 was attenuated. Interestingly, chromatin binding of Rad51 was also affected by FANCD2 depletion, most likely because of the proposed role of FANCD2 in promoting HR.47,48
Consistent with failure of Rad18- and FANCD2-depleted cells to resume DNA synthesis, chromatin levels of replication markers such as Cdc45 and PCNA and Mcm7 also decreased more persistently after CPT treatment in Rad18- or FANCD2-ablated cells relative to siCon-transfected cultures (Fig. 4D). The decrease in chromatin-binding of Cdc45 concomitant with inhibition of DNA synthesis was of the same magnitude that we and others have previously observed following induction of S-phase checkpoint signaling.49,50
We consistently observed an increase in chromatin loading of replication factors after CPT washout in the FANCD2 and Rad18-replete (siCon) cultures. Blow and colleagues have shown that replication stress can lead to firing of otherwise dormant origins.51,52 Therefore, the increased replication factor loading that we observe concomitant with recovery from replicative arrest is potentially consistent with published mechanisms of S-phase DNA damage tolerance. Taken together, our results suggest that persistent CPT-induced replicative arrest in Rad18-depleted cells is associated with defective activation of FANCD2 and HR, leading to reduced processing of CPT induced DNA lesions.
Rnf8 is required for FA pathway activation by CPT.
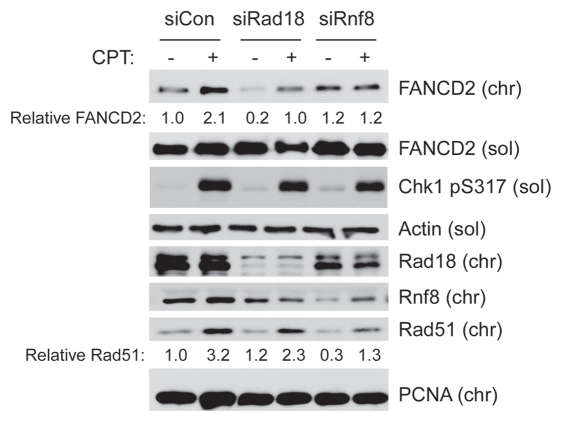
Rad18 is recruited to stalled replication forks due to recognition of RPA-coated DNA structures generated by uncoupling of replicative helicase and polymerase activities.25,53 In contrast, Rad18 recruitment to DSB is dependent on the E3 ligase RNF8 and ubiquitination of histone H2A.23 Because CPT-induced lesions do not cause replicative helicase-polymerase uncoupling or induce PCNA ubiquitination, we predicted that Rad18 activation by CPT would be RNF8-dependent. Therefore, we tested the effects of RNF8-depletion on CPT-induced FA pathway activation. As shown in Figure 5, CPT-induced FANCD2 ubiquitination and levels of Rad18 on chromatin were attenuated in Rnf8-depleted cells. In agreement with the studies of Chen and colleagues, Rnf8-depletion also reduced CPT-induced chromatin-association of Rad51.
Figure 5.
RNF8 mediates FA pathway activation by CPT. Replicate cultures of H1299 cells were transfected with siRad18 and siRnf8 (or non-targeting siCon oligonucleotides). The resulting cells were treated with 100 nM CPT for 1 hr. The resulting cells were separated to give soluble and chromatin fractions that were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
CPT-induced FA pathway activation requires Rad18 E3 ligase activity.
Rad18-mediated TLS is dependent on PCNA-directed E3 ubiquitin ligase activity,18 whereas Rad18-mediated HR is E3 ligase-independent.23 We asked whether Rad18 E3 ligase activity is required for CPT-induced FA pathway activation and tolerance of CPT-induced lesions. Therefore, we depleted endogenous Rad18 using siRNAs targeting the 3′-UTR region of the RAD18 transcript. The resulting cells were complemented with expression vectors encoding siRNA-resistant Wild-Type (WT) Rad18 or an E3 ubiquitin ligase-defective Rad18 harboring a C28F point mutation. Control and Rad18-complemented cells were assessed for CPT-induced chromatin association of FANCD2, Rad51 (using immunoblotting) and CPT sensitivity (using clonogenic survival assays).
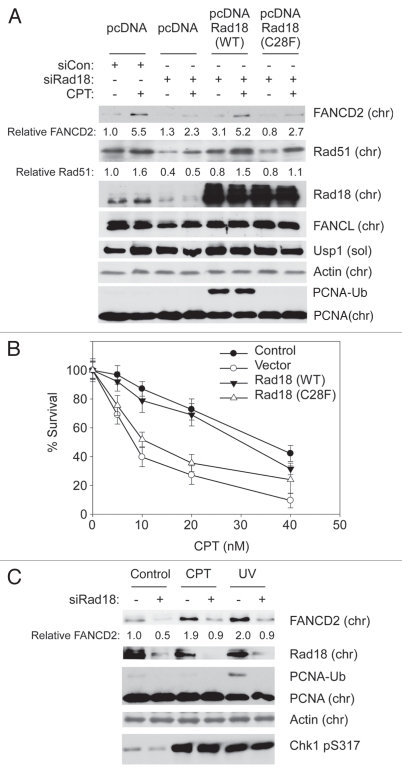
As shown in Figure 6A, CPT-induced FANCD2 ubiquitination was compromised in Rad18-depleted cells. Expression of Rad18 (WT) but not Rad18 (C28F) complemented the defective FANCD2 chromatin binding of Rad18-depleted cells.
Figure 6.
CPT-induced FA pathway activation does not involve PCNA monoubiquitination but requires Rad18 E3 ligase activity. (A) H1299 cells were serially transfected with siRNAs targeting the 3′-UTR region of Rad18 (or with non-targeting siCon oligonucleotides) and then with pcDNA-based expression vectors encoding wild-type Rad18, an E3 ligase defective Rad18-C28F mutant or “empty” pcDNA plasmid for control. Replicate cultures of the resulting cells were treated with 100 nM CPT (or with DMSO for control) for 1 hr. Cell extracts were then separated into soluble and chromatin fractions, which were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (B) In parallel with the experiment described in (A), H1299 cells were depleted of endogenous Rad18, complemented with expression plasmids encoding wild-type Rad18, Rad18-C28F mutant or with “empty” pcDNA vector for control and then tested for CPT-sensitivity using clonogenic survival assays as described under the legend for Figure 1H and Materials and Methods. The graph shows the result of a representative experiment. Each data point represents the mean of triplicate determinations and the error bars represent standard deviations. (C) Replicate plates of H1299 cells were transfected with siRad18 or siCon oligonucleotides. The resulting cultures were treated with 100 nM CPT or were exposed to 20 J/m2 UVC (or were left untreated for controls). After 2 hr, cells were separated into soluble and chromatin fractions, which were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
As expected from a previous report in reference 23, both WT Rad18 and Rad18-C28F corrected the defective chromatin loading of Rad51. Interestingly, in colony survival assays WT Rad18 but not the E3 ligase-inactive Rad18 (C28F) mutant fully complemented the CPT sensitivity resulting from depletion of endogenous Rad18 (Fig. 6B). Therefore Rad18 (C28F) supports chromatin loading of Rad51 and normal HR activity23 but cannot mediate CPT-induced FANCD2 ubiquitination or confer CPT-tolerance.
We note that the levels of ectopically expressed Rad18 achieved by transient transfection exceed physiological Rad18 levels and led to DNA damage-independent PCNA monoubiquitination. Many Rad18-dependent responses to DNA damage are mediated via PCNA monoubiquitination. We considered the possibility that Rad18-dependent PCNA monoubiquitination was involved in FA pathway activation tolerance of CPT-induced DNA damage under our experimental conditions. However, although PCNA was highly ubiquitinated in Rad18 (WT)-transfected cells (even without genotoxin treatment), this level of PCNA ubiquitination alone was clearly insufficient to induce FANCD2 monoubiquitination in the absence of CPT treatment (Fig. 4A). Moreover, we did not detect PCNA ubiquitination in response to CPT, even though the doses of CPT used here induced similar levels of Chk1 phosphorylation to that elicited by 20 J/m2 of UVC (a fluence that elicits both Chk1 phosphorylation and PCNA monoubiquitination, as shown in Fig. 6C). These results are fully consistent with the observations of Niimi et al. who showed that CPT does not induce PCNA monoubiquitination, and that PCNA modification at K164 is necessary for tolerance of UV-induced DNA damage but dispensable for tolerance of CPT-induced lesions. Taken together, these results identify a novel Rad18-mediated mechanism for processing and/or repair of CPT-induced DNA damage. Moreover, because cellular responses to CPT are PCNA ubiquitination-independent,44 we infer that alternative targets of Rad18 E3 ligase activity mediate repair of CPT-induced lesions.
Rad18 status affects chromatin loading of FA core complex proteins but not USP1 expression.
DNA damage-induced FANCD2 monoubiquitination is promoted by the FA core complex (which contains the FANCD2-directed E3 ligase FANCL) and is antagonized by the de-ubiquitinating (DUB) enzyme USP1, which is degraded in response to genotoxins.54 We investigated whether Rad18-dependent FANCD2 ubiquitination is due to increased core complex activation or decreased USP1 levels.
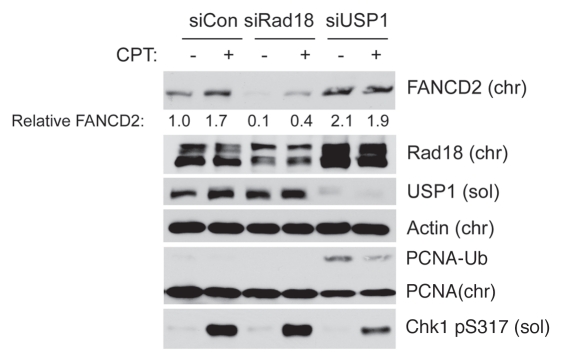
As shown in Figure 7, levels of USP1 were unaffected by CPT treatment in control (Rad18-replete) cultures. Therefore, in contrast with certain other genotoxins,54 FANCD2 ubiquitination in response to low doses of CPT is not a result of USP1 degradation. Moreover, USP1 levels were not affected by Rad18-depletion. We conclude that changes in USP1 levels do not account for CPT-induced and Rad18-mediated FA pathway activation.
Figure 7.
USP1 expression levels are insensitive to CPT and Rad18. Replicate plates of H1299 cells were transfected with siRad18, siUSP1 or siCon oligonucleotides. The resulting cultures were treated with 100 nM CPT. After 2 hr, cells were separated into soluble and chromatin fractions, which were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
In both control and Rad18-depleted cells, chromatin binding of the FA core complex components FANCA and FANCL increased in response to CPT treatment. Moreover, levels of chromatin-associated FANCA and FANCL were reduced in Rad18-depleted cells (Figs. 2A–D and 6A), both basally and following CPT treatment. Therefore, our results suggest that Rad18 regulate FANCD2 monoubiquitination via a mechanism involving increased targeting of the FA core complex to chromatin at sites of CPT-induced lesions.
Discussion
Top1 poisons such as CPT target S-phase cells,55 causing DNA replication-associated DNA damage. Thus, Top1ccs are converted to DSB via a “replication run-off” mechanism, whereby the leading strand is replicated only up to the last nucleotide flanking the 5′ end of the TOP1cc and terminates in a DSB.56 Genetic alterations in DSB-inducible checkpoint signaling pathways (e.g., ATM, CHK2) and DSB-specific DNA repair mechanisms (e.g., NBS1, Rad51C) sensitize mammalian cells to Top1 poisons,9 further consistent with a DSB-mediated mechanism for killing of cells in response to Top1 inhibitors.
While the FA pathway has been studied most extensively in the context of DNA interstrand crosslink repair, FANCs have also been implicated in processing and repair of DSB. In eukaryotic cells, two main pathways exist for repair of DSB: Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR). Studies with cells from mutant Fancd2−/− mice suggested an NHEJ-independent role for Fancd2 in repair of IR-induced DSB.57 Jasin and colleagues showed that monoubiquitinated FANCD2 promotes DSB repair via homologous recombination.47 More recent studies suggest that the FA pathway plays a role in resecting dsDNA ends, thereby directing repair away from abortive NHEJ and toward HR.48
Consistent with a role for the FA pathway in promoting HR, FA cells show sensitivity to CPT (Nomura et al. and this study) and studies using replication-competent Xenopus egg extracts have suggested a requirement for FANCD2 in replication restart following CPT-induced fork collapse.41 However, mechanisms of FA pathway activation and integration of the FA pathway with other DNA replication and repair processes have not been studied.
We show here that efficient CPT-induced FANCD2 activation requires the Rad18 E3 ubiquitin ligase. Consistent with a role for Rad18 in CPT-induced FA pathway activation, we show that both FANCD2 and Rad18 are required for survival of CPT-treated cells. We and others have previously shown that FA pathway activation in response to bulky DNA lesions is Rad18-dependent.27,43 The mechanism of FANCD2 ubiquitination in response to bulky DNA lesions (such as BPDE adducts and CPD) is indirect and occurs secondary to Rad18-induced PCNA monoubiquitination.27 However, in contrast with bulky DNA lesions, CPT does not induce detectable PCNA monoubiquitination as shown in this study and Niimi et al. Moreover, while PCNA monoubiquitination at K164 is important for tolerance of UV- or MMS-induced DNA damage, this PCNA modification does not contribute to CPT-tolerance.44 Thus, Rad18-mediated pathways for repair of Top1cc-induced DSB and bulky DNA lesions are separable based on their differential requirements for PCNA monoubiquitination.
Previous work by the Chen lab also implicated Rad18 in tolerance of CPT- and IR-induced DSB.23 Those elegant studies identified a novel E3 ligase-independent mechanism for Rad18-dependent stimulation of HR, most likely facilitated via direct interactions between Rad18 and a Rad51 paralog, Rad51C.23 Our report identifies an additional mechanism for Rad18-dependent FANCD2 ubiquitination and tolerance of CPT-induced lesions that is dissociable from the pathway described by the Chen lab23 based on its requirement for Rad18 E3 ligase activity.
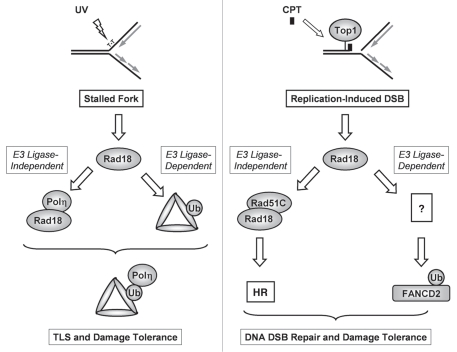
Collectively, these studies indicate that Rad18 plays E3 ligase-dependent as well as E3 ligase-independent roles in tolerance of both bulky DNA lesions (including BPDE adducts, CPD and cis-platin) and DSB (Fig. 8). Our study reveals an interesting analogy between responses to bulky adducts and DSB, whereby Rad18 contributes to tolerance of both forms of damage via separable E3 ligase-dependent and -independent mechanisms: during TLS Rad18 functions as a molecular chaperone that guides Polη to sites of replication fork stalling independently of its E3 ligase activity21,58 and subsequently monoubiquitinates PCNA to promote stable association of TLS polymerases with the replication machinery.19 In response to CPT-induced DSB, Rad18 associates with Rad51C and facilitates its chromatin loading in an E3 ligase-independent manner23 yet also promotes FANCD2 ubiquitination, DNA repair and survival in an E3 ligase-dependent manner (this study).
Figure 8.
E3 ubiquitin ligase-dependent and E3 ligase-independent roles of Rad18 in tolerance of bulky DNA lesions (left) and replication-associated DSB (right). In response to bulky lesions such as UV-induced CPD, Rad18 associates with and chaperones polη to sites of replication stalling in an E3 ligase-independent manner. At the stalled replication fork Rad18 monoubiquitinates PCNA facilitating stable engagement of TLS polymerases with the DNA replication machinery, enabling lesion bypass and DNA damage tolerance. In response to replication-induced DSB, Rad18 associates with Rad51C and facilitates HR in an E3 ligase-independent manner, as demonstrated by Huang et al. Additionally, DSB-induced FA pathway activation and DNA damage tolerance requires Rad18 E3 ligase activity (this study).
Our work prompts further questions regarding the mechanism of Rad18 activation at Top1ccs-induced DSB and cellular targets of Rad18 E3 ligase activity that mediate FA pathway activation. Huang et al. showed that RNF8, an E3 ubiquitin ligase involved in Histone H2A ubiquitination is required for efficient recruitment of Rad18 to DSB.23 In our study, Rnf8-depletion reduced the magnitude of CPT-induced FANCD2 ubiquitination, potentially consistent with a linear activation pathway involving Rnf8, Rad18 and FANCD2. We typically observed a slight reduction of Rnf8 chromatin loading in Rad18-depleted cells (for example see Fig. 5). Chen and colleagues have reported that Rad18 is downstream of RNF8 under certain conditions.23 Our experiments may indicate that Rnf8 and Rad18 do not always operate in a simple linear pathway. This is by no means unprecedented, and participants of DSB signaling pathways do not always have simple upstream/downstream relationships. For example, p95 and ATM have a complex relationship, whereby p95 has roles both upstream and downstream of ATM.
Interestingly, Rad18 deficiency had a greater inhibitory effect on FANCD2 ubiquitination than on chromatin loading of Rad51, whereas Rnf8 depletion was more inhibitory for Rad51 chromatin loading than for FANCD2 monoubiquitination. It is possible, therefore, that additional Rnf8-independent mechanisms contribute to Rad18-mediated FANCD2 ubiquitination. In responses to bulky DNA lesions, recruitment of Rad18 to stalled replication forks and the ensuing PCNA ubiquitination is dependent on RPA-coated ssDNA.53,59 Top1ccs likely generate RPA-coated ssDNA via resection of replication-dependent DSB and at sites of discontinuous DNA replication on the lagging strand.9 Indeed, CPT rapidly induces Chk1 phosphorylation, a signal that is thought to be initiated by recruitment of the ATRIP/ATR complex to RPA-coated ssDNA.60 Thus, it is possible that RPA-coated ssDNA contributes to Rad18 activation at Top1cc-induced DSB.
Potentially, Rad18 might promote CPT-induced FANCD2 monoubiquitination activation via inhibition of FANCD2 deubiquitination or by stimulation of FANCD2-directed E3 Ligase activity. USP1 is a deubiquitinating enzyme for FANCD2, and DNA damage-induced USP1 cleavage has been proposed as a major mechanism for increasing levels of monoubiquitinated FANCD2 following genotoxin treatments.54,61
We show here that Rad18 status does not affect levels of USP1 basally or in CPT-treated cells (Fig. 7). Thus, Rad18 is unlikely to contribute to CPT-induced FA pathway activation via changes in USP1 expression levels. We cannot exclude the possibility that Rad18 affects USP1 activity or that Rad18 influences alternative (putative) FANCD2-directed DUBs. However, since Rad18-depletion reduces the chromatin loading of FA core complex proteins, we consider it most likely that Rad18 contributes to FA pathway activation by promoting FA core complex activation. Precisely how Rad18 E3 ligase activity regulates core complex activation remains unclear. The most direct potential mechanism for Rad18-dependent FA core complex activation involves ubiquitination of one or more members of the core complex. There is a precedent for monoubiquitination of core complex members: Dutta and colleagues demonstrated that UBE2T, an E2 ubiquitin-conjugating enzyme that mediates FANCD2 ubiquitination, is itself monoubiquitinated, although this modification is inhibitory for E2 ligase activity.62 In preliminary experiments, we have not noticed any effects of Rad18 status on electrophoretic mobility of UBE2T or FA core complex members that we have examined. Nevertheless, it is conceivable that reduced ubiquitination of FA core complex proteins in Rad18-deficient cells compromises core complex function. An alternative possibility is that Rad18-mediated modification of other replication fork or DNA damage signaling components in CPT-treated cells contributes to recruitment of the FA core complex. While Rad18 is most commonly studied in the context of PCNA ubiquitination, alternative Rad18 substrates have been described, including RFC2 and 53BP1.25,63 Therefore, it possible that ubiquitinated forms of RFC2, 53BP1 or other unidentified Rad18 targets mediates activation of the FA core complex in response to CPT. Regardless of the precise mechanism(s) by which Rad18 promotes FA pathway activation, this report expands the repertoire of DNA repair mechanisms that are known to be regulated by Rad18 and reveals a new pathway that protects cells from Top1-inhibiting anticancer drugs. Further studies are underway to identify additional substrates of Rad18 that mediate FA pathway activation in response to CPT.
Materials and Methods
Cell culture.
H1299, A549 and HDF cells were maintained as described previously in reference 27. FANCD2-deficient PD20 cells, FANCA-deficient GM6914 and isogenic cell lines stably expressing reconstituted wild-type FANCs were obtained from Dr. Alan D'Andrea, Dana Farber Cancer Institute and were validated for FANC status by immunoblotting as described in recent publications.45,64,65 All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, streptomycin sulfate (100 µg/ml) and penicillin (100 U/ml), 5 mM L-glutamine, and cultures were incubated at 37°C in humidified chambers with 5% CO2.
Antibodies and drugs.
Top1 antibody (Abcam); PCNA; Rad51; FANCD2; FANCL; Rnf8; β-actin (Santa Cruz) Rad18; (Bethyle); Rad6; γH2AX; p-Chk1; p-Chk2; p-ATM (Cell Signaling). Camptothecin (Sigma, St. Louis, MO) was dissolved in anhydrous DMSO at 10 mM concentration and stored at −20°C. The 10 mM CPT stock solution was diluted further in to anhydrous DMSO and added to the cells. All other reagents used in this study were analytical grade and obtained from certified vendors.
RNA interference (RNAi).
All siRNAs used in this study were purchased from Dharmacon (Lafayette, CO) and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols as described previously in reference 27 and 58. The siRNAs used in this study are as follows: siRad18-3, 5′-GAG CAU GGA UUA UCU AUU CAA UU-3′, previously described by Hicks et al. siRad18-3′UTR, 5′-UUA UAA AUG CCC AAG GAA AUU-3′; siFANCD2, 5′-GCA CCG UAU UCA AGU ACA AUU-3′; siFANCA, 5′-CAG CGT TGA GAT ATC AAA GAT-3′; siFANCI, 5′-CTG GCT AAT CAC CAA GCT TAA-3′; siRNF8, 5′-GAG AAG CUU ACA GAU GUU U-3′; non-targeting siControl, 5′-UAG CGA CUA AAC ACA UCA AUU-3′. For all knockdowns, cells were transfected twice (8–10 hr for each transfection) using 100–150 nM siRNA with a 24 hour recovery period between the first and second transfections. Cells were typically subjected to genotoxin treatments 24 hr after the second transfection.
SDS-PAGE and immunoblotting.
Cells were harvested by aspirating the medium and washing twice with cold phosphate buffer saline (PBS). Chromatin (essentially nucleated cytoskeletons) and soluble fractions were prepared according to standard protocols essentially as described by Izumi and colleagues.67 Briefly, cells were resuspended in cytoskeleton (CSK) buffer (10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF and 0.1% Triton X-100, freshly supplemented with protease inhibitor and phos-stop cocktail tablets from Roche), scraped into microfuge tubes and allowed to lyse for 10–20 min on ice. Detergent-insoluble nuclei and soluble fractions were separated by centrifugation at 3,000 g for 5 min. The soluble fractions were removed, and the remaining insoluble nuclear pellets were washed twice with 0.5 ml of lysis buffer. The resulting washed nuclei were resuspended in an approximately equal volume of CSK. Protein concentrations of soluble and nuclear fractions were determined immediately. Extracts were normalized for protein content, dissolved in 4x SDS-PAGE denaturing buffer and heated to 100°C for 10 min. Denatured samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were typically stained with Ponceau to verify transfer and equivalent loading protein loading. Used as Ponceau stain was removed by extensive rinsing with PBS and the resulting membranes were probed with appropriate antibodies. Bound antibodies were detected using chemiluminiscence. Actin (present in soluble and chromatin fractions prepared by this method) was generally used as a loading control. Films were scanned, and relative intensities of protein bands were determined densitometry using Image J software.
In vivo complex of enzyme (ICE) assay.
To assess the formation of Top1ccs in vivo, exponentially growing cells in 60 mm dishes were treated with 1 µM CPT for 30 min and lysed in TE buffer (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA) containing 1% sarkosyl. The cell lysates were fractionated by centrifugation in a CsCl gradient for 22 hr as described previously in reference 68 and 69. For each sample equal volumes of DNA-containing fractions were diluted in 25 mM sodium phosphate buffer and loaded on to a pre-soaked nitrocellulose membrane using a dot-blot apparatus (Bio-Rad). The membranes containing immobilized genomic DNA were immunoblotted with Top1 antibody.
Fluorescence microscopy.
Exponentially growing cells were transfected with indicated siRNAs and plasmid vectors. Transfected cells were trypsinized and seeded in two- or four-well LabTek chamber slides to give a confluence of 40–60% 16–24 hours after replating. The resulting cells were treated with CPT (or DMSO for controls). At various times after CPT treatment, cells were washed with cold PBS and permeabilized with PBS containing 0.2% Triton X-100 (PBS-T) for ∼1 min and then washed twice with PBS. Permeabilized cells were fixed in 3% formaldehyde for 10 min and then in 100% methanol (−20°C) for 10 min at room temperature. Fixed cells were blocked in 10% FBS for 30 min. After three washes with PBS-T, cells were incubated overnight at 4°C with primary antibodies in PBS containing 5% bovine serum albumin (BSA) and 0.1% Triton X-100 (PBST). The slides were washed three times with PBST containing 1% BSA then incubated with anti-mouse IgG-Cy3 antibodies (Sigma-Aldrich) for 2 hr at RT. After washing extensively with PBST to remove unbound secondary antibody, the slides were mounted with DAPI-containing VECTASHIELD® Mounting Medium (Vector Laboratories) and imaged and analyzed with an Olympus BX61 upright fluorescence microscope using a U Plan Fluorite Apo 40x objective lens.
Survival assays.
H1299 or A549 cells were transfected with indicated siRNAs and plasmid vectors. 48 hours post-transfection, 300–500 cells were plated in triplicate in six-well tissue culture dishes. Cells were allowed to attach for 16 hours and treated with indicated concentrations of CPT for 16–20 h. Following CPT treatment, cells were washed three times with PBS, once with (CPT-free) growth medium and placed in fresh complete medium. After 7–10 days, surviving colonies were fixed with methanol for 10 min and stained with crystal violet. Colonies containing at least 50 cells were enumerated.
Reproducibility.
All data shown are representative of experiments that were performed at least three times with qualitatively similar results on each occasion.
Acknowledgements
This work was supported by grants ES09558 and ES12917 from the NIH to C.V. We thank Dr. Tao-Shih Hsieh and Dr. Satoshi Tateishi for help and advice with these studies. We also thank Dr. Alan D'Andrea for kindly providing PD20, GM6914 and their wild-type complemented cells.
Abbreviations
- 3′-UTR
3′-untranslated region
- 53BP1
p53 binding protein 1
- ATM
ataxia telangiectasia mutated kinase
- ATP
adenosine triphosphate
- ATR
ataxia telangiectasia related kinase
- ATRIP
ataxia telangiectasia related kinase interacting protein
- BPDE
benzopyrene-dihydrodiol-epoxide
- BRCA2
breast cancer associated protein 2
- BSA
bovine serum albumin
- Cdc45
cell division cycle 45
- Chk1
checkpoint kinase 1
- Chk2
checkpoint kinase 2
- cis-Pt
cisplatinum
- CPD
cyclobutane pyrimidine dimers
- CPT
camptothecin
- CSK
cytoskeletal buffer
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethylesulfoxide
- DSB
double strand break
- DUB
deubiquitinating enzyme
- EDTA
ethylenediaminotetrachloro acetic acid
- FA
Fanconi anemia
- FANCs
Fanconi anemia proteins
- FANCA
Fanconi anemia A protein
- FANCB
Fanconi anemia B protein
- FANCD1
Fanconi anemia D1 protein
- FANCD2
Fanconi anemia D2 protein
- FANCJ
Fanconi anemia J protein
- FANCL
Fanconi anemia L protein
- FANCN
Fanconi anemia N protein
- FANCI
Fanconi anemia I protein
- FBS
fetal bovine serum
- γ-H2AX
gamma-histone 2AX variant (phosphorylated)
- HDF
human dermal fibroblasts
- HR
homologous recombination
- IB
immunoblotting
- ICE
in vivo complex enzyme assay
- IF
immunofluorescence
- MMS
methyl methanesulfonate
- NBS1
nijmegen breakage syndrome protein1
- NHEJ
non-homologous end joining
- PBS
phosphate-buffered saline
- PCNA
proliferating cell nuclear antigen
- Polη
DNA polymerases eta
- Polι
DNA polymerases iota
- Polκ
DNA polymerases kappa
- RFC
replication factor C
- RPA
replication protein A
- RT
room temperature
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- siRNA
small interference RNA
- ssDNA
single-stranded DNA
- TBE
tris-boric acid-EDTA (buffer)
- TLS
translesion DNA synthesis
- Top1
DNA topoisomerase I
- Top1cc
DNA-topoisomerase I covalent or cleavage complex
- Usp1
ubiquitin-specific processing protease 1
- UV
ultra violet radiation
- WT
wild type
Supplementary Material
References
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Corbett KDBJ. Structure, molecular mechanisms and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:5–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 3.Leppard JBCJ. Human DNA topoisomerase I: relaxation, roles and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 7.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, et al. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J Mol Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 8.Liu LF, D'Arpa P. Topoisomerase-targeting antitumor drugs: mechanisms of cytotoxicity and resistance. Important Adv Oncol. 1992:79–89. [PubMed] [Google Scholar]
- 9.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 10.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 11.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 12.Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjornsti MA. Cancer therapeutics in yeast. Cancer Cell. 2002;2:267–273. doi: 10.1016/s1535-6108(02)00160-5. [DOI] [PubMed] [Google Scholar]
- 15.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, et al. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang TH, Chen HC, Chou SM, Yang YC, Fan JR, Li TK. Cellular processing determinants for the activation of damage signals in response to topoisomerase I-linked DNA breakage. Cell Res. 20:1060–1075. doi: 10.1038/cr.2010.95. [DOI] [PubMed] [Google Scholar]
- 17.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 18.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- 19.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 20.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, et al. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inagaki A, van Cappellen WA, van der Laan R, Houtsmuller AB, Hoeijmakers JH, Grootegoed JA, et al. Dynamic localization of human RAD18 during the cell cycle and a functional connection with DNA double-strand break repair. DNA Repair. 2009;8:190–201. doi: 10.1016/j.dnarep.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, et al. RAD18 promotes DNA doublestrand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 2009;37:2176–2193. doi: 10.1093/nar/gkp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol Cell Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated trans-lesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem. 2010;285:31525–31536. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moldovan GL, D'Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moldovan GL, D'Andrea AD. FANCD2 hurdles the DNA interstrand crosslink. Cell. 2009;139:1222–1224. doi: 10.1016/j.cell.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang TT, D'Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 33.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL and FANCI. Mol Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Alpi AF, Patel KJ. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair. 2009;8:430–435. doi: 10.1016/j.dnarep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pommier Y. Camptothecins and topoisomerase I: a foot in the door. Targeting the genome beyond topoisomerase I with camptothecins and novel anticancer drugs: importance of DNA replication, repair and cell cycle checkpoints. Curr Med Chem Anticancer Agents. 2004;4:429–434. doi: 10.2174/1568011043352777. [DOI] [PubMed] [Google Scholar]
- 38.Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nomura Y, Adachi N, Koyama H. Human Mus81 and FANCB independently contribute to repair of DNA damage during replication. Genes Cells. 2007;12:1111–1122. doi: 10.1111/j.1365-2443.2007.01124.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Fan Q, Ren K, Auerbach AD, Andreassen PR. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma. 2010;119:637–649. doi: 10.1007/s00412-010-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang LC, Stone S, Hoatlin ME, Gautier J. Fanconi anemia proteins stabilize replication forks. DNA Repair. 2008;7:1973–1981. doi: 10.1016/j.dnarep.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 43.Geng L, Huntoon CJ, Karnitz LM. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol. 2010;191:249–257. doi: 10.1083/jcb.201005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood. 2011 doi: 10.1182/blood-2010-10-311761. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–2255. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 47.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 49.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, et al. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 50.Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 51.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, et al. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 55.Horwitz SB, Horwitz MS. Effects of camptothecin on the breakage and repair of DNA during the cell cycle. Cancer Res. 1973;33:2834–2836. [PubMed] [Google Scholar]
- 56.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houghtaling S, Newell A, Akkari Y, Taniguchi T, Olson S, Grompe M. Fancd2 functions in a double strand break repair pathway that is distinct from non-homologous end joining. Hum Mol Genet. 2005;14:3027–3033. doi: 10.1093/hmg/ddi334. [DOI] [PubMed] [Google Scholar]
- 58.Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, et al. Phosphorylated Rad18 directs DNA Polymerase {eta} to sites of stalled replication. J Cell Biol. 2010;191:953–966. doi: 10.1083/jcb.201006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, et al. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 60.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 61.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 62.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 63.Tomida J, Masuda Y, Hiroaki H, Ishikawa T, Song I, Tsurimoto T, et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J Biol Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song IY, Barkley LR, Day TA, Weiss RS, Vaziri C. A novel role for Fanconi anemia (FA) pathway effector protein FANCD2 in cell cycle progression of untransformed primary human cells. Cell Cycle. 9:2375–2388. doi: 10.4161/cc.9.12.11900. [DOI] [PubMed] [Google Scholar]
- 65.Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem. 285:31525–31536. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, et al. Differential roles for DNA polymerases eta, zeta and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Izumi M, Yatagai F, Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J Biol Chem. 2001;276:48526–48531. doi: 10.1074/jbc.M107190200. [DOI] [PubMed] [Google Scholar]
- 68.Subramanian D, Kraut E, Staubus A, Young DC, Muller MT. Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res. 1995;55:2097–2103. [PubMed] [Google Scholar]
- 69.Daroui P, Desai SD, Li TK, Liu AA, Liu LF. Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J Biol Chem. 2004;279:14587–14594. doi: 10.1074/jbc.M311370200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.