Abstract
BRAF, a cellular oncogene and effector of RAS-mediated signaling, is activated by mutation in ~60% of melano-mas. Most of these mutations consist of a V600E substitution resulting in constitutive kinase activation. Mutant BRAF thus represents an important therapeutic target in melanoma. In an effort to produce a pre-clinical model of mutant BRAF function in melanoma, we have generated a mouse expressing BRAF V600E targeted to melanocytes. We show that in these transgenic mice, widespread benign melanocytic hyperplasia with histolo-gical features of nevi occurs, with biochemical evidence of senescence. Melanocytic hyperplasia progresses to overt melanoma with an incidence dependent on BRAF expression levels. Melanomas show CDKN2A loss, and genetic disruption of the CDKN2A locus greatly enhances melanoma formation, consistent with collaboration between BRAF activation and CDKN2A loss suggested from studies of human melanoma. The development of melanoma also involves activation of the Mapk and Akt signaling pathways and loss of senescence, findings that faithfully recapitulate those seen in human melanomas. This murine model of mutant BRAF-induced melanoma formation thus provides an important tool for identifying further genetic alterations that cooperates with BRAF and that may be useful in enhancing susceptibility to BRAF-targeted therapeutics in melanoma.
Keywords: BRAF, p53, CDKN2A, AKT, melanoma, senescence
Introduction
The most prevalent oncogenic mutations that pose a potential therapeutic target in human melanoma occur in BRAF (Pollock and Meltzer, 2002). This protein is a serine–threonine specific protein kinase that is activated by RAS and that functions to transduce signals intracellularly. BRAF mutations have been detected in 60–70% of malignant melanomas and in a variety of other cancer types, including thyroid, lung and colon cancers, with most mutations comprising a single substitution (V600E) that results in constitutive activation of the kinase function (Pollock and Meltzer, 2002; Goel et al., 2006). BRAF mutation is not sufficient to induce malignancy: it has been shown that up to 80% of benign nevi carry the same mutation in BRAF as is observed in melanoma (Pollock et al., 2003). Yet the high prevalence of BRAF mutation in this disease, and earlier successes in using small-molecule inhibitors to treat other solid tumors with activating kinase mutations (Lynch et al., 2004), suggests that BRAF is an appealing therapeutic target in melanoma, although in clinical trials initial attempts to target BRAF with the small-molecule inhibitor sorafenib have yielded only rare tumor responses (Bardeesy et al., 2001). The observed mutation rate of BRAF in benign nevi, and the small fraction of nevi that transform to melanoma, underscores the fact that genetic changes in addition to those found in BRAF are necessary for the development of overt melanoma. Understanding these additional alterations may be a prerequisite for successful BRAF-targeted treatments.
Perhaps the most important additional genetic changes identified in melanoma are in the Cdkna2a locus, which encodes both p16INK4A and p14ARF (p19Arf in the mouse). Germline mutations in p16INK4A confer a familial predisposition for melanoma (Haluska et al., 2006; Laud et al., 2006), and recent findings implicate p16INK4A in the senescent response to oncogene mutation (Gray-Schopfer et al., 2006). In melanocytes, BRAF mutation alone leads to nevus formation with initial cell proliferation and later growth arrest and senescence with concomitant induction of both p16INK4A and senescence-activated β-galactosidase (SA-β-gal). Subsequent loss of p16INK4A occurs with tumor progression (Michaloglou et al., 2005). Several murine models that introduce activated H-RAS into a background deficient in Cdkn2a or p16Ink4a alone support the role of this locus in melanoma development (Chin et al., 1997).
The p53 pathway is also abrogated in melanoma-genesis. Mutations in p53 itself are rare. However, as alternative splicing of the CDKN2A mRNA yields two distinct messages, encoding p16INK4A and the p53-modulator p14ARF, loss of the locus results in reduction of p53 pathway activity. Recent studies suggest a critical role for the function of p14ARF as a tumor suppressor. It serves to arrest cell cycle progression or promote cell death after oncogenic stimulation, loss of pRb or DNA damage, and participates in the control of p53 through its interaction with the MDM2 protein (Muthusamy et al., 2006; Chang et al., 2007). p14ARF deficiency has been shown to abrogate oncogene-induced senescence and to increase susceptibility to transformation. Loss or mutation of p14ARF facilitates the progression of melanoma, and it is thought that the low frequency of p53 mutations in melanoma is related to the frequent loss of p14ARF, which renders p53 mutation or loss superfluous (Castresana et al., 1993). As is the case for p16INK4A, murine studies support the role of the p53 pathway in melanoma in studies showing that p19Arf−/−/Tyr-Hras and p53−/−/Tyr-Hras mutant mice develop melanoma with high penetrance (Bardeesy et al., 2001; Sharpless et al., 2003).
Murine models of the above genetic alterations that incorporate recent findings on BRAF will prove useful in dissecting the common and distinct role of activated signaling and CDKN2A and p53 pathways in vivo in the melanocyte lineage. To better understand the biology of BRAF mutation in melanoma and its therapeutic implications, we constructed a mouse model in which the BRAF V600E is specifically expressed in melanocytes using the mouse tyrosinase enhancer and promoter. Mice carrying a transgenic mutant BRAF showed benign melanocytic hyperplasia histologically reminiscent of human nevi. We then crossed the BRAF V600E transgenic mice with Cdkn2a-null (affecting both p16Ink4a and p19Arf) and p53-null mice. We show here that mice carrying the mutant BRAF allele develop melanoma, and that the genetic features of the melanoma suggest that senescence is overcome concurrent with the loss of p16Ink4a expression, in some cases Cdkn2a locus deletion, and activation of Akt. In backgrounds heterozygous for either Cdkn2a or p53 deficiency, melanoma develops with an enhanced incidence, a shorter latency to tumor formation, and metastases to lung and lymph nodes. This murine system faithfully recapitulates the genetic changes seen in human melanoma.
Results and discussion
Transgenic mutant BRAF V600E expression from a Tyr-promoter leads to melanocytic and Schwann cell hyperplasia with evidence of senescence
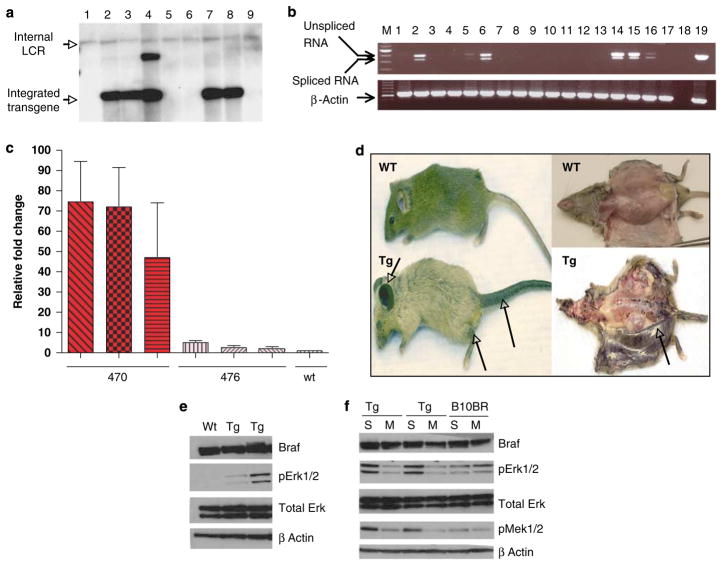
The mutant BRAF expression construct was designed such that the BRAF V600E gene, cloned from a human melanoma cell line, was inserted into the vector (pNEB 193) downstream of the murine tyrosinase locus control region (promoter enhancer) (Supplementary Figure 1a). This construct was injected into single-cell embryos of F1 C57BL/J6XCBA mice and the offspring were genotyped by Southern blotting as shown in Figure 1a. Four independent founder lines that expressed the transgene in the germline were propagated for study and the sequence of the transgene was verified. As shown in Figure 1b, RT–PCR showed that the transgene was expressed in the skin and brain and very weakly in the lung (Figure 1b, lanes 2, 6 and 5, respectively), but not in the heart, kidney or liver, with the expression in the skin resulting at least in part from expression in melanocytes (Figure 1b, lanes 14, 15 and 16).
Figure 1.
Genotype and phenotype of the mutant BRAF transgenic mouse. (a) Southern blot detection of transgene. A strong extra band in lane 4 might be because of partial digestion of genomic DNA. LCR, locus control region (part of tyrosinase enhancer). (b) RT–PCR analysis of BRAF expression. The bands of 500 and 435 bp represent unspliced and spliced mRNA, respectively. The spliced form represents about 65 bp of 3′ untranslated intron, which is about 65 bp in size. 1–6, tissues from BRAF V600E mice: 1, heart; 2, brain; 3, kidney; 4, liver; 5, lung; 6, skin. 7–13, corresponding tissues from C57BL6 animal. 14–16, transgenic isolated melanocytes. 17, wild-type melanocytes. 18, negative control. 19 DNA vector (not spliced). Lower panel, β-actin controls; 19, positive control ACTB. (c) Real-time RT-PCR quantitative expression analysis of BRAF V600E expression. Three whole-skin samples from 470 animals, three from 476, and wild type (wt). Each analysis was carried out in triplicate. (d) Phenotype of BRAF V600E Tg mouse line 470 compared with C57BL6. (e) Western blot of whole-cell lysate from cultured melanocytes of wt and transgenic (Tg) animals. (f) Western blot of the effect of human BRAF V600E region-specific siRNA on mouse melanocytes from BRAF V600E Tg mice and B10BR wt mouse melanocyte cell lines. S: scrambled (irrelevant) siRNA, M: BRAF V600E mutant region-specific siRNA. pErk1/2 is significantly and specifically decreased by human BRAF V600E specific siRNA as compared with control. pMek is also decreased.
Two founder lines, denoted 470 and 476, were studied in detail. These lines differed in the extent of their phenotype, with line 470 being more penetrant. Real-time RT–PCR examination of the level of BRAF V600E transcript expression in the two lines (Figure 1c) found that the level of expression of transgene was three-fold higher in line 470 as compared with line 476, whereas there is no transgene expression in wild-type skin samples. To determine the relative expression levels of the transgene, immunoblotting was carried out to detect both murine endogenous and human exogenous BRAF (which are not distinguishable by available antibodies). The results showed that there were no discernible differences in total BRAF protein (we used total cell lysates from cultured mouse melanocytes and also whole-skin lysate from both wild-type and transgenic animals for the analysis), suggesting the transgenic protein level is low compared with endogenous levels (Figure 1e and Supplementary Figure 1b). Using qRT–PCR, we did not see any difference between wild-type and transgene expression, suggesting transgene expression is low compared with endogenous BRAF. However, we cannot discount the possibility that a small percentage of melanocytes have a high level of expression of the transgene, which in turn drives the neoplastic phenotype.
The phenotype of line 470 compared with C57BL6 is shown in Figure 1d. The animals that carried mutant BRAF had hyperpigmented skin, most evident in the ear pinnae and the limbs, their coat color varied from normal to light, their tail was hypertrophied, and the eyes of some animals were small. Gross examination at necropsy (Figure 1d) showed that the dermis is uniformly hyperpigmented.
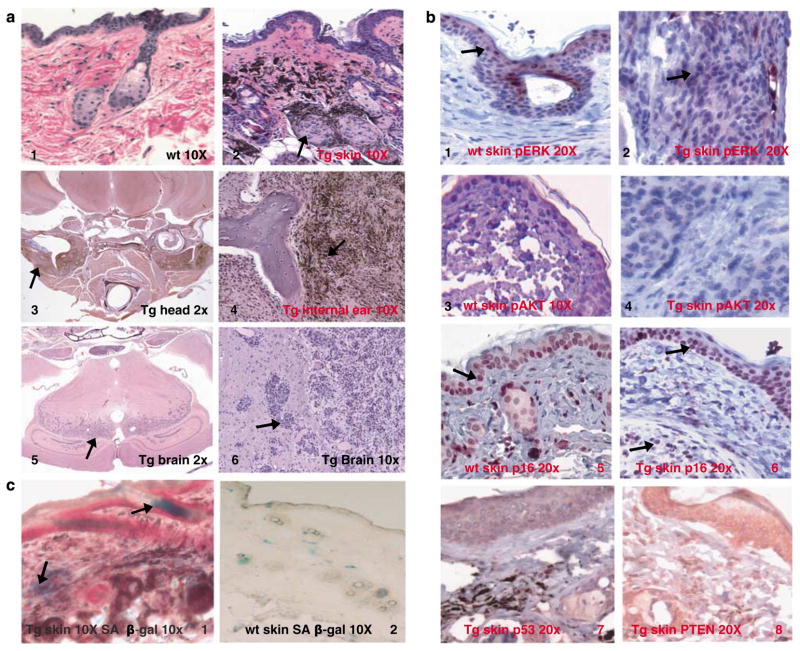
Histopathology showed that the hyperpigmentation was consequent to a benign melanocytic hyperplasia similar to nevi. Two patterns of melanocytic proliferation were observed. The most prevalent pattern showed abundant spindle cells in the dermis and subcutaneous tissue, frequently termed ‘Schwannian’ differentiation of melanocytes, visualized most prominently in line 470 (Figure 2a, panel 2). The second pattern, more characteristic of the 476 line, showed large nests of epithelioid cells, generally located in the deep dermis and subcutis (Supplementary Figure 2a, panel 4). Both patterns are of melanocytic origins as evident by S-100 staining (data not shown). No mitotic figures were identified in either pattern (data not shown).
Figure 2.
Histopathology and biochemistry of transgenic animals. (a) Panel 1, H&E of skin of wild-type animal; panel 2, H&E of skin of BRAF V600E animal (line 470) showing dermal melanocytic proliferations. Panel 3, section through the base of the skull showing vestibular schwannian melanocytic tumors, which are shown in high magnification in panel 4. Panel 5, the midbrain showing melanocytic infiltration. Panel 6, high-magnification image of the same tumor (all sections from line 470). (b) Immunohistochemistry of wild-type skin (panels 1, 3 and 5) and Tg skin (line 470, panels 2, 4 and 6) are for pErk, pAkt and p16Ink4a, whereas panels 7 and 8 are staining for p53 and PTEN, respectively, in the skin of line 470. (c), SA-β-gal is induced in the skin of BRAF V600E (line 470, panel 1) animals as compared with wild-type skin (panel 2), which is consistent with a senescent response to mutant BRAF.
The central nervous tissue was involved most prominently in line 470 (Figure 2a, panels 3, 4, 5 and 6). Pigmented spindle cell proliferations were identified in the retro- and peri-bulbar areas of the eye, the meninges, the temporal and parietal bones of the cranium, bone marrow cavity, and in the surrounding soft tissues (Figure 2a, panels 5 and 6). The infiltrative spindle cell tumors were variably pigmented with rare mitoses, and these cells showed neural crest differentiation representing Schwannian differentiation as in the skin (compare Figure 2a, panels 2 and 4). Electron microscopy showed the characteristic lamellar morphology of pigmented cells consistent with Schwann cells (Supplementary Figure 2b). Ultrastructurally there were groups of spindled pigmented cells that exhibited features of Schwannian differentiation, including prominent external lamina with lamina lucida and lamina densa, numerous pinocytotic vesicles, and abundant interdigitating cell processes with infoldings of membranes forming pseudomesaxons around collagen bundles. The presence of melanosomes at varying stages of development showed that these tumor cells are capable of melanogenesis. The majority of these proliferations seem benign; however, the extension into the cortex showed the capacity for at least locally aggressive behavior.
Our observations are consistent with the known expression of tyrosinase in the brain. Tyrosinase transcriptional function has been reported earlier in several regions of the mouse brain, including retinal pigment epithelial cells (Tief et al., 1998). Tyrosinase Cre-recombinase mice made by (Tonks et al. 2003) showed that tyrosinase is also expressed in other neural crest and neuroepithelial derived cells. In their mouse model, the reporter gene under the control of the tyrosinase enhancer and promoter is expressed in the glia of the optic nerve (neuroepithelial derived cells), basal forebrain, hippocampus, olfactory bulbs and the granule cell layer of the lateral cerebellum cortex. Functionally, it has been proposed that tyrosinase may serve a role in neuromelanin synthesis within some of these neural tissues (Tief et al., 1998). Indeed, a proportion of catecholaminergic neurons of substantia nigra in tyrosine hydroxylase knockout mice were able, through tyrosinase-mediated pathways, to produce norepinephrine and dopamine at levels of 8 and 2–3% of control mice, respectively.
Examination of the liver, lung, spleen, heart, pancreas, kidney and gastrointestinal tissue showed no involvement
We concentrated on the cutaneous melanocytic phenotype. Immunohistochemistry showed that the transgenic melanocytes expressed moderate levels of pErk1/2 as compared with wild-type skin (compare Figures 2b, panels 1 and 2), but not pAkt (Figure 2b, panel 4), and expression of p16Ink4a (Figure 2b, panel 6), p53 (Figure 2b, panel 7) and Pten (Figure 2b, panel 8) was observed. Wild-type skin showed little p16 expression, which suggests that p16 is induced by mutant BRAF in transgenic skin (Figure 2b, panel 5). The hyperplastic melanocytes also expressed senescence-associated β-galactosidase (SA-β-gal), a marker for senescence (Figure 2c, panel 1), and the senescent melanocytes did not form soft agar colonies. The co-expression of mutant BRAF, p16 and SA-β-gal suggests a senescence mechanism. Indeed (Michaloglou et al. (2005) have shown in vitro that BRAF V600E expression in melanocytes induces senescence, and that human nevi carrying BRAF mutations express SA-β-gal. This group and others have also suggested that the senescent mechanism is mediated through p16Ink4a in melanocytes (Sviderskaya et al., 2002; Michaloglou et al., 2005; Gray-Schopfer et al., 2006; Courtois-Cox et al., 2006). Our data are consistent with these reports.
We sought to examine whether these changes are specifically attributable to mutant BRAF, and to understand the biochemical consequences of signaling in these tissues. siRNA specific for the mutant human BRAF was introduced into cultured melanocytes from transgenic animals (Figure 1f). As shown, the specific human BRAF V600E specific siRNA, but not irrelevant control siRNA, decreased levels of phosphorylated Mek and Erk (Sharma et al., 2005), suggesting that the transgene-encoded mutant BRAF is responsible for the altered signaling observed in transgenic mouse skin.
Transgenic animals carrying mutant BRAF develop melanoma
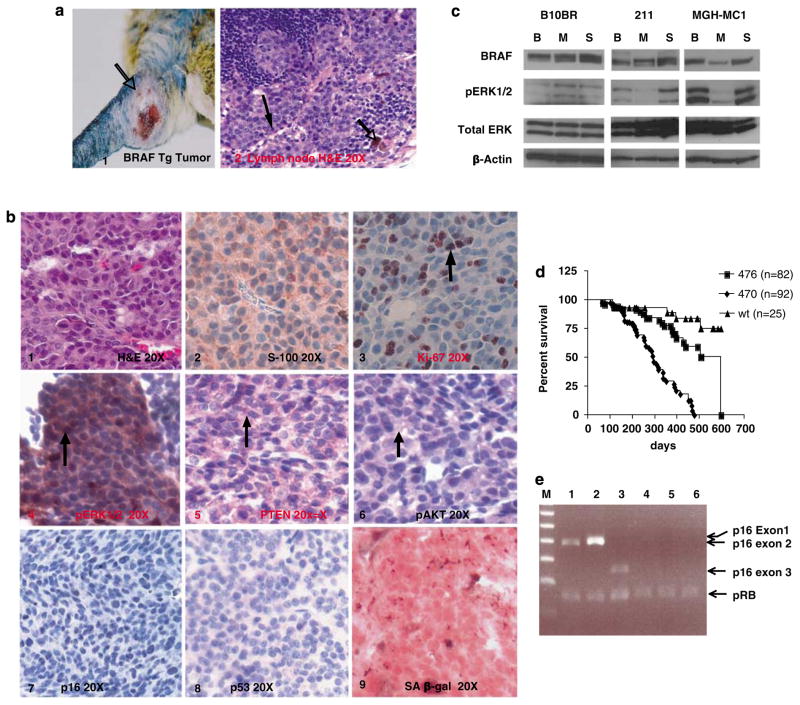
Melanomas developed in the animals carrying trans-genic mutant BRAF with an incidence dependent on BRAF expression levels, and survival was affected concomitantly (Supplementary Table 1). Figure 3a (panels 1 and 2) shows a melanoma from the 470 line, and a metastasis to a lymph node draining the region. Tumors were located in the dermis, subcutis and soft tissues, and were composed of pigmented epithelioid cells with nuclear pleomorphism and mitotic figures. All melanocytic proliferations showed cytoplasmic and nuclear staining for S100 protein (Figure 3b, panel 2). The benign melanocytic proliferations showed only rare cells staining for Ki-67, constituting <1% of the cells, whereas in the melanomas Ki-67 was identified in up to 32% of the cells (Figure 3b, panel 3). About 10% of animals from the 470 line, which expressed the highest levels of mutant BRAF mRNA, developed melanoma; the median survival for this line was 295 days. In line 476, melanomas were rare, with a median survival of 595 days. Figure 3d shows the Kaplan–Meier survival curves for these lines generated in this study. The mortality of animals dying without tumors was largely due to CNS involvement. Supplementary Table 1 tabulates the tumor incidence for the strains, and the incidence in the genetic backgrounds discussed below.
Figure 3.
Melanomas in mice carrying mutant BRAF alone metastasize and show pAkt activation and loss of the Ink4a locus. (a) Phenotype of Tg mouse melanoma tumor, and histology of a metastasis to a draining lymph node (line 470). (b) Immunohistochemistry of one of the representative melanomas from BRAF V600E Tg mice. Panel 1, H&E; 2, S100; 3, Ki-67; 4, pErk; 5, Pten; 6, pAkt; 7, p16; 8, p53; 9, SA-β-gal. (c) Specificity of signaling alterations to mutant BRAF activity. B10BR, wild-type mouse melanocyte cell line; 211, tumor cell line derived from BRAF V600E mouse melanoma; MGH-MC1, human melanoma cell line carrying BRAF V600E mutation; B: mouse BRAF-specific siRNA; M: human BRAF V600E-specific siRNA; S: luciferase (irrelevant) siRNA. pErk level was significantly and specifically downregulated by human BRAF V600E specific siRNA as compared with mouse Braf siRNA. (d) Kaplan–Meier plot of mortality of BRAF V600E transgenic lines. (e) Multiplex PCR of Cdkn2a exons 1α, 2 and 3 in control and in tumor from BRAF V600E animal. Lower band in all lanes, pRb control. Lanes 1–3 contain tail DNA and lanes 4–6 contain DNA from tumor sample #211. Lanes 1 and 4, exon 1α; 2 and 5, exon 2; 3 and 6, exon 3.
We next sought to determine the contribution of endogenous murine Braf versus human mutant trans-genic BRAF to abnormal signaling in these melanomas. siRNA to both the murine transcript and the human mutant transcript was introduced into murine control melanocytes, a melanoma cell line derived from the 470 mice, and a human melanoma cell line. As shown in Figure 3c, and consistent with our results derived from non-neoplatic skin (Figure 1f), phosphorylation of ERK is inhibited only by human mutant siRNA in the cell lines derived from the transgenic 470 mice and human melanomas.
We examined the tumors arising in the mutant BRAF background for additional biochemical and genetic changes. Phospho-Erk was uniformly overexpressed in the melanomas; expression levels by IHC seemed higher than in transgenic melanocytes (compare Figures 2b, 2 and 3b, panel 4). This increase in pErk level parallels that seen in the transition from human nevi to melanoma and may not be solely due to mutant BRAF (Uribe et al., 2006). pAkt was also uniformly strongly expressed in the murine melanomas compared with melanocytes (compare Figures 2b, panel 4 and 3b, panel 6). In human melanomas, the PI3K-AKT pathway is almost always activated in conjunction with the MAPK pathway, with activation of AKT through phosphoryla-tion in human melanomas conveying prognostic sig-nificance (Dai et al., 2005). One mechanism for AKT activation is that melanomas frequently accompany BRAF activation with loss of PTEN (Wu et al., 2003; Tsao et al., 2004; Goel et al., 2006). Additional mechanisms include NRAS mutation and AKT amplification (Dhawan et al., 2002; Stahl et al., 2004). None of the tumors that arose in these mice showed Pten loss by IHC (Figure 3b, panel 5), and no Pten or Nras mutations were detected by sequencing. Although the mechanism is not yet clear, our observations here support the hypothesis that activation of these pathways is necessary for melanoma development and progression.
Human sporadic melanomas occur with additional alterations in the p16Ink4a-pRB pathway. Here immuno-histochemistry for p16Ink4a showed heterogeneous or total loss of p16 Ink4a expression in melanomas compared with benign hyperplastic melanocytes (compare Figures 2b, panel 6 and 3b, panel 7), illustrating the complete loss of p16Ink4a expression by IHC. Multiplex PCR of this tumor DNA showed homozygous deletion of the entire Cdkn2a locus (Figure 3e), and this deletion included exon 2 and therefore resulted in both p16Ink4a and p19Arf loss. Of the six analysed tumors, a second tumor showed exon 2 and 3 homozygous deletion, and all six showed absent or reduced p16 Ink4a expression by IHC. No other p16Ink4a mutations were detected by sequencing, and no p53 mutations were detected. The loss of p16Ink4a results in loss of senescent phenotype of neoplastic BRAF V600E melanocytes (Figure 3b, panel 9).
The observations above emphasize that almost universal alterations in the p16INK4A-CDK4-RB and p14ARF-p53 pathways accompany activation of signaling pathways in human melanomas. This occurs most frequently through concurrent loss of p16INK4A and p14ARF via mutation or deletion of the Cdkna2a locus, as observed in this model. In contrast, mutations in p53 are relatively rare in melanoma (Castresana et al., 1993). The loss of the p14ARF product of the Cdkna2a locus (Pomerantz et al., 1998; Zhang et al., 1998; Christophorou et al., 2006) renders p53 mutation superfluous, as supported by cell line data, which show that most p53 mutations occur in melanoma when the Cdkna2a locus is normal (Haluska et al., 2006). Still it has been shown that p53 loss contributes to the capacity of H-RAS to promote murine melanoma (Bardeesy et al., 2001) and with mutant BRAF to promote zebra fish melanoma (Patton et al., 2005). Thus, we also sought to test the hypotheses that the combination of mutant BRAF and an introduced deficiency for either Cdkn2A or p53 would augment the development of melanomas.
Introduction of the mutant BRAF allele into Cdkn2a- and p53-deficient backgrounds leads to an increase in tumor incidence
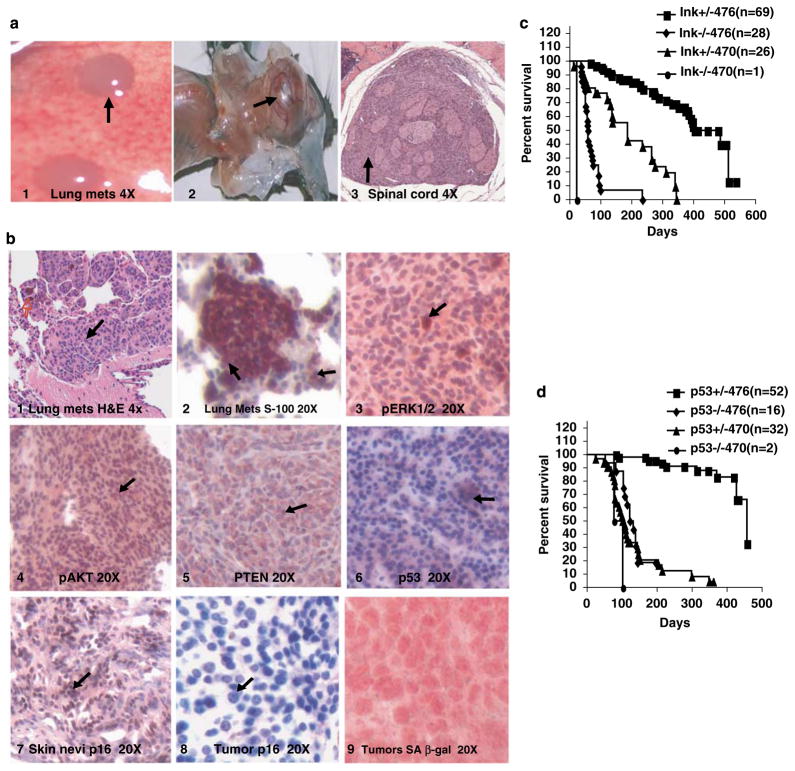
Concentrating on the 470 and 476 lines, for which we had determined BRAF expression levels, we crossed these lines with Cdkn2a-null mice (Serrano et al., 1996; Chin et al., 1997) (a generous gift of Lynda Chin), which carry a deletion of Cdkn2a exons 2 and 3 and therefore abrogate both p16Ink4a and p19Arf expression, and with p53-null mice (Jackson Laboratory, Bar Harbor, ME, USA). Murine strains that carry Cdkn2a deletions develop a variety of tumors but fail to develop melanoma (Serrano et al., 1996). Our results showed that the incidence of melanomas increased with Cdkn2a or p53 deficiency, that the latency of tumor formation is markedly lessened, and that the lifespan of populations carrying these concurrent genetic defects is substantially reduced. Figure 4a (panel 1) illustrates metastatic melanoma in the lung arising in a transgenic BRAF V600E Cdkn2a+/− background, with other mice also showing metastatic clusters of cells in the lymph nodes. Primary tumors as well as metastatic tumors showed loss of p16Ink4a by IHC, retention of Pten expression and overexpression of pAkt (Figure 4b, panels 8, 5 and 4, respectively). Retained p53 expression was found to have a strong cytoplasmic pattern (Figure 4b, panel 6). SA-β-gal expression is lost, consistent with abrogation of senescence (Figure 4b, panel 9). Results in the p53-deficient background were similar histopathologically and immunohistochemically (Supplementary Figure 4).
Figure 4.
Melanomas in Cdkn2a- and p53-deficient mice show the Ink4a locus and abrogation of senescence. (a) Panel 1, lung metastases in a tumor from BRAF V600E (470) × Cdkn2a+/− animal; 2, hydrocephalus in the same genotype; 3, spinal cord transverse section showing melanocytic infiltration of spinal cord. (b) Immunohistochemistry of primary melanoma arising in BRAF V600E × Cdkn2a+/− animal. Panel 1, pErk; 2, pAkt; 3, Pten; 4, skin p16Ink4a; 5, tumor p16Ink4a; 6, p53; 7 and 8, p16 and 9, SA-B-gal senescence staining on one of the BRAF+Cdkn2a tumors. (c) Kaplan–Meier plot of Cdkn2a crosses showing the relationship of mortality to mutant BRAF dose and inversely to Cdkn2a dose. (d) Similar plot of p53 crosses.
In the Cdkn2a-null background, which is deficient for both p16Ink4a and p19Arf, and which closely replicates the mutations seen in many human metastatic melanomas and melanoma cell lines, mortality was uniform and very early. Mice developed hydrocephalus and paralysis within days to weeks, as shown in Figure 4a, panel 2. Histology showed a pigmented cell proliferation within the CNS and especially the spinal cord, as seen in Figure 4a (panel 3), the result of which is obstruction of CNS outflow, hydrocephalus and death within 22–60 days (Supplementary Figure 3a, panel 2 and Figure 3b, panels 1–4, showing spinal cord tumors in BRAF Cdkn2a+/−background).
We conclude that the BRAF-V600E mutation is not sufficient for tumor formation and loss of the Cdkn2a locus is a common and perhaps necessary step in the progression of tumor development in this background. This is not surprising, as the loss of p16Ink4a, or the loss of p53 and the predicted subsequent reduction in p21 both result in the deregulation of Cyclin and CDK complexes and control of cellular proliferation. This also has a direct implication with relation to human melanoma genetics, and this mouse model faithfully recapitulates the human disease.
The understanding of the genetic changes contributing to the development of melanoma has been poorly exploited therapeutically. With the discovery of BRAF mutations in a large proportion of melanomas, our comprehension of the mechanistic steps giving rise to these tumors has been dramatically improved. Recent data shed light on the genetic alterations of a subset of tumors that do not carry BRAF mutations (Curtin et al., 2005), but the most appealing therapeutic target remains mutated BRAF. Pharmacologic inhibition of the signaling intermediate BRAF by sorafenib alone does not lead to clinical responses (Eisen et al., 2006), in contrast to inhibition in other tumor types of upstream signal-initiating molecules such as EGFR (Lynch et al., 2004). This emphasizes the need for understanding concurrent genetic alterations that occur in melanoma development. The MAPK pathway is driven both by RAS and by BRAF in almost all cell types. In melanoma, RAS is mutated in <20% of cases, and usually mutation favors N-RAS. Two mouse models that carry alterations in the RAS signaling cascade and develop melanoma have been reported and studied. One is a murine model carrying activated H-RAS on a tyrosinase promoter. This model shows melanocytic hyperplasia with intense skin pigmentation. Placement of Tyr-RAS into a Cdkn2a- or p53-null background resulted in the development of highly vascularized but amelanotic melanoma and no metastases (Chin et al., 1997). A second RAS mouse model is more faithful to human melanoma genetics: it carries the melanocytic expression of NRAS Q61K under the control of the tyrosinase promoter. Tyr-NRAS results in a hyperpig-mented skin phenotype and the formation of melanomas and favors the acquisition of a metastatic behavior on a Cdkn2a −/− background (Ackermann et al., 2005). In our Tyr-BRAF mouse, metastatic melanoma develops both in the wild-type background and in Cdkn2a+/− background. Mutation in BRAF is probably serving as an early event favoring the initiation of melanoma, but because oncogenic BRAF induces senescence the melanocytes progress to nevi and require a second alteration in the genome to circumvent senescence and allow melanoma formation (Table 1).
Table 1.
Changes that occur during progression of melanocytes to melanoma
| Melanocyte | Human nevus/Tg Braf model | Melanoma | |
|---|---|---|---|
| BRAF status | Normal | V600E | V600E |
| pERK level | Low | Moderately increased | High |
| pAKT level | Low | Low | High |
| p16 expression | Low | Induced | Lost |
| SA-B-gal expression | Low | Induced | Lost |
| Phenotype | Quiescent | Senescent | Malignant |
On the basis of melanoma formation consequent to spontaneous or engineered loss of CDKN2a in combination with BRAF V600E expression in melanocytes, our mouse model is the first of its kind that recapitulates human melanoma with its genetic features. The precise nature of additional steps that occur as melanomas develop in the context of mutant BRAF, such as activation of AKT and the mechanisms that control them, is not yet understood. This mouse model will enable us to use genetic methods to understand this progression and to test putative therapeutic strategies in vitro and in vivo.
Materials and methods
Construction of transgenic mice
The BRAF transgene consisted of a 2.3-kb fragment of a BRAF V600E cDNA cloned from human melanoma cell line A375 by RT–PCR. The mouse tyrosinase enhancer, promoter and SV40 poly-A tail were generous gifts from Dr Louis Montilou (CNB, Spain). BRAF V600E was inserted downstream of the tyrosinase enhancer promoter and the SV40 poly-A tail was inserted downstream of BRAF. The DNA construct was gel purified (Gene Clean Turbo kit from Qbiogen, Carlsbad, CA, USA), sequenced and microinjected into single-cell embryos of F1 C57BL/J6XCBA mice. We identified the transgene-positive founder pups by Southern blot and PCR.
Southern blots
Purified genomic DNA of 20–30 μg obtained from tail biopsy was digested overnight with XbaI and KpnI enzymes and Southern blotted with standard methods. Probe LCR1 was amplified from the tyrosinase enhancer region and cloned into a TA vector. The LCR1 fragment was digested and gel purified, and labeled randomly with a random primer kit (Amersham Biosciences, Piscataway, NJ, USA) using 32P. The membrane was incubated with the radioactively labeled probe and washed by standard protocols. The membrane was exposed to an X-ray film at −80 ºC.
RT–PCR analysis
Total RNA was isolated from various major organs, such as the skin, heart, brain, lung, liver and spleen, using the Ribopure kit (Ambion, Austin, TX, USA). Total RNA was used for single-step RT–PCR with the Superscript one-step RT–PCR kit (Invitrogen, Carlsbad, CA, USA). The primer pair used for RT-PCR were; forward primer for the BRAF cDNA (5′-GGA TAC CTG TCT CCA GAT CTC AGT AAG-3′) and the reverse primer from the SV40 poly-A Tail (5′-TAG AAT GTT GAG AGT CAG CAG TAG CCT-3′). These primer pairs amplify two bands, one among them being 65 bp shorter, which represents the spliced 3′ untranslated intron. For real-time PCR the following primers (these are human BRAF specific primers) were used: BRAFRTU1 (5′-GTGGATGGCAACAGAAGTC-3′) and BRAFRTL1 (5′-GAAACCAGCCCGATTCAAGGA-3′). For internal control we use 18S rRNA primer sets (Ambion).
Histology and immunohistochemistry
Routine histology was carried out on 10% neutral buffered formalin-fixed tissues. For mouse brain, whole skull was fixed in Bouin’s fixative for about 14–18 days. All immunohisto-chemistry was carried out on formalin-fixed paraffin-embedded 5-μm sections using standard protocols. Tissue sections were incubated at 4 ºC overnight with primary antibodies: pMapk antibody (cat#4376, 1:100; Cell Signaling, Danvers, MA, USA), pAkt (cat#3787, 1:50; Cell Signaling), S-100 (1:150; AbCam, Cambridge, MA, USA), Pten and p53 (1:100; ab23694 and ab32049, respectively, AbCam), Ki-67 (rabbit monoclonal clone Sp6, 1:250; Labvision, Fremont, CA, USA), except for p16Ink4a (1:100, F-12; Santa Cruz Biotechno-logy, Santa Cruz, CA, USA), which was incubated for 45 min at room temperature. A biotinylated goat anti-rabbit secondary antibody (1:200; Vector Laboratories, Burlingame, CA, USA) was incubated for 1 h at room temperature. For p16Ink4a staining we used the mouse on mouse tissue staining kit (Vector Laboratories) as recommended by the manufacturer. Avidin-biotin peroxidase complexes were then incubated for 30 min (Elite ABC reagent; Vector Laboratories). Nova-Red (Vector Laboratories) was used as the final chromogen and hematoxylin was used as the nuclear counter stain. Staining for SA-β-gal was carried out as described (Dimri et al., 1995) on fresh frozen skin samples from both wild-type and BRAF transgenic animal.
Isolation of primary epidermal melanocytes and siRNA transfection
Epidermal melanocytes were harvested from 1-day-old pups as described earlier (Tamura et al., 1987). For siRNA transfection studies we harvested about 2 × 106 melanocytes (about 16–18 days-old culture) and transfected them with 100 pmol of siRNA targeting either human BRAF V600E (stealth siRNA synthesized from Invitrogen) or mouse BRAF (Dharmacon, Lafayette, CO, USA) using nucleofection technology (NHEM kit from Amaxa Biosystem, Cologne, Germany). As a control, we used Medium GC content negative-control siRNA obtained from Invitrogen. To control transfection efficiency we used GFP plasmid vector. For controls we used BIOBR wild-type melanocytes (Tamura et al., 1987; Yale University, Yale, CT, USA). At 48 h after siRNA transfection the whole cell protein was harvested. The total protein was estimated using DC protein assay reagent (Bio-Rad, Hercules, CA, USA). In all, 20 μg of total protein was loaded onto 4–12% Bis–Tris Gel (Invitrogen) and run in 1 × MOPS buffer. The protein was transferred to nitrocellulose membrane (Invitrogen) and western blotting was carried outusing BRAF antibody (cat# sc-55522, 1:4000; Santa Cruz), pERK1/2 (cat# 9101, 1:1000; Cell Signaling), total ERK (cat# 4695, 1:1000; Cell Signaling), pMek1/2 (cat#9121, 1:1000; Cell Signaling), total Mek (cat# 9122, 1:1000; Cell Signaling) and β-actin (1:6000; Sigma Aldrich, St Louis, MO, USA). The sequence of BRAF V600E specific siRNA used was as published earlier, using sense strand GGUCUAGCU ACAGAGAAAUCUCGAU (Hingorani et al., 2003; Sharma et al., 2005) and mouse Braf-specific siRNA sense strand GGAGUUACAUGUUGAAGUAUU (Dharmacon).
Supplementary Material
Acknowledgments
This work was supported by NIH grant CA095798 to FGH. We thank Mohammad Miri for technical assistance, Dr Louis Montoliu for the tyrosinase promoter, Dr Lynda Chin for the mouse strains, and Dr Philip Tsichlis and Dr Trevor Pemberton for useful discussions.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res. 2005;65:4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana JS, Rubio MP, Vasquez JJ, Idoate M, Sober AJ, Seizinger BR, et al. Lack of allelic deletion and point mutation as mechanisms of p53 activation in human malignant melanoma. Int J Cancer. 1993;55:562–565. doi: 10.1002/ijc.2910550407. [DOI] [PubMed] [Google Scholar]
- Chang DL, Qiu W, Ying H, Zhang Y, Chen CY, Xiao ZX. ARF promotes accumulation of retinoblastoma protein through inhibition of MDM2. Oncogene. 2007;26:4627–4634. doi: 10.1038/sj.onc.1210254. [DOI] [PubMed] [Google Scholar]
- Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol. 2005;23:1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Singh AB, Ellis DL, Richmond A. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 2002;62:7335–7342. [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen T, Ahmad T, Flaherty KT, Gore M, Kaye S, Marais R, et al. Sorafenib in advanced melanoma: a phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12:2301s–2307s. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198–5202. [PubMed] [Google Scholar]
- Laud K, Marian C, Avril MF, Barrois M, Chompret A, Goldstein AM, et al. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. J Med Genet. 2006;43:39–47. doi: 10.1136/jmg.2005.033498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Muthusamy V, Hobbs C, Nogueira C, Cordon-Cardo C, McKee PH, Chin L, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2006;45:447–454. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Meltzer PS. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell. 2002;2:5–7. doi: 10.1016/s1535-6108(02)00089-2. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, et al. p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- Tamura A, Halaban R, Moellmann G, Cowan JM, Lerner MR, Lerner AB. Normal murine melanocytes in culture. in vitro Cell Dev Biol. 1987;23:519–522. doi: 10.1007/BF02628423. [DOI] [PubMed] [Google Scholar]
- Tief K, Schmidt A, Beermann F. New evidence for presence of tyrosinase in substantia nigra, forebrain and midbrain. Brain Res Mol Brain Res. 1998;53:307–310. doi: 10.1016/s0169-328x(97)00301-x. [DOI] [PubMed] [Google Scholar]
- Tonks ID, Nurcombe V, Paterson C, Zournazi A, Prather C, Mould AW, et al. Tyrosinase-Cre mice for tissue-specific gene ablation in neural crest and neuroepithelial-derived tissues. Genesis. 2003;37:131–138. doi: 10.1002/gene.10242. [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe P, Andrade L, Gonzalez S. Lack of association between BRAF mutation and MAPK ERK activation in melanocytic nevi. J Invest Dermatol. 2006;126:161–166. doi: 10.1038/sj.jid.5700011. [DOI] [PubMed] [Google Scholar]
- Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.