Abstract
Several methods are presently available for gene expression analysis. However, few of them are suitable for detection of moderate numbers of genes in thousands of samples with high speed and low cost. There is great demand for such a method for use in diagnostics and screening. To address this need, we have developed an assay for gene expression analysis using microspheres and a fluidic instrument made by Luminex. The assay is named Beads Array for the Detection of Gene Expression (BADGE). BADGE can monitor up to 100 genes in a single reaction, and it takes only 1 h to hybridize and <20 sec to read the results of all 100 genes in a sample for the detection process. For the genes detected in five independent replicate experiments, the standard deviation was <35% of the mean. We have monitored multiple pathogenesis-related genes simultaneously in chemical-treated and control Arabidopsis samples employing the BADGE assay. The data were compared with those obtained from an established technology, Affymetrix GeneChip. The changes in expression profiles were very similar. Our study showed that the BADGE assay was capable of profiling expression of multiple genes at affordable cost and rapid speed.
Current techniques for analysis of gene expression can be divided into three classes. The first class is hybridization-based techniques such as Northern blotting (Alwine et al. 1977), subtraction cloning (Hedrick et al. 1984; Swaroop et al. 1991; Diatchenko et al. 1996; Lavery et al. 1997), and DNA microarrays (Schena et al. 1995). The second class is PCR-based techniques, including differential display (Liang and Pardee 1992) and quantitative PCR. The third class is sequence-based techniques including SAGE (Velculescu et al. 1995), ESTs (Adams et al. 1991; Adams et al. 1995; Lee et al. 1995, Vasmatzis et al. 1998), and mass-spectrometry sequencing (Murray 1996).
Traditional Northern blotting requires several days to obtain results, and one can only study a single gene in a few samples each time. Laborious and time-consuming processes such as electrophoresis and blotting are necessary in Northern blotting, which makes the development of a high-throughput method of Northern blotting impractical. The introduction of microarrays tremendously advanced the study of gene expression profiles and genomic compositions. The technology involves attaching probes such as oligonucleotides derived from ESTs, cloned cDNA, or PCR products to the surface of nylon filters, glass slides, or silicon chips at high density. To determine gene expression level, labeled cDNAs or cRNAs are hybridized to the capture probes on the arrays, and the hybridized signals are scanned and measured as the fluorescence intensity at the site of each capture probe. The Affymetrix GeneChip allows detection of >7000 genes in one array—approximately equal to the complete genome of yeast (Lockhart et al. 1996; Wodicka et al. 1997). GeneChip is therefore a powerful tool for genomic profiling. However, limitations do exist for GeneChip technology for certain applications. First, the composition of the genes represented by a GeneChip is usually fixed. Investigators do not have much freedom to select genes of their own interest under most circumstances, and many studies often require exploring just a few genes instead of thousands of genes. Having a customized array for GeneChip technology is extremely difficult in practice, and requires a tremendous amount of work from computer specialists, engineers, chemists, bioinformatic scientists, and biologists. Second, Affymetrix GeneChip technology uses expensive pre-made GeneChips and also requires expensive instruments (hybridization station and scanner) that are not affordable for many laboratories. The enormous amount of data necessary for accurate and systematic analysis needs to be further analyzed by costly software. Making self-designed cDNA microarrays, as an alternative to buying pre-made Affymetrix GeneChips, also requires an expensive spotter and a high quality scanner. Third, the hybridization step for GeneChip and cDNA microarrays requires incubation for 16 h. This long hybridization step increases the difficulties for high-throughput screening on the scale of thousands of samples per day.
Real-time PCR methods like TaqMan permit the analysis of thousands of samples per day with good sensitivity but are limited in multiplicity because of the PCR amplification involved and the multiple fluorescent probes required. Most studies are only analyzing two genes per reaction using TaqMan (Lie and Petropoulos 1998). Owing to the above limitations, GeneChip technology, cDNA microarrays, and TaqMan are not well suited to studying small sets of genes (e.g., 10–100) in a large number of samples (up to thousands). For many applications such as diagnostic analysis it is often necessary to detect several specific genes from large numbers of samples. Other technologies for gene expression analysis also have their limitations. For instance, EST requires a great deal of sequencing, subtraction cloning and differential display need additional cloning, and SAGE involves complicated sample preparation.
Nanogen (http://www.nanogen.com) and Xanthon (http://www.xanthon.com) are working on different tools for gene expression diagnostic analysis. In this study, we present a new technology platform, BeadsArray, which delivers multiplexed, fast, accurate, and cost-effective bioassays. The assay uses the commercially available Luminex100 LabMAP system and multiple color-coded microspheres (Luminex Corp.) in a rapid assay for gene expression. Using the LabMAP system, several applications for immunoassays and SNP detection have been published (Fulton et al. 1997; Gordon and McDade 1997; Oliver et al. 1998; Smith et al. 1998; Chen et al. 2000; Iannone et al. 2000). However, no study concerning gene expression analysis has been reported using this system. Taking advantage of the affordable Luminex100 system and the inexpensive microspheres, we have established a gene expression assay on this BeadsArray platform. We present here a model study of monitoring expression of pathogenesis-related (PR) genes in Arabidopsis RNA samples. Arabidopsis thaliana is a model plant widely used for genetic studies, and the sequence of its genome has been determined. To assay gene expression, oligonucleotide capture probes derived from target genes are synthesized and immobilized to fluorescent microspheres via a simple chemical coupling reaction. The beads are hybridized to labeled cRNA and passed through a counting device, which records the identity of each microsphere and the probe intensity associated with it. Up to 100 sets of microspheres with different fluorescence color codes are available from Luminex, which potentially enables the analysis of 100 genes in a single assay. This assay is named BeadsArray for the Detection of Gene Expression (BADGE). The advantages of the BADGE assay include high flexibility, affordability, multiplicity, rapid process, and high-throughput format. The method is well suited for diagnostic detection in clinical samples and for identification of marker genes in crop breeding and compound screening.
RESULTS
Evaluation of Specificity of Hybridization between Targets and Capture Probes Coupled to Microspheres
Oligonucleotides with unique sequences of 25 bases were selected from each of 20 Arabidopsis genes and used as capture probes in the first experiment (Table 1). Because NTSR is a human gene that is absent from the Arabidopsis genome, it was chosen as a negative control. Among the 20 genes, most are disease-related genes, whereas UBQ5 and ACT2 are constitutively expressed genes that serve as internal controls (Table 1).
Table 1.
Capture Probe Sequences
| Genea | Putative gene function | Capture sequence |
|---|---|---|
| PR1 | pathogenesis-related Pr-1–like protein | 1. AGTGAGACTCGGATGTGCCAAAGTG |
| 2. ACTACAACTACGCTGCGAACACGTG | ||
| 3. GTGAACATGTGGGTTAGCGAGAAGG | ||
| putative antifungal protein | 1. TGGTGCTAAATCGTGTGTATTTTAC | |
| 2. TCTACCACTAATCTTTGGTGCTAAA | ||
| 3. ACAAGTGTATCTGTTACGTCCCATG | ||
| PAD3 | a member of the cytochrome P450 family | 1. GTAGGAGAATATGTCCAGGGATGAC |
| 2. CAGAATGTTCTTAACGAGCATCTTA | ||
| 3. CTCCGGTCAGAACAGGAGATTGAAG | ||
| UBQ5 | ubiquitin 5 | 1. ATCTGTTGTAGCGGTAGATCGATCC |
| 2. GTGTGGACTCACCTACGTTTACCAG | ||
| 3. GTCATTTTGATCGCCATTACTGTGG | ||
| ACT2 | actin 2 | 1. TCCCATTGTTTTGTAGCTCTGAGTG |
| 2. TGGGGTTTTATGAATGGGATCAAAG | ||
| 3. GTCCAGGAATCGTTCACAGAAAATG | ||
| NTSR | Homo sapiens neurotensin receptor 1 | TCAACGTGGCCAGCCTGAGTGTGGA |
| PR5 | thaumatin-like protein | 1. CTACGCTTATGACGACGAAACGAGC |
| 2. AAACTTGTCCTCCCACGGACTACTC | ||
| 3. TCAATATTGCTGCCGTGGAGCTAAC | ||
| THI2 | thionin | 1. GGCCGCCTAAACTGTTGTAATGGTC |
| 2. GTAAAGCGAACCTCCTTAAGGTGTC | ||
| 3. TGCAAGTTAGGGTGTGAAACTTCTG | ||
| AOS | allene oxide synthase | 1. AATCTGTTGCTGCTCTACATGTTCG |
| 2. TTTCTCGTCGTTAAGGAAAGCTAGC | ||
| 3. AGCTTTTGAGGCATGTGTTGTGGTC | ||
| PAL | phenylalanine ammonia lyase | 1. AGCTACTGGTCCCAACGGTGAAGCT |
| 2. CGTGGTACAATCACCGCCTCCGGAG | ||
| 3. CAGATTCCTTAACGCCGGAATATTC | ||
| PAD4 | putative protein | 1. CCAGAGGAGATCTTTGTTACGGGAA |
| 2. TGGTTGGATGAGGCGAGAAAAGAGA | ||
| 3. AGATACGCGAGCACAACGCAAGATA | ||
| GST1 | glutathione S-transferase | 1. TACGATCGCTAAAGCTNACAAAGAG |
| 2. GGAGAATCACCTCGTTCGCCATTGT | ||
| 3. TCGTCAATGTACTGAACCTGGATGA | ||
| LOX1 | lipoxygenase | 1. TCACGTGTTGTTCTATATCCCATGG |
| 2. GGGAATTCCAAATAGCGTCTCTATC | ||
| 3. TCTAAAGAATGGGCGGCTGAGAAAG | ||
| EDS1 | EDS1 has simlarity to eukaryotic lipases | 1. gccgctacaaatacgctcagagagg |
| 2. GAGCAGAGACTGGATCAATTTAGCA | ||
| 3. GCCGGTATTTTCGACGAGGTGCTTG | ||
| ELI3 | a novel plant defense gene | 1. GTGATTGATGTTGCCAACACGATGA |
| 2. CTTGCAAAGGCTGACGTTAAGTACC | ||
| 3. GCGGATTATGTCAACACCGCCATGG | ||
| AIG1 | Novartis proprietary database | 1. TGGCCTGTTATGAGTCAGCTTTTGC |
| 2. TTCTATGTTCCTCGTTTGAGCATGC | ||
| 3. AGACTTTGTGTTTCTAACGCGGAAC | ||
| ATHcor | coronatine-induced protein 1 | 1. GTAGGTGGAATTGTGGTTGCGTTTC |
| 2. TGGATATGTTGGACGATGATTTGCC | ||
| 3. ACAAAGAGTGTAAGGCGACGAAAGC | ||
| RaR047 | similar to sulfotransferase-like flavonol | 1. AGAGATTGGAGGATGGAGAGATAGC |
| 2. GAGTTCTTGGAATGTGGCTTTATTG | ||
| 3. TATTGGAGTACTGGTATGCAAGCCG | ||
| AIG2 | Novartis proprietary database | 1. TCCGAATGGGAGAAGTAGGGAAGAG |
| 2. TTTAATTCTCGAATGTGCTCCGGTC | ||
| 3. GTCTACGGCAGTTTCCAAGAACCAG | ||
| PRX1 | peroxidase | 1. AACTCCAACTCTCTGCTCCATGATG |
| 2. TGCATTTGTGGAGGCAATGAATAGG | ||
| 3. ACTGACACAATCCCCTTGGTGAGAG |
All the genes are derived from Arabidopsis thaliana except NTSR which is a Homo sapiens gene.
The capture probes derived from the 20 Arabidopsis genes and the human gene NTSR were coupled to different color-coded microspheres in separate reaction tubes and then mixed together for multiplexed assays. Targets (nucleotides to be analyzed) were oligonucleotides complementary to capture probes and tagged with biotin on the 5′ end. The targets were mixed in four tubes in various amounts (5, 10, 20, or 40 fmoles) as indicated in Table 2. In the hexaplexed assay (Fig. 1A), only the first six targets in Table 2 (PR1, PDF, PAD3, UBQ5, ACT2, and NTSR) and their corresponding capture probes coupled to microspheres were added to the reaction. Phycoerythrin (PE)-conjugated streptavidin was added to the reaction to detect bound targets that were biotinylated. The signal of each target hybridized to its specific capture probe coupled to microspheres was determined by the fluorescence intensity of phycoerythrin. A reaction tube with all the components except the targets was used as a negative control. At least 200 microspheres of each set were analyzed by the Luminex100 system. The Mean Fluorescence Intensity (MFI) of the counted microspheres, which corresponds to the number of targets bound, was calculated and displayed by the Luminex software. The icosaaplexed assay (Fig. 1B) was conducted the same way as the hexaplexed assay except that all 20 targets and 20 sets of microspheres were mixed in the reaction, as indicated in Table 2. These two experiments were designed to address two questions: Are the hybridization signals linearly related to the added targets in the multiplexed assay? Does hybridization between capture probes and targets remain specific as multiplicity increases? As shown in Figure 1A, the hybridization signals from the six targets varied with the added target amounts in a linear fashion, and the signal from the negative control NTSR was not detectable above background. In the experiment shown in Figure 1B, 20 targets were added to the assay. Again, linear signals were obtained, and the signal detected from the negative control NTSR was at the background level. The results from the two experiments showed that the specificity of hybridization did not change while increasing multiplicity from 6 to 20 in the assays. The hybridization between each capture probe and target was specific and linearly proportional. However, when the targets of some genes (e.g., PR5 and AIG2) were present at high concentrations, the signals reached a plateau, as shown in Figure 1B. It is possible that this problem could be avoided by coupling capture probes to microspheres with better efficiencies.
Table 2.
Fmols of Gene-Specific Nucleic Acid Fragments Added in the Multiplexed Assays
| Tube # | PR1 | PAD3 | UBQ5 | ACT2 | NTSR | PR5 | THI2 | AOS | PAL | PAD4 | GST1 | LOX1 | EDS1 | ELI3 | AIG1 | ATH1 | RaR1 | AIG2 | PRX1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 5 | 10 | 20 | 40 | 5 | 0 | 10 | 20 | 40 | 5 | 10 | 5 | 10 | 20 | 40 | 5 | 5 | 10 | 20 | 40 |
| #2 | 10 | 20 | 40 | 5 | 10 | 0 | 20 | 40 | 20 | 10 | 5 | 10 | 20 | 40 | 20 | 10 | 10 | 20 | 40 | 20 |
| #3 | 20 | 40 | 5 | 10 | 20 | 0 | 40 | 5 | 10 | 20 | 40 | 20 | 40 | 5 | 10 | 20 | 20 | 40 | 5 | 10 |
| #4 | 40 | 5 | 10 | 20 | 40 | 0 | 5 | 10 | 5 | 40 | 20 | 40 | 5 | 10 | 5 | 40 | 40 | 5 | 10 | 5 |
Capture probes containing 25-mer oligonucleotides were chosen from 19 Arabidopsis genes as shown in the table and coupled with different color-coded microspheres. NTSR is a sequence with 25-mer oligonucleotides chosen from the human genome that is absent in the Arabidopsis genome and serves as negative control in these experiments. The complementary sequences to the capture probes of the 20 genes were synthesized with biotin tagged on the 5′-end and used as targets in the assay. The target oligonucleotides were mixed at various amounts in four tubes as indicated in the table and measured in the multiplexed assays as shown in Figure 1. In the experiment of hexaplexed assay, only the first six targets from the table were added, the results are presented in Figure 1A. In the experiment of dodecaplexed assay, all twenty targets were added and the corresponding signal of each gene was measured (Fig. 1B).
Figure 1.
Specificity of hybridization was determined in the hexaplexed assay (A) and the icosaplexed assay (B). Oligonucleotides representing 20 genes were coupled to 20 different color-coded microspheres. Various amounts of the complementary target oligonucleotides tagged with biotin were mixed in four tubes as indicated in Table 2. NTSR served as a negative control in the experiment. (A) Only the first six targets (see Table 2) and their corresponding capture-probe-coupled microspheres were added to the assay. (B) All 20 targets and their corresponding capture-probe-coupled microspheres were added to the assay. In both experiments A and B the background fluorescence of each set of microspheres was subtracted.
Spiking Experiments in a Whole Population of Labeled Arabidopsis cRNA
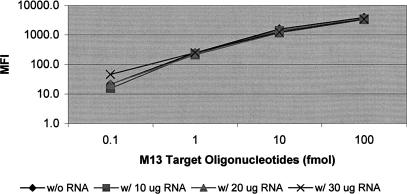
Next, we tested the specificity of hybridization in a whole population of biotin-labeled Arabidopsis cRNA. In this experiment, we also determined the range of target amount over which hybridization signals were linearly correlated so that the levels of target genes could be accurately quantified in a population of cRNA. To this end, M13 target oligonucleotides were added to a population of biotin-labeled Arabidopsis cRNA (Fig. 2). The M13 capture probe is a 25-mer oligonucleotide derived from the M13 phage that is absent in the Arabidopsis genome, and the M13 target is another oligonucleotide complementary to the M13 capture probe and tagged with biotin at the 5′ end. The M13 target oligonucleotide was spiked into 10, 20, or 30 μg of cRNA at 0.1, 1.0, 10.0, and 100.0 fmoles, respectively. The tubes from these spiked reactions plus the control tubes (without cRNA) were hybridized at 48°C for 1 h. The signal of the M13 target was detected by measuring the bound streptavidin-phycoerythrin on the microspheres coupled with the M13 capture probe. Assuming that the average size of each gene in a population of Arabidopsis cRNA is 1000 bases and the average molecular weight of each base is 300, then 0.1 fmole would correspond to 30 pg of a gene. Therefore, spiking 0.1, 1.0, 10.0, and 100.0 fmoles of the known target into 10 μg of cRNA is equivalent to the molecular ratios of 1:300,000, 1:30,000, 1:3,000, and 1:300, respectively. As shown in Figure 2, the hybridization signals of M13 target in the absence of cRNA (control) and in the presence of 10, 20, or 30 μg of cRNA were almost identical. This result showed that specific hybridization between the M13 capture probe and its target was not affected by the presence of a large population of biotin-labeled Arabidopsis cRNA, indicating the feasibility of using the assay for analyzing specific hybridization reactions in biological samples. Furthermore, as indicated in Figure 2, the hybridization signal was proportional to the target amount between the frequencies (ratio of a specific target gene to a population of genes) of 1:30,000 and 1:300 (with addition of 1–100 fmoles of target). The minimum amount of detectable target in the linear range is approximately 1.0 fmole for this BeadsArray system under this condition, which corresponds to the level of moderately abundant transcripts in a total population of 10 μg of RNA. We define a moderately abundant transcript as one with 10–30 copies per cell. This experiment showed that the BADGE assay seems capable of measuring transcripts with moderate to abundant expression levels in cells.
Figure 2.
Specific hybridization in the presence of biotin-labeled wild-type Arabidopsis cRNA. The M13 capture probe was coupled to microspheres and the complementary sequence tagged with biotin was used as a target in this experiment. The M13 target oligonucleotide was added as 0.1, 1.0, 10.0, and 100.0 fmoles to the assay in the absence or presence of the indicated amount of biotin-labeled cRNA prepared from wild-type Arabidopsis leaves. In the presence of 10 μg of cRNA, the addition of 0.1, 1.0, 10.0, and 100.0 fmoles of target was approximately equal to a spiking ratio of 1:300,000, 1:30,000, 1:3,000, and 1:300, respectively.
The Expression Levels of Pathogenesis-Related Genes in Arabidopsis as a Function of Time Following BTH Treatment Were Profiled by the BADGE Assay
In response to pathogen attack or chemical exposure, a series of pathogenesis-related (PR) genes is activated in many plant species. The expression pattern of these PR genes is important for understanding the defense mechanism in plants (Glazebrook et al. 1997; Glazebrook 1999). Transgenic plants that overexpress various PR genes show enhanced disease resistance against certain pathogens (Broglie et al. 1989; Alexander et al. 1993; Liu et al. 1995). Therefore, a method for monitoring expression levels of a group of PR genes simultaneously is helpful for elucidation of signal transduction networks in plants and useful for screening induced and repressed PR marker genes in mutant plants. Benzothiadiazole (BTH) is a well-studied chemical activator of PR genes in many plants including Arabidopsis, tobacco, and other important agricultural plants (Gorlach et al. 1996; Lawton et al. 1996). We studied the induction of a group of PR genes in Arabidopsis as a function of time following BTH treatment.
In our experiments, total RNA was isolated from Arabidopsis leaves that were harvested at various times (1–9 d) after the application of BTH or water (control). The total RNA was transcribed into cDNA by reverse transcription using oligo(dT). Double-stranded cDNA was synthesized and biotin-labeled cRNA was subsequently obtained through an in vitro transcription reaction using T7 RNA polymerase and biotinylated UTP. The labeled cRNA was hybridized to premixed sets of 20 microspheres at 48°C for 1 h. Then, the expression levels of the 20 genes were measured simultaneously in the multiplexed assay through the detection of streptavidin-conjugated phycoerythrin (Fig. 3). The results obtained from BTH-treated samples are shown in the left panels of Figure 3 (Fig. 3A,C,E,G) and the corresponding results obtained from water-treated samples (controls) are shown in the right panels (Fig. 3B,D,F,H). The 20 genes measured in these experiments were organized into four groups according to their properties and expression patterns.
Figure 3.
Simultaneous detection of 20 genes from either BTH- or water-treated Arabidopsis samples as a function of time (1, 3, 5, 7, and 9 d). The left panels (A,C,E,G) are the results detected from BTH-treated samples, and the right panels (B,D,F,H) are the results detected from water-treated samples (control). The 20 genes were organized into four groups in this figure for presentation according to their properties and their expression levels affected by BTH treatment. The first group contained BTH-induced genes (A,B), the second group had unchanged PR genes (C,D), the third group included BTH-repressed genes (E,F), and the fourth group consisted of control genes (G,H).
The genes in the first group (Fig. 3A,B) including PR1, PR5, and PRX1 were induced by BTH. The expression levels of these genes reached their peaks on day 3 following BTH treatment and remained elevated until day 5 (Fig. 3A), after which the signals declined. In contrast, the expression levels of these genes in all the control samples (water-treated Arabidopsis) remained low and unchanged (Fig. 3B). Compared with the controls, the message levels of PRX1, PR1, and PR5 were increased ∼3-fold to 10-fold in these samples by 3 d after treatment with BTH.
The genes in the second group (Fig. 3C,D) did not change their expression levels significantly in either BTH- (Fig. 3C) or water-treated (Fig. 3D) samples. Throughout the whole experiment, very stable expression levels were observed for this group of genes, and no elevated signals were detected in the BTH-treated samples. This indicated that the regulation of the pathogenesis-related genes in the second group is different from that of the three genes in the first group.
In contrast with the genes in the first group, the expression levels of the genes in the third group (Fig. 3E,F) were repressed by BTH. As shown in the figure, the expression levels of PAL, ELI3, and AOS decreased ∼2-fold to 4-fold in the BTH-treated samples compared with the controls on day 3. After day 5, the expression levels of the three genes began to recover.
The genes in the fourth group (Fig. 3G,H) were constitutively expressed and served as internal controls. As expected, their expression levels remained unchanged in both BTH- (Fig. 3G) and water-treated (Fig. 3H) samples. The signal from M13 stayed at the background level. Measured by another established technology, TaqMan, the expression levels of TRX3 and APX3 in the induced and noninduced Arabidopsis samples were generally unchanged.
The Validation of the BADGE Assay
To evaluate the validity and feasibility of an assay, it is necessary to examine reproducibility and linearity of the assay, and to compare data with that obtained by other established technologies. To address reproducibility, all of the experiments described in the previous sections were repeated three to five times. Based on the five separate experiments described in Figure 3, the maximum standard deviation of expression signals in these assays was 35% of the mean. The standard deviation for most of the genes ranged from 5% to 20% of the mean. For the 20 genes studied in Figure 3, only three genes showed standard deviations of 30%–35%. Therefore, we concluded that for the genes analyzed in these experiments, the hybridization signals differed by no more than 35%. Different labeling efficiency, random fragmentation, and other factors could contribute to the variation. We considered this level of variation acceptable because the reproducibility of many microarray systems is such that only differences greater than twofold are significant.
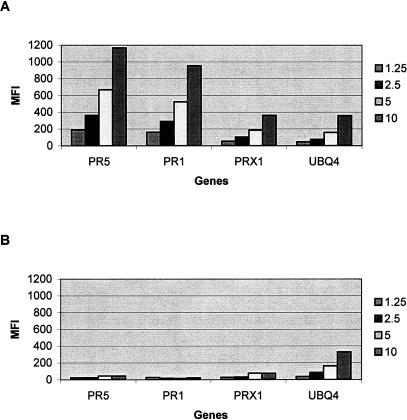
To determine if the signals are linearly related to the amounts of target cRNA, we varied cRNA amounts (1.25 μg, 2.5 μg, 5.0 μg, and 10 μg) in separate assays using the Arabidopsis samples from 3 d of treatment with BTH or water. Again, we simultaneously measured the expression levels of 20 genes as discussed in the experiment presented in Figure 3. To make the graph clear, here we only present the results of the genes from the first group (induced by BTH) and the internal control UBQ4 in these assays. As shown in Figure 4A, linear signals were detected from 2.5 μg to 10 μg of biotin-labeled cRNA that was prepared from samples treated by BTH. The signals from 1.25-μg samples were low for the genes PRX1 and UBQ4 (∼2× higher than background), indicative of the sensitivity limit of the assay. If 2 μg of cRNA is the minimal amount to get reasonable signals, it means that for genes expressed with moderate abundance (e.g., UBQ4), their expression levels should be easily detectable from samples prepared from 1–2 μg of total RNA using this BADGE assay. Again, if one assumes that the average size of all mRNAs is 1000 bases in the Arabidopsis genome and the average molecular weight of the four bases is 300, a gene with moderate abundance in 2 μg of a total population of RNA is at the level ∼0.7 fmole. For a second time, this experiment showed that the minimal amount of detectable target gene for this BADGE assay was ∼1 fmole, which is consistent with the result obtained from the spiking experiment shown in Figure 2. The results shown in Figure 4B are the expression levels of PR1, PR5, PRX1, and UBQ4 from control Arabidopsis samples that were treated with water. The signals for PR1, PR5, and PRX1 remained at low levels as anticipated (Fig. 4B). In contrast, the expression level of the internal control gene UBQ4 was always linearly correlated to the added target amount (Fig. 4A,B).
Figure 4.
Different amounts of cRNA (1.25, 2.5, 5.0, and 10 μg) prepared from Arabidopsis samples at 3 d after treatment with either BTH (A) or water (B) were used in this experiment to test if the signals were linear with the target amount. The results of the three PR genes from the first group classified in Figure 3 (PR5, PR1, and PRX1, induced by BTH) and the results from an internal control UBQ4 are presented in this figure. Linear signals were detected from 1.25 μg to 10 μg of biotin-labeled cRNA for the three PR genes in the sample that was treated with BTH (A). In contrast, the expression levels of PR5, PR1, and PRX1 in the control sample remained low (B). However, the expression signal of the internal control UBQ4 was always linearly related to the added target cRNA amount in both BTH-treated and control samples (A,B).
GeneChip technology from Affymetrix is widely accepted as a reliable method for monitoring expression levels of several thousand of genes simultaneously. A GeneChip specifically made for Arabidopsis was used to measure the expression of genes in Arabidopsis leaves after treatment with BTH for 3 d. Aliquots of the same samples were also analyzed by the BADGE assay using the Luminex100 system. The results from the BADGE assay were compared with that represented on the Arabidopsis GeneChip (Fig. 5). GST1 and PRX1 were not included in this figure because the two genes were not found in the pre-made Arabidopsis GeneChip.
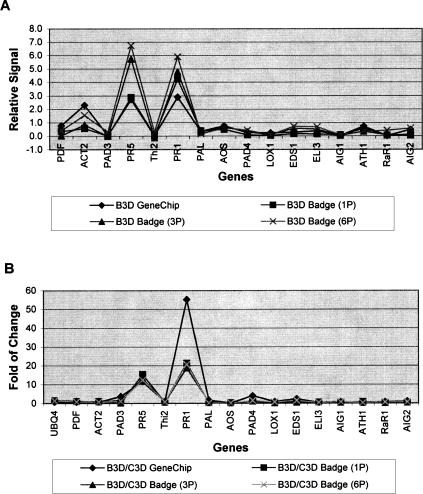
Figure 5.
Biotinylated cRNA samples prepared from Arabidopsis leaves at 3 d after BTH treatment (B3D) were used for this experiment. The cRNA was hybridized to either microspheres (coupled with single-probe, 1P; three-probe, 3P; and six-probe, 6P; respectively) made by Luminex or Arabidopsis GeneChip made Affymetrix in separate experiments. The hybridization signals of the genes obtained from the two technologies were normalized to the expression level of UBQ4 as shown in A. The fold change in hybridization signal of the BTH-treated sample to the control sample is presented in B.
We normalized the hybridization signals of all the genes detected by GeneChip and BADGE against the expression level of UBQ4. The relative hybridization signals were plotted versus their corresponding genes (Fig. 5A). As shown in Figure 5A, the trends of the curves obtained by the two technologies were very similar. Although the actual values of the relative hybridization signals to UBQ4 were not exactly the same for the two technologies, signals detected from genes with high expression levels by GeneChip were always high as detected by BADGE, and signals detected from genes with low expression levels appeared consistently low. The data obtained from GeneChip technology strongly supported the data from the BADGE assay.
Our experiments showed that different capture probes chosen from the same gene gave similar results. To check if a single capture probe is sufficient, we also designed multiple capture probes from each gene and then we mixed either three or six capture probes together in equal molar ratios and coupled them together to the same set of microspheres. We hybridized cRNA from an Arabidopsis sample to the three-probe (3P) and six-probe (6P) microspheres, respectively. The data were normalized against the signal of UBQ4 as described above and also plotted together in Figure 5A. The relative expression signals for all the genes were very similar. According to the information provided by Luminex, the diameter of a single microsphere is 5.6 μm and can be coupled to 100,000–500,000 molecules of capture probes on the surface. Multiple capture probes were supposed to occupy the surface of a microsphere randomly and evenly when they were mixed in equal molar ratio in the coupling reactions. The use of multiple capture probes coupled to microspheres could avoid the problem of possible inefficient binding that was associated with a single capture probe.
The hybridization signals obtained from experiments using one-, three-, and six-probe microspheres were all different. Theoretically, for the BADGE assay, the probability of hybridizing to either single- or multiple-probe microspheres for target genes (cRNA) should be equal because the densities of capture probes per microsphere were the same (100,000–500,000 molecules per microsphere regardless of whether they were mono or multiple probes) if we assumed the association and dissociation constants of each capture probe to its target sequence were the same. However, many factors could result in different hybridization signals in practice. For instance, the random fragmentation of target genes in cRNA samples, the different hybridization efficiencies of various capture probes, and the different coupling efficiencies of multiple capture probes to microspheres. In spite of these factors, the expression profiles of the genes studied in these experiments (using 1P, 3P, and 6P microspheres) agreed well between experiments and were consistent with the results obtained from the GeneChip experiment. Our experiments indicated that single and multiple capture probes worked equally well. Although none of our designed capture probes failed in the BADGE assay, to avoid the potential problem of possible inefficient binding arising from a single capture probe, we recommend using multiple capture probes, say, three, for the experiments.
We next compared the fold changes in hybridization signals of BTH-treated samples to the control samples (Fig. 5B). The fold change in hybridization signal for each gene was calculated as the signal from the BTH-treated sample over the signal from the water-treated sample. Consistently detected by BeadsArray (1P, 3P, and 6P microspheres) and GeneChip, the expression levels of PR5 and PR1 were significantly induced by BTH at 3 d after treatment (Fig. 5B), but the expression levels of other genes were not changed dramatically in the two samples.
DISCUSSION
We present a new method using BeadsArray for the quantitative determination of multiple genes. This approach quantifies gene expression levels based on hybridization of mRNA populations to the probes derived from genes of interest that are coupled to different color-coded microspheres. A flow cytometer is used to simultaneously measure the hybridization signal associated with the surface of the microspheres and to categorize the color-coded microspheres. The method allows performance of up to 100 assays simultaneously in a small volume (10–100 μL) of fluid. This BeadsArray method offers affordable cost, rapid speed, and high flexibility with capability for the high-throughput assays that are increasingly needed. It is ideal for applications such as diagnostic detection of disease genes from clinical samples and screening of characteristic marker genes from many biological systems.
TMAC buffer was chosen for the assay because the hybridization conditions in this buffer are based on the length of nucleic acid fragments instead of the compositions of G + C and A + T (or A + U). Therefore, it is easier to design and select capture probes from different genes without considering the compositions of probes when TMAC buffer is used for multiplexed hybridization assays. In our case, every capture probe was a 25-mer that is close to the 3′ end of each gene. Other criteria for the selection of capture probes include unique sequence and minimum secondary structures.
Different hybridization times were tested in our experiments. For example, the hybridization between capture probes coupled to microspheres and labeled cRNA populations was conducted at 48°C for 30 min, 60 min, 150 min, and 16 h in different experiments. The prolonged hybridization times of 150 min and 16 h did not enhance the hybridization signal compared with 60 min, indicating that the hybridization was complete within 60 min under our conditions. The hybridization signal from the experiment with a 30-min incubation was 10% lower than that with a 60-min incubation. As a result, the 60-min hybridization time was chosen for our experiments. The hybridization between oligonucleotides and capture-probe-coupled microspheres was faster. Incubation as brief as 15 min was sufficient. Therefore, the hybridization time required for this BADGE assay was much shorter than that required for other technologies. For instance, the hybridization time for Affymetrix GeneChip is 16 h, for cDNA microarray is 4 h or longer, and for Northern blotting is overnight. The long hybridization time for these technologies is one of the barriers to adapting them to high-throughput screening systems.
The next step is labeling by addition of the fluorescent probe streptavidin-PE to the reaction. Usually, after labeling, it is required that the excess free fluorescent probe be removed from reactions to decrease background before detection. For example, extensive washes with one or several different buffers are necessary for Affymetrix GeneChip, cDNA microarray, and Northern blotting, but not for the BADGE assay. In our experiments, the results were unaffected by washes after the addition of streptavidin-phycoerythrin (data not shown). This feature shortens the whole process and greatly simplifies the assay. In addition, multiplicity is easily changed by just adding or reducing the types of capture probes coupled to microspheres in the reaction. All these features make the BADGE assay easy to adapt to a high-throughput format. Presently, the Luminex100 system is able to read samples from a 96-well plate, and it takes less than 1 h to finish analyzing 96 samples. Therefore, it is feasible to screen 1000 samples per day using the current model. Luminex is developing a new robotic model, LuminexHTS, which will be available on the market soon. Even higher throughput will be possible when the new LuminexHTS model is used for the assay.
As shown by the results, reasonable signals were obtained from 1 in 30,000 molecules (Fig. 2). We are presently working on other labeling methods to further increase the sensitivity of this assay and decrease the cost. The speed of the assay, the short hybridization time, and the simple procedures of this BADGE assay make it superior to any other technologies.
In summary, the BADGE and microarray technologies are equivalent for the application of gene expression analysis but suited for different applications. Owing to the high cost, microarray technology is not appropriate for studying large numbers of samples but is excellent for getting information from the whole genome of an organism. TaqMan is good for screening large numbers of samples with great sensitivity and accuracy, but only two genes can be studied in each assay. The BADGE assay, on the other hand, is ideal for detection of 10–100 genes from enormous numbers of samples with high efficiency and affordable cost. For that reason, it is well suited for diagnostic screening of disease genes and marker genes expressed at or above moderate abundance. The rapid speed and simple operation of the assay make it easy to adapt to a high-throughput format. We predict that this technology will have a wide application in clinical, pharmaceutical, and agricultural studies.
METHODS
Capture Probe and Its Coupling to Microspheres
A unique sequence of 25 bases within a region of 600 bases from the 3′ end of a target gene was chosen as a capture probe. Multiple capture probes could be selected from the same gene at different positions. The Tm of the chosen capture probe usually ranged from 50°C to 65°C and minimal secondary structure was preferred. Capture probes were chosen using commercial software (Vector NTI). All capture-probe oligonucleotides were synthesized with 5′-amino uni-linker (Oligos Etc.) and then covalently linked to carboxylated fluorochrome microspheres (Luminex). Specifically, 5 × 106 carboxylated microspheres were pelleted in a microcentrifuge for 1 min at maximum speed, and the supernatant was carefully removed using a pipette without disturbing the microspheres. The microspheres were resuspended in 50 μL of a buffer containing 0.1 M MES (Sigma) at pH 4.5. The amino-substituted capture probe was dissolved in ddH2O at a concentration of 1 mM, and 1 μL of the solution (containing 1 nmole of capture-probe oligonucleotides) was added to the microspheres for the coupling reaction. The coupling reaction was initiated by adding 2.5 μL of freshly made 10 mg/mL 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloride (EDC; Aldrich). The mixture of microspheres, capture probes, and EDC was vortexed briefly and incubated at room temperature for 30 min in the dark. After the 30-min incubation, a second 2.5 μL of freshly prepared EDC solution (10 mg/mL) was added to the reaction and incubated for an additional 30 min. This step was repeated for a total of three times (three EDC additions). During the incubations, the reaction was occasionally mixed by finger flicking the tube to keep the microspheres in suspension. After the coupling reaction, 1 mL of 0.02% Tween 20 (BioRad) was added to the microspheres. The solution was mixed well and centrifuged in a microcentrifuge for 1 min at maximum speed. The supernatant containing free capture-probe oligonucleotides and excess EDC was carefully removed. The microspheres were washed again in 1 mL of 0.1% SDS (Ambion) to ensure the removal of free capture probe and EDC. In the end, the capture-probe-conjugated microspheres were resuspended in 100 μL of a buffer containing 0.1 M MES at pH 4.5. The coupled microspheres were stored at 4°C in a dark box and stayed stable for at least 6 mo. The microspheres were diluted in TE buffer and counted using a cell counter slide under 100× magnification. For a single hybridization assay, about 7500 coupled microspheres of each set were used. The coupling efficiencies and hybridization specificity were evaluated by hybridizing the coupled microspheres to their corresponding biotinylated complementary oligonucleotides.
Plant Materials and Chemical Application
Wild-type Arabidopsis thaliana ecotype Columbia (Col) plants were grown in soil at 22°C in growth chambers programmed for a 12-h-light (8000–10,000 lx) and 12-h-dark cycle for 3 wk. Benzothiadiazole (BTH) was freshly added in sterile water to reach a final concentration of 300 μM of active ingredient and then applied as a fine mist to 3-week-old Arabidopsis, ∼0.5–1.0 mL per plant. Another set of control Arabidopsis grown in the same conditions was sprayed with a fine mist of sterile water. Leaves were harvested from plants 1, 3, 5, 7, and 9 d after BTH or water treatment and immediately frozen in liquid nitrogen. Leaf samples were stored at −80°C. For each experiment, three pots of Arabidopsis were treated and collected. The water- or BTH-treated plants were placed in different growth chambers under identical conditions to avoid cross induction.
Total RNA Extraction and cRNA Labeling
Frozen tissue samples were ground to a fine powder in liquid nitrogen using a mortar and pestle. The tissue powder in liquid nitrogen was transferred to a 50-mL sterile tube. We did not allow the samples to thaw. The samples were uniformly homogenized in either TRIzol reagent (GIBCO BRL) or RNAwiz reagent (Ambion) for about 30 sec using a rotor-stator homogenizer Tissue Tearor (Biospec Products). Total RNA was extracted from the tissue homogenates following the manufacturers' protocols and quantitated by absorbance at 260 nm. Total RNA (10 μg) of each sample was converted to double-stranded cDNA using an oligo(dT) primer plus a 5′-end T7 RNA polymerase promoter sequence via the Superscript Choice System for cDNA Synthesis (GIBCO BRL). Double-stranded cDNA was purified by phenol–chloroform extraction, phase lock gel (5 Prime–3 Prime, Inc.), and ethanol precipitation. In vitro transcription was applied on the cDNA samples using T7 RNA polymerase (RNA Transcript Labeling Kit; Enzo Diagnostics). The cRNA was labeled by incorporating biotinylated UTP (Boehringer Mannheim) using a ratio of 1:3 of labeled to unlabeled UTP in the reaction. Next, the labeled cRNA was purified by ethanol precipitation and quantitated by taking absorbance at 260 nm. Before hybridization to capture-probe-coupled microspheres, the cRNA was randomly fragmented by incubating at 94°C for 35 min.
Hybridization of Targets to Capture-Probe-Coupled Microspheres
The 1× hybridization buffer contained 3 M Tetramethylammonium-chloride (TMAC; Sigma), 0.1% SDS, 50 mM Tris-chloride (pH 8.0), and 4 mM EDTA (pH 8.0). The stock hybridization solution was prepared as 1.5× and stored at 50°C to prevent precipitation. In the first step, target samples containing 10 μg of labeled cRNA or a desired amount of nucleic acid fragments in 20 μL were denatured by heating at 95°C for 10 min. Capture-probe-conjugated microspheres (∼7500 beads per color) were mixed in 40 μL of 1.5× hybridization buffer and subsequently added to the denatured target samples. The hybridization mixture was quickly vortexed and incubated at 48°C for 1 h in an Eppendorf microtube incubator (Eppendorf Scientific) with a shaking speed of 300 rpm. The hybridization conditions for DNA fragments were slightly different. For example, for the two experiments shown in Figure 1, A and B, the hybridization temperature was set at 55°C and the time was 15 min. After incubation, the hybridization mixture was spun for 1 min at 14,000 g in a microcentrifuge. Supernatant was carefully removed with a pipette without disturbing the microspheres. The microspheres were washed by adding 50 μL of 1× hybridization solution, mixed by finger flicking, and incubated at 48°C for 5 min without shaking. Microspheres were collected by centrifugation for 1 min at maximum speed and the supernatant was removed. The microspheres were washed a total of three times, then 50 μL of 1× TMAC and 0.5 μL of 1 mg/mL streptavidin-conjugated R-phycoerythrin (Molecular Probes) were added to the microspheres. The solution was briefly vortexed and incubated in the dark at room temperature for 10 min. The microspheres (35 μL) were analyzed on the Luminex100 system, and at least 200 events of each set of microspheres were counted.
Flow Cytometry Analysis
The Luminex100 system integrates lasers, optics, fluidics, electronics, and signal processing to identify each set of color-coded microspheres from every other set, and to measure the total fluorescence at the microsphere's surface for each set to quantify the amount of bound target. Each set of microspheres was distinguished by assigned fluorescence color code (different percentage of red and orange) inside the microspheres. The fluorescence associated on the surface of each microsphere corresponding to amounts of targets was detected and measured by a laser detector. The MFI of each subset of microspheres was calculated and displayed on a monitor by a digital signal processor and the Luminex proprietary software. The Luminex100 system detects fluorescent dyes with an excitation wavelength of ∼532 nm and an emission wavelength ∼580 nm. For each experiment, 200 events of each subset of microspheres were analyzed on the Luminex100 system to obtain an MFI that was representative of the whole population of each set of microspheres.
Acknowledgments
We thank Syngenta R & T employees Jane Glazebrook, Fumiaki Katagiri, and their groups for selecting pathogenesis-related genes for this study; Vaijayanti Gupta for providing mutant Arabidopsis; Devon Brown for providing BTH; and Kay Lawton for providing Arabidopsis samples. We are especially grateful to Sherman Chang, Bin Han, Yen Tran, and Tong Zhu for their help on the GeneChip results. Finally, we thank Peng Luan, Jane Glazebrook, Fumiaki Katagiri, and Steve Whitham for critical comments on this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL li.yang@syngenta.com; FAX (858) 812-1097.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.190901.
REFERENCES
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377 (3547 suppl):3–174. [PubMed] [Google Scholar]
- Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci. 1993;90:7327–7331. doi: 10.1073/pnas.90.15.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglie KE, Biddle P, Cressman R, Broglie R. Functional analysis of DNA sequences responsible for ethylene regulation of a bean chitinase gene in transgenic tobacco. Plant Cell. 1989;6:599–607. doi: 10.1105/tpc.1.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Iannone MA, Li MS, Taylor JD, Rivers P, Nelsen AJ, Slentz-Kesler KA, Roses A, Weiner MP. A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res. 2000;10:549–557. doi: 10.1101/gr.10.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis. Curr Opin Plant Biol. 1999;2:280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Use of Arabidopsis for genetic dissection of plant defense responses. Annu Rev Gene. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- Gordon RF, McDade RL. Multiplexed quantification of human IgG, IgA, and IgM with the FlowMetrix system. Clin Chem. 1997;43:1799–1801. [PubMed] [Google Scholar]
- Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry. 2000;39:131–140. [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Fleury-Olela F, Schibler U. Selective amplification via biotin- and restriction-mediated enrichment (SABRE), a novel selective amplification procedure for detection of differentially expressed mRNAs. Proc Natl Acad Sci. 1997;94:6831–6836. doi: 10.1073/pnas.94.13.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Lee NH, Weinstock KG, Kirkness EF, Earle-Hughes JA, Fuldner RA, Marmaros S, Glodek A, Gocayne JD, Adams MD, Kerlavage AR, et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lie YS, Petropoulos CJ. Advances in quantitative PCR technology: 5′ nuclease assays. Curr Opin Biotech. 1998;9:43–48. doi: 10.1016/s0958-1669(98)80082-7. [DOI] [PubMed] [Google Scholar]
- Liu D, Raghothama KG, Hasegawa PM, Bressan RA. Osmotin overexpression in potato delays development of disease symptoms. Proc Natl Acad Sci USA. 1995;91:1888–1892. doi: 10.1073/pnas.91.5.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart D, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotech. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Murray KK. DNA sequencing by mass spectrometry. J Mass Spectrom. 1996;31:1203–1215. doi: 10.1002/(SICI)1096-9888(199611)31:11<1203::AID-JMS445>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Oliver KG, Kettman JR, Fulton RJ. Multiplexed analysis of human cytokines by use of the FlowMetrix system. Clin Chem. 1998;44:2057–2060. [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Smith PL, WalkerPeach CR, Fulton RJ, DuBois DB. A rapid, sensitive, multiplexed assay for detection of viral nucleic acids using the FlowMetrix system. Clin Chem. 1998;44:2054–2056. [PubMed] [Google Scholar]
- Swaroop A, Agarwal N, Gruen JR, Bick D, Weissman SM. Differential expression of novel Gs alpha signal transduction protein cDNA species. Nucleic Acids Res. 1991;19:4725–4729. doi: 10.1093/nar/19.17.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasmatzis G, Essand M, Brinkmann U, Lee B, Pastan I. Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc Natl Acad Sci. 1998;95:300–304. doi: 10.1073/pnas.95.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotech. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]