Abstract
Human replication protein A (RPA), a heterotrimeric protein complex, was originally defined as a eukaryotic single-stranded DNA binding (SSB) protein essential for the in vitro replication of simian virus 40 (SV40) DNA. Since then RPA has been found to be an indispensable player in almost all DNA metabolic pathways such as, but not limited to, DNA replication, DNA repair, recombination, cell cycle and DNA damage checkpoints. Defects in these cellular reactions may lead to genome instability and, thus, the diseases with a high potential to evolve into cancer. This extensive involvement of RPA in various cellular activities implies a potential modulatory role for RPA in cellular responses to genotoxic insults. In support, RPA is hyperphosphorylated upon DNA damage or replication stress by checkpoint kinases including ATM (ataxia telangiectasia mutated), ATR (ATM and Rad3-related) and DNA-PK (DNA-dependent protein kinase). The hyperphosphorylation may change the functions of RPA and, thus, the activities of individual pathways in which it is involved. Indeed there is growing evidence that hyperphosphorylation alters RPA-DNA and RPA–protein interactions. In addition, recent advances in understanding the molecular basis of the stress-induced modulation of RPA functions demonstrate that RPA undergoes a subtle structural change upon hyperphosphorylation, revealing a structure-based modulatory mechanism. Furthermore, given the crucial roles of RPA in a broad range of cellular processes, targeting RPA to inhibit its specific functions, particularly in DNA replication and repair, may serve a valuable strategy for drug development towards better cancer treatment.
Keywords: Replication protein A, DNA damage response, Phosphorylation, DNA damage, Checkpoint, DNA repair
INTRODUCTION
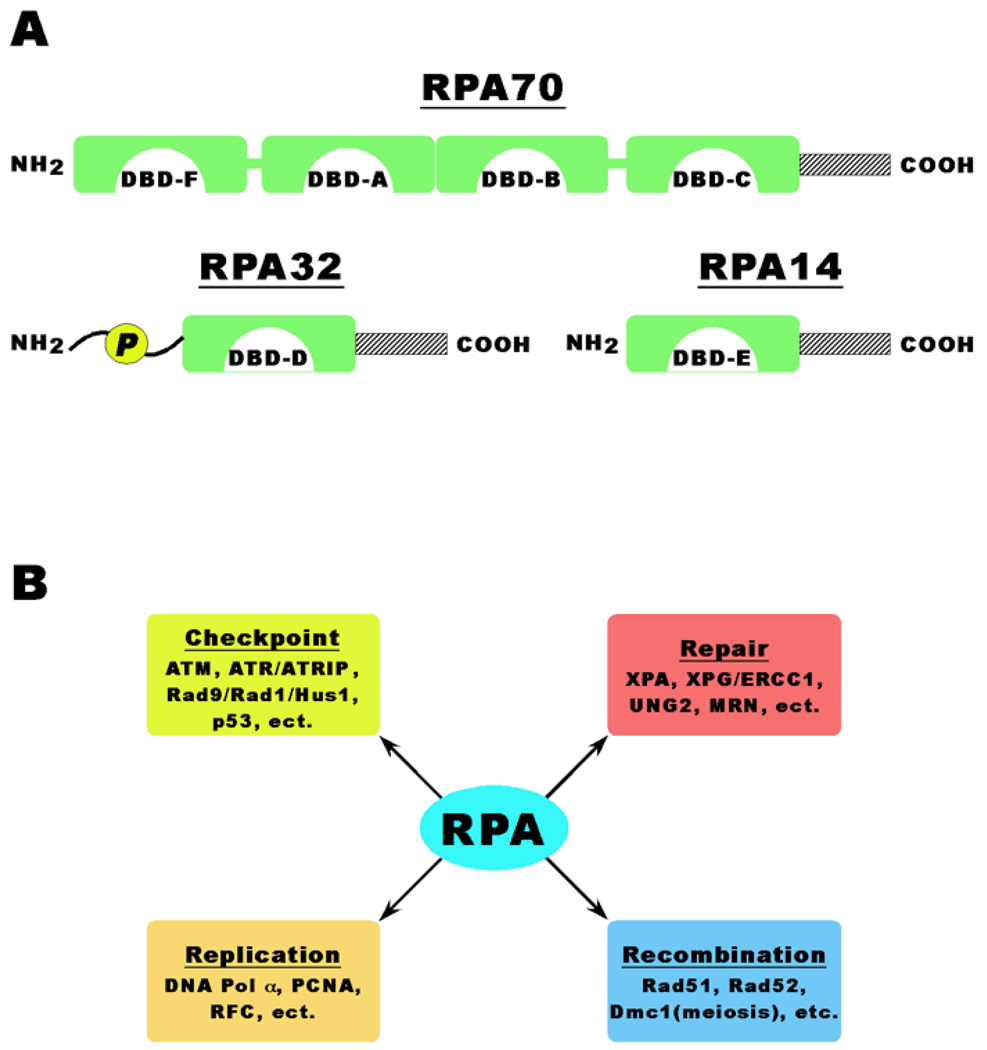
Single-stranded DNA (ssDNA) is perhaps one of the most ubiquitous and important biological intermediate structures formed throughout the life of cells. Thus, it is critical that ssDNA is protected from unwanted attack by endonucleases and that its unwound state is maintained for important DNA metabolic reactions and the assembly of various related biological apparatuses. Replication protein A, the main eukaryote single-stranded DNA binding protein, is a protein of heterotrimer composed of three tightly associated subunits of ~70, 32, and 14 kDa (referred as to RPA70, RPA32, and RPA14, respectively) (Figure 1). Consistent with the importance of ssDNA formation, RPA is required for almost all aspects of cellular DNA metabolism such as DNA replication, recombination, DNA damage checkpoints, and all major types of DNA repair including nucleotide excision, base excision, mismatch and double-strand break repairs. RPA participates in such diverse pathways through its ability to interact with DNA and numerous proteins involved in these processes (Figure 1B).
Figure 1.
Panel A: RPA exhibits a modular construction designed around the OB fold. Human RPA contains 6 OB folds, four on RPA70 subunit (DBD-A, DBD-B, DBD-C, and DBD-F), and one each on RPA32 (DBD-D) and RPA14 (DBD-E). Each subunit also contains a conserved C-terminal α-helix (shaded boxes), which interact to form the trimerization core. The N-terminal domain of RPA32 exists as an extended unstructured domain and contains most of the phosphorylation sites for the trimer. The N-terminal domain is phosphorylated in both a cell cycle and DNA damage-dependent manner.
Panel B: RPA is required for all major DNA metabolism pathways including DNA replication, repair, and recombination. RPA directly interacts with a variety of protein factors in each pathway to facilitate DNA metabolism including (but not limited to) the protein factors listed. In addition, RPA is involved in the initiation of DNA damage checkpoints and hyperphosphorylated in response to genotoxic stress.
In addition to the essential role of RPA in DNA replication initiation and elongation (Fairman and Stillman, 1988; Wobbe et al., 1987; Wold and Kelly, 1988), recently RPA is found to be potentially involved in cell cycle checkpoints and DNA damage checkpoints, from initiation of DNA damage checkpoint signaling through execution and activation of checkpoints. The checkpoint signaling cascades consist of damage sensors, signal transducers, mediators and effectors that, if activated, eventually inhibit cell cycle progression to stabilize stalled replication forks, and to promote DNA repair or trigger apoptosis (Kastan and Bartek, 2004; Sancar et al., 2004; Zhou and Elledge, 2000). Given that RPA undergoes extensive phosphorylation in cells responding to DNA damage or genetic stress (Liu and Weaver, 1993; Zernik-Kobak et al., 1997), involvement of RPA in the diverse cellular activities suggests a modulatory role for RPA in cellular DNA damage responses.
Genomes of cells are constantly under attack by various DNA-damaging agents. In response, cells launch an array of biological reactions to meet the challenges and to maintain genome stability. These processes include damage detection and signaling, DNA repair, transcriptional responses, DNA damage checkpoints, and apoptosis (Li and Zou, 2005; Sancar et al., 2004). Removal of DNA damage prevents the mutagenic potentials of the lesions from ultimately being expressed, but requires the action of DNA repair and the activation of DNA damage checkpoints which delay or arrest cell cycle progression to allow sufficient time for repair and to prevent error-prone replication of DNA. Induction of a programmed cell death process eliminates the cells with devastating DNA damage. Failure of such responses may lead to catastrophic biological consequences such as genomic instability, potentially causing human diseases including cancers. The extensive involvement of RPA in DNA damage response pathways together with the fact that RPA is hyperphosphorylated upon DNA damage underscores the importance in understanding the molecular mechanism of RPA in these processes.
In this review, discussion will be focused on the recent advances in understanding the roles of RPA in cellular DNA damage responses and the underlying molecular and biochemical basis that governs the functions of RPA.
STRUCTURE OF RPA AND ITS BINDING TO ssDNA
All known cellular functions of RPA rely on or are mediated by its binding affinity for ssDNA. The heterotrimeric protein RPA binds ssDNA in a sequential binding manner with a 5’ to 3’ polarity (Bochkarev and Bochkareva, 2004; de Laat et al., 1998; Iftode et al., 1999; Wold, 1997). Although no complete structure for RPA has been solved, combination of biochemical and structural analyses has yielded much insight into the domain organization of RPA. The central structural and functional element of RPA is the oligosaccharide/oligonucleotide binding fold (OB-fold). RPA contains six OB-folds, each of which consists of five β-strands arranged in a β-barrel, a structure common among ssDNA binding proteins (Bochkarev and Bochkareva, 2004; Gomes et al., 1996). The RPA70 subunit contains four OB-folds denoted DBD-A (DNA binding domain A), DBD-B, DBD-C, and DBD-F, while the RPA32 subunit contains DBD-D and RPA14 has DBD-E (Figure 1A). Biochemical analyses have localized the major ssDNA binding affinity to the tandem DBD-A and DBD-B of RPA70. The binding is initiated by an interaction of DBD-A and DBD-B with a length of 8–10 nucleotides (nt) at the 5’-side of ssDNA (Bochkarev et al., 1997). A more stable intermediate binding of 13–22 nt mode occurs with the additional involvement of DBD-C (Brill and Bastin-Shanower, 1998; Iftode et al., 1999). Finally, the cooperative binding of all four RPA DBDs (A–D) occludes a size of ~30 nt of ssDNA (Bastin-Shanower and Brill, 2001; Blackwell et al., 1996; Kim et al., 1992). The association constant of the binding ranges from 108 to 1011 M−1 depending on the sequence and length of the substrate (Kim et al., 1994; Kim and Wold, 1995; Liu et al., 2005b). The DBD-F has a low affinity for ssDNA and is involved primarily in interactions with other DNA metabolism proteins. RPA14 does not exhibit affinity for ssDNA but is required for the stable heterotrimer formation (Iftode et al., 1999; Wold, 1997). In addition to the OB-fold and the C-terminal α-helix domain for many protein interactions (Mer et al., 2000), RPA32 also contains an unstructured Nterminal phosphorylation domain.
RPA IN DNA REPAIR
RPA is required for each of the four major DNA repair pathways: nucleotide excision repair (NER), base excision repair (BER), DNA mismatch repair (MMR), and DNA double strand break (DSB) repair (Wold, 1997). In NER, RPA is believed to play a role in DNA damage recognition (Burns et al., 1996; Costa et al., 2003; He et al., 1995; Sancar et al., 2004; Thoma and Vasquez, 2003) and in recruiting and positioning of XPG and ERCC1-XPF endonucleases to the lesion site for incision reactions (de Laat et al., 1998; Matsunaga et al., 1996). In the later stage of NER, RPA participates in the gap-filling reaction, along with PCNA, RFC, and DNA polymerase δ or ε (Aboussekhra et al., 1995). RPA was implicated in BER via interaction with human uracil-DNA glycosylase (UNG2) and its stimulatory effect in long-patch BER (DeMott et al., 1998; Nagelhus et al., 1997). The involvement of RPA in the MMR process was revealed recently (Ramilo et al., 2002). In the homologous recombinational repair of DSBs, RPA has been shown to interact with two members of the RAD52 epistasis group proteins, Rad51 and Rad52, and to modulate their activities (Park et al., 1996; Raderschall et al., 1999; Stauffer and Chazin, 2004; Sugiyama et al., 1998; Van Komen et al., 2002). In particular, Rad52 recognizes RPA-bound ssDNA, (Sugiyama and Kowalczykowski, 2002; Sung et al., 2003). Human RPA also has been reported to interact with breast cancer susceptibility proteins, BRCA1 and BRCA2, two probable recombination mediators, as well as tumor suppressor p53 (Bochkareva et al., 2005; Choudhary and Li, 2002; Wong et al., 2003).
RPA AND DNA DAMAGE CHECKPOINT RESPONSES
DNA damage checkpoints are biochemical pathways that delay or arrest cell cycle progression in response to DNA damage (Nyberg et al., 2002). In mammalian cells the DNA damage checkpoints are activated upon DNA damage via the ATR (-ATRIP), ATM, and Rad9-Rad1-Hus1/Rad17-Rfc2–5 signaling pathways. Both ATM and ATR proteins are the members of phosphatidylinositol 3-kinase-like kinase (PIKK), while Rad9-Rad1-Hus1 and Rad17-Rfc2–5 are the counterparts of PCNA and RFC, respectively (Burtelow et al., 2001; Caspari et al., 2000; Parrilla-Castellar et al., 2004; Venclovas and Thelen, 2000). The ATM kinase seems to be involved in the detection of DNA DSB via Mre11/Rad50/Nbs1 complex (MRN) (Chan et al., 2000; Gately et al., 1998; Lee and Paull, 2004; Lee and Paull, 2005; Paull and Lee, 2005), whereas ATR kinase is critical for cellular responses to a variety of DNA damage including DSB. When activated, these protein kinases eventually phosphorylate and modulate the cellular activities of downstream factors in DNA damage responses (e.g. Chk1 and Chk2) (Abraham, 2001; Bartek et al., 2004). The 9-1-1/ Rad17-Rfc2–5 complex appears to be translocated to the sites of DNA damage independently of the ATR and ATM, but may also be essential for activation of the downstream kinase of ATR (i.e. Chk1) and initiation of the checkpoint responses (Bao et al., 2004; Lowndes and Murguia, 2000; O'Connell et al., 2000; Zou et al., 2002).
There is accumulating evidence to support the involvement of RPA in cellular checkpoint activation after DNA damage. In both budding and fission yeasts, several mutations in RPA caused the hypersensitivity of cells to DNA damaging agents, and defective G1/S and intra-S checkpoints (Lee et al., 1998; Longhese et al., 1996; Pellicioli et al., 2001). A particular mutation (L45E) in the large subunit of yeast RPA, rfa1-L45E, led to the decreased Rad53 phosphorylation and the cells failed to arrest at the G2/M checkpoint when DSBs were introduced (Umezu et al., 1998). Using Xenopus oocytes system, it has been shown that RPA was necessary for chromatin association of ATR and suppression of DNA synthesis in response to DNA strand breaks, and immunodepletion of RPA abrogated an aphidicolin-induced DNA replication checkpoint (Costanzo et al., 2003; You et al., 2002). A recent report further demonstrated that the uncoupling of helicase and polymerase activities, which leads to formation of long regions of RPA-coated ssDNA at replication forks, is necessary for ATR checkpoint signaling in Xenopus extracts (Byun et al., 2005). In mammalian cells, the chromatin association and nuclear foci formation of ATR after exposure to genotoxic agents are dependent on RPA (Dart et al., 2004; Zou and Elledge, 2003). Specifically, RPA is required for localization of ATR to DNA damage sites and for activation of ATR-mediated phosphorylation of Chk1 and Rad17, most likely through the recognition of ATRIP, the interacting partner of ATR, to the RPA-ssDNA complex (Ball et al., 2005; Zou and Elledge, 2003).
RPA-coated ssDNA is also an important intermediate structure recognized by the Rad17-Rfc2–5 complex, which facilitates the recruitment of the 9-1-1 complex to the gapped and primed DNA structures in vitro (Zou et al., 2003). In human cells, RPA interacts with 9-1-1 complex, mediated by its binding to Rad9 (Wu et al., 2005a). The cellular interaction and nuclear co-localization of these two complexes are significantly stimulated by DNA damage, supporting the notion that RPA and 9-1-1 complexes work cooperatively to activate checkpoint signaling (Wu et al., 2005a). Consistently, knockdown of the RPA expression in cells by small interference RNA (siRNA) blocks the DNA damage-dependent chromatin association of 9-1-1 (Wu et al., 2005a). These results suggest that RPA may serve as an upstream regulator for the activity of the 9-1-1 complex in the cellular checkpoint network. Taken together, RPA may play a role in initiation of DNA damage checkpoints through the binding of RPA to the long stretches of ssDNA resulting from replication fork stalling under replication stress or at DNA damage sites. This extended ssDNA when bound by RPA serves as a common intermediate structure for the assembly of two independent checkpoint apparatuses, 9-1-1/Rad17-Rfc2–5 and ATR-ATRIP complexes, at the sites of DNA damage (Zou and Elledge, 2003; Zou et al., 2003).
Despite these observations, however, whether formation of the RPA-ssDNA intermediate complex is essential for activation of ATR kinase cascade remains in dispute (Cortez, 2005). Ball et al. demonstrated recently that a mutant of ATRIP that does not bind to the RPA-ssDNA complex still supports the ATR phosphorylation of Chk1, suggesting that the interaction of ATRIP with RPA-ssDNA is not absolutely required for ATR activation (Ball et al., 2005). Using siRNA, it also has been shown that neither RPA70 nor RPA32 is essential for the hydroxyurea- or UV-induced phosphorylation of the ATR substrate Chk1 (Dodson et al., 2004). Interestingly, ATR or the ATR-ATRIP complex binds to both naked and RPA-covered ssDNA with comparable affinities (Unsal-Kacmaz and Sancar, 2004). These data suggest that activation of ATR may occur through RPA-independent pathways.
It remains unclear whether RPA participates in the ATM-dependent checkpoint pathway. RPA has been reported to interact and co-localize with MRN complex, which appears to function as a damage sensor upstream of ATM activation in cellular DNA damage responses (Lee and Paull, 2005; Paull and Lee, 2005; Robison et al., 2004; Robison et al., 2005). Moreover, the unwinding of DNA ends by MRN was essential for stimulation of ATM activity towards its downstream cellular targets p53 and Chk2, suggesting a possible involvement of RPA in ATM signaling (Lee and Paull, 2005).
HYPERPHOSPHORYLATION OF RPA IN RESPONSE TO DNA DAMAGE
RPA is phosphorylated in a cell cycle-dependent manner (Din et al., 1990; Dutta and Stillman, 1992; Oakley et al., 2003). It is also undergoes hyperphosphorylation in response to a variety of DNA damage agents such as UV or ionizing irradiation (Binz et al., 2004; Liu and Weaver, 1993). The unstressed cell cycle-dependent phosphorylation occurs during G1/S transition and in M-phase, primarily at the conserved cyclin-CDK phosphorylation sites of Ser-23 and Ser-29 in the unstructured N-terminus of RPA32 subunit (Din et al., 1990; Dutta and Stillman, 1992; Fang and Newport, 1993; Niu et al., 1997; Pan et al., 1994; Zernik-Kobak et al., 1997). In contrast, the stress-induced hyperphosphorylation of RPA is much more extensive. Nine potential phosphorylation sites have been suggested within the unstructured N-terminal domain of RPA32 (RPA32N), including Ser-4, Ser-8, Ser-11/Ser-12/Ser-13, Thr-21, Ser-23, Ser-29 and Ser-33 in UV-irradiated human cells (Niu et al., 1997; Nuss et al., 2005; Zernik-Kobak et al., 1997). Although it remains unknown how many and which of these sites are concurrently phosphorylated on a single RPA molecule upon DNA damage, a recent study by Nuss et al. (2005) showed that at least four of those sites can be concurrently phosphorylated in vitro and in vivo with human cells treated with DNA damage agents. Interestingly, the same study also reported several new DNA damage-induced phosphorylation sites in the RPA including Thr-98 in RPA32 subunit and the sites within residues 112–163 and 569–600 of RPA70 subunit (Nuss et al., 2005). Since the Thr-98 residue is completely buried in the crystal structure of the trimerization core of RPA (Bochkareva et al., 2002), it was suggested that the phosphorylation of Thr-98 may imply an in-solution dynamic nature of the regions of RPA p14 that bury the residue (Nuss et al., 2005).
In contrast to the cell cycle-dependent phosphorylation of RPA, the role of RPA hyperphosphorylation remains elusive. The DNA damage-induced hyperphosphorylation of RPA is believed to be carried out by the members of PIKK kinase family including DNA-PK, ATM and ATR (Binz et al., 2004; Block et al., 2004). The hyperphosphorylation also occurs in a ssDNA-binding- and replication-dependent manner in cells (Bartrand et al., 2004; Oakley et al., 2001; Rodrigo et al., 2000). Replication is probably necessary for conversion of the unrepaired DNA lesions or intermediates to DSBs for subsequent formation of ssDNA (Dunkern and Kaina, 2002; Robison et al., 2005). These are consistent with the involvement of RPA-ssDNA binding in the initiation of the checkpoints by these kinases. However, the relative contribution of these kinases to RPA hyperphosphorylation and the different potential roles of the hyperphosphorylation by these kinases have not been defined. Given that these kinases are involved in the initiation of DNA damage checkpoints, it is possible that the RPA hyperphosphorylation is required for regulation of the cellular activities controlled by these checkpoints in response to different genetic stresses.
It has been suggested that the RPA hyperphosphorylation may reduce the role of RPA in DNA replication while shifting a fraction of the pool of cellular RPA to DNA repair reactions based on the following observations (Binz et al., 2004): (i) cellular extracts prepared from DNA damage cells have a reduced ability to support in vitro SV40 DNA replication while replication activity can be restored to the extracts by addition of purified RPA (Carty et al., 1994; Iftode et al., 1999; Liu et al., 2000; Patrick et al., 2005; Wang et al., 1999); and (ii) RPA hyperphosphorylation appears to have no effects on NER activity in vitro with cellular extracts or a purified reconstituted system (Ariza et al., 1996; Pan, 1995; Patrick et al., 2005). Consistently, the RPA32 mutant that mimics the hyperphosphorylation by substitution of the phosphorylatable residues with aspartic acid in RPA32N, is unable to localize to the replication centers in cells, but is competent to associate with DNA damage foci (Vassin et al., 2004); also the hyperphosphorylation disrupts the RPA interaction with DNA polymerase α in vitro (Patrick et al., 2005). By contrast, there is a controversy over the hyperphosphorylation effect on RPA binding to ssDNA in vitro as both no change (Binz et al., 2003; Oakley et al., 2003) and a decrease (Fried et al., 1996; Patrick et al., 2005) in the binding have been reported. A possible explanation for this discrepancy is that the effect is oligonucleotides sequence-dependent (Patrick et al., 2005).
On the other hand, since RPA is involved in all major DNA repair pathways including BER, NER, MMR, and DSB repair through its DNA binding and its interactions with various repair proteins (Mer et al., 2000), an interesting question is whether hyperphosphorylated RPA is preferentially engaged in specific repair pathways. Indeed, a recent report shows that the cellular interaction of RPA with two DSB repair factors, Rad51 and Rad52, is predominantly mediated by the hyperphosphorylated forms of RPA after UV or camptothecin (CPT) treatments (Wu et al., 2005b). It is likely that hyperphosphorylated RPA is preferentially recruited to DSB repair in a checkpoint-dependent manner likely because DNA double strand breaks are the most devastating DNA damage.
BIOCHEMICAL BASIS OF MODULATORY ROLE OF RPA
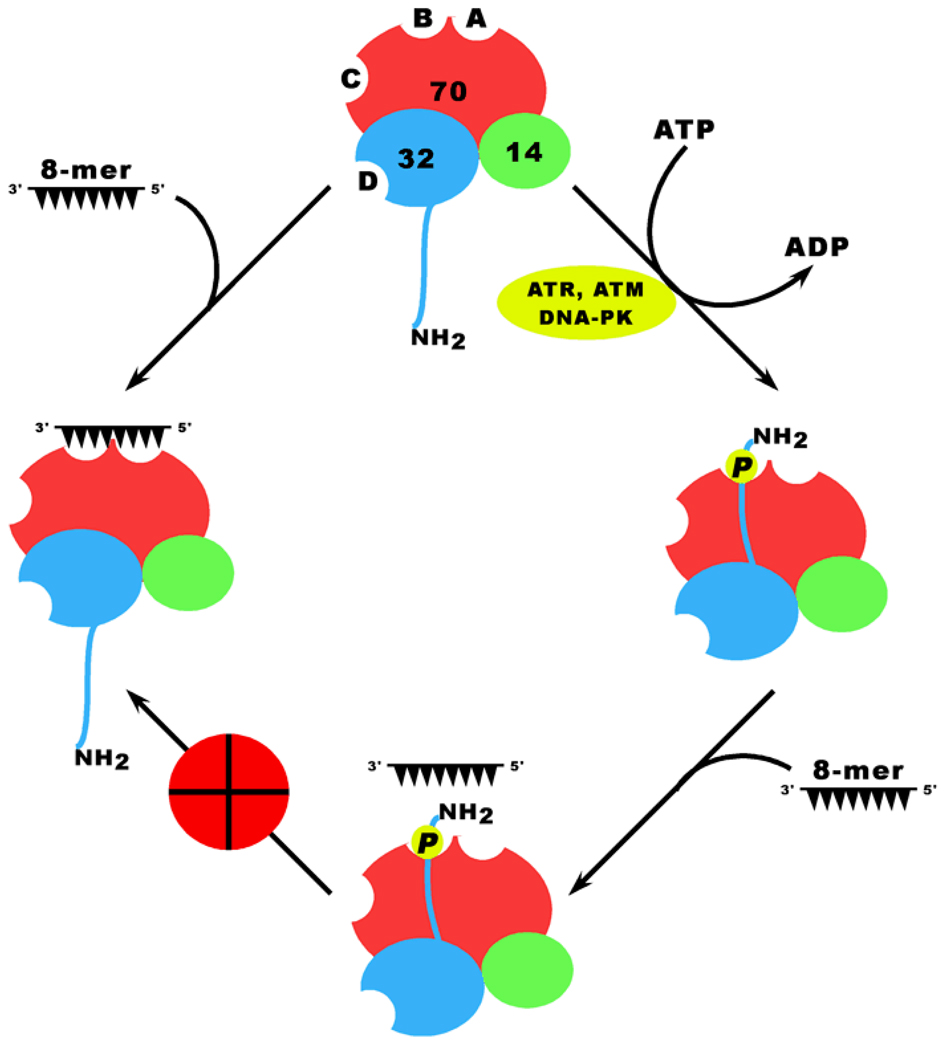
One of the most challenging issues regarding the role of RPA in DNA damage responses and RPA hyperphosphorylation is how the functions of RPA are modulated by hyperphosphorylation. There are possibly two mechanisms by which the modulation can be achieved: (i) the recognition of the hyperphosphorylated domain of RPA by hyperphospho-binding proteins; and (ii) a hyperphosphorylation-induced structural transformation of RPA leading to the disruption of RPA-DNA and RPA-protein interactions. Using purified RPA70 fragment (RPA701–168) and a synthetic acidic peptide mimicking the hyperphosphorylated N-terminus of RPA32, a NMR study shows that electrostatic interactions occur between the basic cleft of DBD-F (RPA70N) and the mimicking acidic peptide (Binz et al., 2003). However, the study is based on protein fragments rather than the full-length RPA heterotrimer. The structure-based mechanism for the full length of RPA has been revealed in a recent study (Liu et al., 2005a), demonstrating that upon hyperphosphorylation RPA undergoes a subtle structural change involving the ssDNA binding cleft of DNA binding domain B of RPA70 subunit. This is likely due to an invoked direct interaction of the hyperphosphorylated N-terminus of RPA32 (hyp-RPA32N) with DBD-B through electrostatic contacts between the two domains, which are highly negatively and positively charged, respectively (Liu et al., 2005a) (Figure 2). This potential inter-domain interaction in RPA (with an association constant in the order of 107 M−1) may enable phosphorylation to modulate the cellular activities of RPA, which is critical for cellular responses to genotoxic stresses. Indeed, such structural alteration or inter-domain interaction results in a significant decrease in the binding affinity of RPA to short ssDNA or partial DNA duplexes containing short ssDNA tails (8–11 nt), which is most likely due to the competitive blocking of ssDNA binding to DBD-B by hyp-RPA32N (Figure 2). In contrast, no substantial effect occurs for binding with longer ssDNA. The negligible effect is probably due to the much higher affinity (Ka is in the order of 109–11 M−1) for RPA binding to the long length of ssDNA than that for the hyp-RPA32N and DBD-B interaction.
Figure 2.
Proposed mechanism for RPA modulation by hyperphosphorylation of the RPA32 N-terminal domain. RPA binds an 8-mer oligonucleotide with DNA binding domains A and B of RPA70 subunit, which is likely essential for replication origin firing. Upon DNA damage, however, the N-terminal domain of RPA32 subunit is hyperphosphorylated by ATR, ATM, and/or DNA-PK, inducing a subtle structural shift featured with a direct interaction of the negatively charged N-terminal domain with the positively charged DNA binding cleft of DBD-B. This interaction (Ka ≥ ~107 M−1) is sufficient to compete with and displace the 8-mer oligonucleotides, subsequently blocking RPA from binding short ssDNA. However, the interaction of RPA with 30-mer oligonucleotides (Ka ≥ ~109 M−1) is unaffected by this interaction. This proposed mechanism provides a means for down-regulation of DNA replication by preventing RPA association with replication origins without affecting its ability to participate in DNA repair pathways.
Binding of RPA to short ssDNA is of biological significance particularly in replication initiation at origins where RPA binds to the melted DNA bubble containing a DNA single-stranded region of about 8-nt during replication initiation (Blackwell and Borowiec, 1994; Borowiec and Hurwitz, 1988; Parsons et al., 1990). Since it is the replication origin firings that is targeted by DNA damage checkpoints to induce S-phase cell cycle arrest upon DNA damage (Shechter et al., 2004), it is possible that the checkpoint-induced RPA hyperphosphorylation may play an important role in downregulating the initiation of DNA replication. In support, hyperphosphorylation-mimicking RPA fails to associate with replication centers in vivo (Vassin et al., 2004); and the hyperphosphorylated RPA is considerably less supportive of SV40 DNA replication than native RPA (Carty et al., 1994; Iftode et al., 1999; Liu et al., 2000; Patrick et al., 2005; Wang et al., 1999). For the first time, the study by Liu et al. reveals a potentially important structural basis for RPA to play a modulatory role in DNA damage responses.
It is also worth noting that although RPA hyperphosphorylation occurs primarily in the N-terminal domain of RPA32 subunit, the recent identification of the damage-induced phosphorylation sites on RPA70 suggests a potential involvement of these sites in modulation of RPA functions. Since these sites are located in DBD-C of RPA70 and in the linker region of DBD-A and DBD–F, a possible role of the RPA70 phosphorylation is to destabilize or facilitate RPA binding to duplex DNA or proteins (Nuss et al., 2005). However the details of the effects remain to be defined.
DRUG DEVELOPMENT BY TARGETING RPA
The essential role of RPA in DNA metabolisms, particularly in DNA replication and damage responses, makes it a worthful target for drug development in cancer treatment. This is because rapid division of cancer cells depends on replication of genomes and also the increased cell ability in DNA repair is believed to be one of the causes to drug resistance acquired in chemotherapeutic treatment of cancers. In an effort to identify inhibitors of RPA, a homogeneous high-throughput screening assay using a fluorescent reporter has been developed to measure RPA-DNA binding activity in the presence of a collection of 2,000 small chemicals (Andrews and Turchi, 2004). The effect of these chemicals on the RPA-DNA interaction has been determined. As the result, several positively-scored candidates for inhibition of RPA binding activity have been identified.
CONCLUSIONS
RPA is intimately involved in cellular DNA metabolism ranging from DNA replication, recombination, to DNA damage/stress responses. Defects in these processes are associated with a score of human diseases. This underscores the importance in understanding the molecular and biochemical mechanisms of the potential modulatory functions of RPA. While many details still remain unknown, recent efforts focusing on RPA hyperphosphorylation and the role of RPA in DNA damage checkpoints have started to resolve the mystery. In general, current evidence appears to document RPA’s involvement in the initiation of replication-associated DNA damage checkpoints (Ward et al., 2004), and also RPA’s role as a hyperphosphorylation-dependent downstream checkpoint effector for regulation of DNA metabolic pathways. In addition to its conventional role in DNA metabolism, RPA may serve as a mediator for cross-talk between the pathways regulated by its hyperphosphorylation. A possible scenario is that upon DNA damage the RPA binds to long stretches of ssDNA that resulted from various DNA damage-related events such as collapse and stalling of the replication fork and DNA repair (e.g. DSB processing); The RPA-ssDNA complex recruits DNA damage checkpoint protein kinases (ATR-ATRIP, ATM, and 9-1-1 complex) to the damage sites, which triggers the checkpoint signaling. After the recruitment for checkpoint activation, the checkpoint kinases in turn hyperphosphorylate RPA mediated by proteins such as 53BP1 (Yoo et al., 2005) for subsequent modulation of its cellular activities essential for DNA replication and repair. However, the details of the molecular mechanism for elucidating the effects of hyperphosphorylation of RPA on DNA metabolic pathways remain to be defined in the future.
ACKNOWLEDGMENTS
We thank Dr. Phillip Musich for critical reading of this manuscript.
Contract grant sponsor: National Cancer Institute (NCI), National Institutes of Health (NIH); Contract grant number: CA86927 (to Y.Z.)
LITERATURE CITED
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80(6):859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Turchi JJ. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Mol Cancer Ther. 2004;3(4):385–391. [PubMed] [Google Scholar]
- Ariza RR, Keyse SM, Moggs JG, Wood RD. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 1996;24(3):433–440. doi: 10.1093/nar/24.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D. ATRIP Binding to Replication Protein A-Single-stranded DNA Promotes ATR-ATRIP Localization but Is Dispensable for Chk1 Phosphorylation. Mol Biol Cell. 2005;16(5):2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Lu T, Wang X, Zheng H, Wang LE, Wei Q, Hittelman WN, Li L. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23(33):5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5(10):792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bartrand AJ, Iyasu D, Brush GS. DNA stimulates Mec1-mediated phosphorylation of replication protein A. J Biol Chem. 2004;279(25):26762–26767. doi: 10.1074/jbc.M312353200. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J Biol Chem. 2001;276(39):36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Lao Y, Lowry DF, Wold MS. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J Biol Chem. 2003;278(37):35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3(8–9):1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol Cell Biol. 1994;14(6):3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA, Masrangelo IA. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol Cell Biol. 1996;16(9):4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32(3):997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14(1):36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385(6612):176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. Embo J. 2002;21(7):1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec JA, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. Embo J. 1988;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Bastin-Shanower S. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol Cell Biol. 1998;18(12):7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Guzder SN, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271(20):11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276(28):25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee M-c, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19(9):1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. Embo J. 1994;13(9):2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitrioux K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20(4):1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Son SC, Block W, Ye R, Khanna KK, Wold MS, Douglas P, Goodarzi AA, Pelley J, Taya Y, Lavin MF, Lees-Miller SP. Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J Biol Chem. 2000;275(11):7803–7810. doi: 10.1074/jbc.275.11.7803. [DOI] [PubMed] [Google Scholar]
- Choudhary SK, Li R. BRCA1 modulates ionizing radiation-induced nuclear focus formation by the replication protein A p34 subunit. J Cell Biochem. 2002;84(4):666–674. doi: 10.1002/jcb.10081. [DOI] [PubMed] [Google Scholar]
- Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19(9):1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Chigancas V, Galhardo Rda S, Carvalho H, Menck CF. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85(11):1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11(1):203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Dart DA, Adams KE, Akerman I, Lakin ND. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J Biol Chem. 2004;279(16):16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12(16):2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMott MS, Zigman S, Bambara RA. Replication protein A stimulates long patch DNA base excision repair. J Biol Chem. 1998;273(42):27492–27498. doi: 10.1074/jbc.273.42.27492. [DOI] [PubMed] [Google Scholar]
- Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4(6):968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- Dodson GE, Shi Y, Tibbetts RS. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J Biol Chem. 2004;279(32):34010–34014. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- Dunkern TR, Kaina B. Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol Biol Cell. 2002;13(1):348–361. doi: 10.1091/mbc.01-05-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. Embo J. 1992;11(6):2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman MP, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. Embo J. 1988;7(4):1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport JW. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J Cell Sci. 1993;106((Pt 3)):983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- Fried LM, Koumenis C, Peterson SR, Green SL, van Zijl P, Allalunis-Turner J, Chen DJ, Fishel R, Giaccia AJ, Brown JM, Kirchgessner CU. The DNA damage response in DNA-dependent protein kinase-deficient SCID mouse cells: replication protein A hyperphosphorylation and p53 induction. Proc Natl Acad Sci U S A. 1996;93(24):13825–13830. doi: 10.1073/pnas.93.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gately DP, Hittle JC, Chan GK, Yen TJ. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol Biol Cell. 1998;9(9):2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Henricksen LA, Wold MS. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry. 1996;35(17):5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374(6522):566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34(3):141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33(47):14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- Kim C, Snyder RO, Wold MS. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12(7):3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wold MS. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34(6):2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304(5667):93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94(3):399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Li L, Zou L. Sensing, signaling, and responding to DNA damage: organization of the checkpoint pathways in mammalian cells. J Cell Biochem. 2005;94(2):298–306. doi: 10.1002/jcb.20355. [DOI] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, McHugh MM, Beerman TA, Melendy T. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J Biol Chem. 2000;275(2):1391–1397. doi: 10.1074/jbc.275.2.1391. [DOI] [PubMed] [Google Scholar]
- Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13(12):7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y. Modulation of Replication Protein A Function by Its Hyperphosphorylation-induced Conformational Change Involving DNA Binding Domain B. J Biol Chem. 2005a;280(38):32775–32783. doi: 10.1074/jbc.M505705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang Z, Utzat CD, Liu Y, Geacintov NE, Basu AK, Zou Y. Interactions of human replication protein A with single-stranded DNA adducts. Biochem J. 2005b;385:519–526. doi: 10.1042/BJ20041151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Neecke H, Paciotti V, Lucchini G, Plevani P. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 1996;24(18):3533–3537. doi: 10.1093/nar/24.18.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10(1):17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Park CH, Bessho T, Mu D, Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem. 1996;271(19):11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103(3):449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, Krokan HE. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem. 1997;272(10):6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- Niu H, Erdjument-Bromage H, Pan ZQ, Lee SH, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272(19):12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry. 2005;44(23):8428–8437. doi: 10.1021/bi0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Walworth NC, Carr AM. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10(7):296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- Oakley GG, Loberg LI, Yao J, Risinger MA, Yunker RL, Zernik-Kobak M, Khanna KK, Lavin MF, Carty MP, Dixon K. UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein. Mol Biol Cell. 2001;12(5):1199–1213. doi: 10.1091/mbc.12.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley GG, Patrick SM, Yao J, Carty MP, Turchi JJ, Dixon K. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry. 2003;42(11):3255–3264. doi: 10.1021/bi026377u. [DOI] [PubMed] [Google Scholar]
- Pan Z, Park CH, Amin AA, Hurwitz J, Sancar A. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. PNAS. 1995;92(10):4636–4640. doi: 10.1073/pnas.92.10.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZQ, Amin AA, Gibbs E, Niu H, Hurwitz J. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1994;91(18):8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271(31):18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3(8–9):1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Parsons R, Anderson ME, Tegtmeyer P. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J Virol. 1990;64(2):509–518. doi: 10.1128/jvi.64.2.509-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick SM, Oakley GG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 2005;44(23):8438–8448. doi: 10.1021/bi048057b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4(6):737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7(2):293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A. 1999;96(5):1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilo C, Gu L, Guo S, Zhang X, Patrick SM, Turchi JJ, Li GM. Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol Cell Biol. 2002;22(7):2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279(33):34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- Robison JG, Lu L, Dixon K, Bissler JJ. DNA lesion-specific co-localization of the Mre11/Rad50/Nbs1 (MRN) complex and replication protein A (RPA) to repair foci. J Biol Chem. 2005;280(13):12927–12934. doi: 10.1074/jbc.M414391200. [DOI] [PubMed] [Google Scholar]
- Rodrigo G, Roumagnac S, Wold MS, Salles B, Calsou P. DNA replication but not nucleotide excision repair is required for UVC-induced replication protein A phosphorylation in mammalian cells. Mol Cell Biol. 2000;20(8):2696–2705. doi: 10.1128/mcb.20.8.2696-2705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6(7):648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J Biol Chem. 2004;279(24):25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277(35):31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci U S A. 1998;95(11):6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278(44):42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog. 2003;38(1):1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148(3):989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Sancar A. Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol Cell Biol. 2004;24(3):1292–1300. doi: 10.1128/MCB.24.3.1292-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J Biol Chem. 2002;277(46):43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24(5):1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28(13):2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou XY, Wang H, Huq MS, Iliakis G. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J Biol Chem. 1999;274(31):22060–22064. doi: 10.1074/jbc.274.31.22060. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J Biol Chem. 2004;279(11):9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- Wobbe CR, Weissbach L, Borowiec JA, Dean FB, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci U S A. 1987;84(7):1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wold MS, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988;85(8):2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Ionescu D, Ingles CJ. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene. 2003;22(1):28–33. doi: 10.1038/sj.onc.1206071. [DOI] [PubMed] [Google Scholar]
- Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005a;24(29):4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang Z, Liu Y, Zou Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem J. 2005b;391((Pt 3)):473–480. doi: 10.1042/BJ20050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo E, Kim BU, Lee SY, Cho CH, Chung JH, Lee CH. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene. 2005;24(35):5423–5430. doi: 10.1038/sj.onc.1208710. [DOI] [PubMed] [Google Scholar]
- You Z, Kong L, Newport J. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J Biol Chem. 2002;277(30):27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J Biol Chem. 1997;272(38):23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16(2):198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100(24):13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]