Abstract
MicroRNAs are endogenous small RNA molecules that regulate gene expression. Although the biogenesis of microRNAs and their regulation have been thoroughly elucidated, the degradation of microRNAs has not been fully understood. Here by using the pulse–chase approach, we performed the direct measurement of microRNA lifespan. Five representative microRNAs demonstrated a general feature of relatively long lifespan. However, the decay dynamic varies considerably between these individual microRNAs. Mutation analysis of miR-29b sequence revealed that uracils at nucleotide position 9–11 are required for its rapid decay, in that both specific nucleotides and their position are critical. The effect of uracil-rich element on miR-29b decay dynamic occurs in duplex but not in single strand RNA. Moreover, analysis of published data on microRNA expression profile during development reveals that a substantial subset of microRNAs with the uracil-rich sequence tends to be down-regulated compared to those without the sequence. Among them, Northern blotting shows that miR-29c and fruit fly bantam possess a relatively rapid turnover rate. The effect of uracil-rich sequence on microRNA turnover depends on the sequence context. The present work indicates that microRNAs contain sequence information in the middle region besides the sequence element at both ends.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNA molecules that specifically regulate eukaryotic gene expression. The biogenesis of miRNAs has been thoroughly elucidated. MiRNA genes are transcribed by either RNA polymerase II or III (1–3), and the primary transcripts are subject to multiple processing steps for maturation into the functional form of miRNAs. These steps include processing of primary transcripts into precursor by Drosha/DGCR8 complex (4,5), exporting the precursors from nucleus to cytoplasm by exportin-5 and then cleaving the precursors into mature miRNAs by Dicer (6–9). It has been revealed that regulation of miRNA expression level occurs at each steps of the constitutive maturation pathway (10,11).
Comparing to the comprehensive studies on the miRNA generation, the destruction of mature miRNA remains elusive. First, on the whole, the stability of miRNAs has not been unambiguously clearly defined. Previous studies showed the heterogeneity of miRNA lifespan, ranging from many days in vivo to a few hours in cell culture. MiR-208 sustains 3 weeks after its transcriptional inhibition and miR-29b is as short-lived in HeLa cells as several hours (12,13). Additionally, it is impressive that miRNAs in neuron are much more instable (14–16). Most recently a study carried out the global measurement of miRNA lifespan in which the microarray technique was employed to profile miRNA expression after the transcriptional shutoff in HEK293 cells (17). The observation revealed that a majority of miRNAs does not appreciably change during transcriptional arrest for 8 h. The processing of primary transcripts and precursors of miRNAs, however, is expected to continue and result in replenishing the mature miRNA pool when the transcription has been inhibited. Taking it into consideration, this measurement probably overestimated the miRNA lifespan. Hence further clarifying the stability of miRNAs in a general way is in need. Second, while the trans-acting factors have been extensively screened, very little is known about cis-elements that direct the specificity of miRNA degradation. RNA binding proteins AGO2 and translin were reported to protect the resident miRNA from degradation (18,19). On the other hand, the nucleases SDN1 and XRN2 were identified to cleave miRNAs in the plant Arabidopsis thaliana and invertebrate Caenorhabditis elegans, respectively, and both nucleases cut miRNAs in the sequence independent manner (20,21). In addition, it has been well known that modifications at the 3′-end of RNA molecules mediate the stability of message RNAs (22). Until recently it has been discovered that among a number of miRNAs, miR-122 alone lends the mechanism from mRNAs to decrease their vulnerability to nucleases by 3′-end addition of adenosine (23). The inherent characteristic of miR-122 for selectively directing its 3′-end modification remains unknown. More interesting is that although miRNAs are as short as ∼22 nt, they contain abundant sequence information to determine their individual property. A 6 nt element at miR-29b 3′-tail controls its cellular localization (13). Most recently it was documented that a 7 nt element at miR-382 3′-end is necessary for its degradation (17). This raises the further interest to explore the sequence content in miRNAs themselves.
In the present work, we began with a direct measurement of miRNA lifespan in cell culture, and then analyzed the sequence content of the decayed miRNAs. Our result showed that uracil at the middle region in miR-29 family is required for their accelerated turnover.
MATERIALS AND METHODS
MiRNA mimickers
Synthetic duplex of miRNA mimickers in our assay are completely complementary. The sequences of miRNA mimickers are listed in the Supplementary Table S2. Mimickers were synthesized by ShangHai GenePharma Co., Ltd.
Cell culture and transfection
HeLa cells were maintained in Dulbecco’s Modified Eagle Media (DMEM, Invitrogen) supplemented with 10% fetal bovine serum. For half life measurement, 2.5 × 105 HeLa cells were seeded into each well of 6-well plate. Twenty hours later, 40 nM miRNA mimickers were transfected by Lipofectamine 2000 (Invitrogen). Four hours after transfection, the medium was removed, cells were washed twice with PBS and were cultured with fresh medium. The time point at which the medium was replaced was set as 0 h and then cells were harvested at the time points indicated.
Real-time PCR
About 100 ng of total RNA was reverse transcribed by Improm II (Promega) with both a miRNA-specific stem-loop primer and random primer. The reaction was carried out at 42°C for 1 h. The reaction mixture was 100-fold diluted and real-time PCR with SYBR green was performed. Expression of miRNAs was normalized to U6 and ΔΔCt analysis was used to calculate the relative expression of miRNAs. The products were analyzed by melting curve and manual inspection by electrophoresis. Real-time PCR for each miRNA was repeated for three times.
Dual luciferase reporter assay
The multiple cloning sites of plasmid pGL3 (Promega) in the promoter region were removed and filled in, and then a synthetic oligonucleotide containing KpnI and XhoI sites was inserted at XbaI site which is in 3′-UTR region of luciferase coding sequence. Cdc42 3′-UTR was amplified by RT–PCR and then inserted into the modified pGL3 by KpnI and XhoI digestion. The forward primer is GATGGTACCTCTCCAGAGCCCTTTCTGC and reverse primer GTCCTCGAGCAAAGAATTGAGACATGAGAAAGC. About 100 nM miRNA mimickers or negative control RNAs, 80 ng pRL plasmid and 1 μg pGL3-cdc42 3′-UTR plasmid were transfected into HeLa cells by Lipofectamine 2000 (Invitrogen), with triple replications for each transfection. Twenty-four hours after transfection, cells were harvested and reporter activity was examined with the dual luciferease reporter assay kit (Promega).
Northern blotting
RNA was isolated by TRIZOL (invitrogen) and 6 μg RNA for each sample was analyzed in 15% polyacrylamide gel with 8 M urea. RNAs were transferred onto positively charged Nylon membrane (Ambion) and immobilized by ultraviolet light for 2 min. Oligonucleotide probes were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Takara). All probe sequences are listed in Supplementary Table S3. The membranes were probed at 39°C with the hybridization solution PerfectHyb (Toyobo) overnight. The membranes were washed by 2× SSC and 0.5% SDS twice at 37°C. Radioactive signals were quantified using Quantity ONE (Bio-Rad). MiRNA abundance was normalized to U6 and the half life was then calculated according to the signals at five time points (0, 2, 4, 8 and 12 h). Lifespan of miR-29 family (including the wild-type and mutants) was examined by three independent experiments and other miRNAs two independent experiments.
Analysis of developmental expression profiles of miRNAs
Expression profiles of worm, fly and fish miRNAs were collected from published data. Data during all developmental stages in worm, the egg stage in fly and the first 2 days embryonic stage of fish were analyzed. MiRNAs with no expression signal were excluded. Either continuous or transient decrease of expression was classed as ‘down-regulated’ and others were classed as ‘not down-regulated’. Each of both classes in three species was summed up. The difference between miRNAs with and without U-rich sequence was examined by the Fisher’s exact test.
RESULTS
Pulse–chase assay to examine the miRNA lifespan
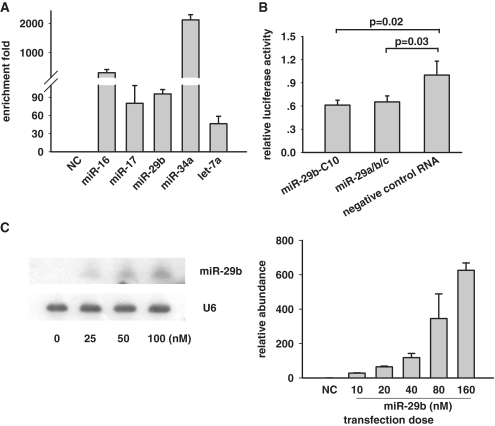
Both transcription and post-transcriptional processing influence miRNAs generation, and hence transcriptional shutoff may not completely terminate the generation of miRNAs. We therefore chose to employ the pulse–chase assay with synthetic RNA duplex as miRNA mimickers to examine the miRNA turnover in cell culture (24). Synthetic duplex of miRNA mimickers in our assay are completely complementary. First, we examined whether these miRNA mimickers could steadily accumulate in cells and therefore minimize the interference by endogenous miRNA species. Five microRNAs with different endogenous expression level were selected (25), and their mimickers were transfected into HeLa and analyzed by real-time PCR. A synthetic duplex RNA, which does not target any RNAs, served as the negative control (NC). Each miRNA increased substantially and therefore allow a long-term chase (Figure 1A). Previously it has been proven that this type of synthetic miRNA mimickers could incorporate into the RNA-induced silencing complex (RISC) (13). Here to further validate whether these mimickers could simulate miRNAs in a cellular context, we examined the silencing effect of synthetic miRNA mimickers on target mRNAs. MiR-29 family, including miR-29a, b and c, was reported to directly target cdc42 mRNA in HeLa cells (26). Our reporter assay showed that the synthetic miR-29 mixture (equal amount of miR-29a/b/c) was able to repress the luciferase reporter fused to the cdc42 3′-UTR by 40% (Figure 1B). A miR-29 mutant with the substitution of uracil at nucleotide position 10 with cytosine (miR-29b-C10) was as effective as the wild-type.
Figure 1.
Synthetic double-strand RNAs as miRNA mimickers for studying the stability of mature miRNAs. (A) 40 nM synthetic miRNA mimickers were transfected into HeLa cells for 4 h and the cellular abundance of each miRNA was assayed by real-time PCR. Relative abundance shows the relative enrichment fold of miRNA quantity after transfection of miRNA mimickers to with a negative control RNA (NC). (B) miRNA mimickers for miR-29 family (miR-29a/b/c) were able to suppress the luciferase reporter with 3′-UTR of its target mRNA cdc42 in HeLa cells. (C) Left: miR-29b mimickers with indicated concentration were transfected into HeLa cells and quantified by either northern blotting (left) or qPCR (right). Cellular abundance of miRNA is proportionally to the transfection dose at the range indicated. Luciferase assay and PCR was repeated independently three times, and northern blotting two times.
MiRNAs compete in loading into the RISC. Saturated RISC may leave miRNAs free and therefore make them more vulnerable to degradation. To exclude this effect in our assay, we titrated the transfection dose of miRNA mimickers at different concentrations. Northern blotting showed that cellular abundance had a good linear correlation with the transfection dose for miR-29b mimicker (Figure 1C, left). As high as 100 μM of miR-29b mimicker, the abundance of cellular miR-29b paralleled with the transfection dose. In contrary, negative control RNA was hardly detected in the same assay (Supplementary Figure S1). Real-time PCR verified this correlation (Figure 1C, right). In the subsequent assays, we therefore adopted the transfection dose of 40 nM miRNA mimickers in the measurement of miRNA lifespan. We noted that due to global increase of miRNA abundance in response to cell contact as reported previously (27), cells in pulse–chase assay should be appropriately administrated to <80% confluence in order to avoid the artificial interference.
Lifespan of miRNAs are long but diverse
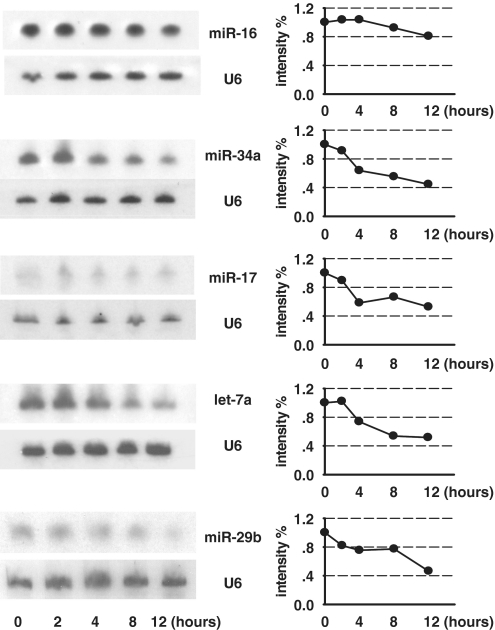
Five miRNAs with various expression levels in HeLa were selected for lifespan measurement, including miR-16 with high abundance, let-7a, miR-17 and miR-29b with mediate expression, and miR-34a with no detection (25). Pulse–chase assay was used to measure lifespan of these representative miRNAs mentioned above. Generally the miRNA half life was 12 h or longer (Figure 2). Furthermore, these representative miRNAs possessed distinct characteristic in their dynamic change. MiR-16 did not have apparent decrease in their abundance until the time duration limit (12 h); MiR-34a and miR-17 possessed a moderate turnover rate, with a half life of about 12 h; Let-7a and miR-29b rapidly disappeared, with a half life of 11 and 7 h, respectively (Figure 2). The diversity of decay dynamics of miRNAs raises a question of sequence determinant of their stability.
Figure 2.
The stability of mature miRNAs varies individually in HeLa cells. MiRNA mimickers were transfected into HeLa cells for 4 h and Northern blotting was used to measure the abundance of miRNAs at the indicated time points.
Uracils at nucleotide position 9–11 of miR-29b are required for its rapid decay
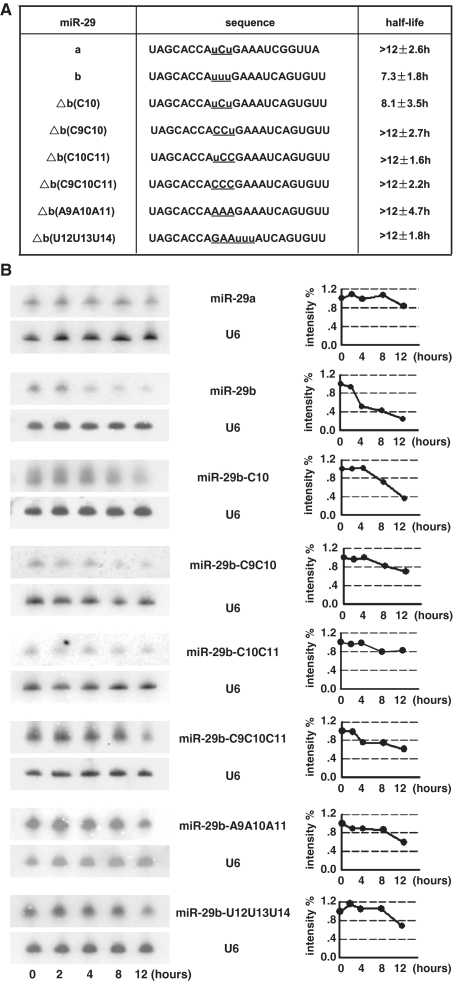
MiR-29a is stable, while miR-29b is rapidly decayed in HeLa cells although their sequence are highly similar (13), thus we took miR-29 family as a paradigm to study the sequence dependence of miRNA degradation. MiR-29a and miR-29b differ at two regions. One is at their 3′-end and the other is at nucleotide position 10, with cytosine in miR-29a and uracil in miR-29b (Figure 3A). It has already been proven that the sequence at miR-29b 3′-end alone is not responsible for its rapid decay (13). We asked whether the variance at position 10 together with near nucleotides is involved in miRNA inherent stability. At the beginning, uracil was mutated into cytosine for miR-29b (miR-29b-C10), and this mutation did slightly enhance miRNA stability (Figure 3B). We further questioned whether uracils near position 10 had a synergic effect with the central uracil (U10) to function. Mutation of uracils in both position 9 and 10 (miR-29b-C9C10) into cytosine prolonged the half life of miR-29b (Figure 3B). This effect was observed for mutation in both position 10 and 11 (miR-29b-C10C11) to a more significant degree. All three uracils from position 9 to 11 into cytosine (miR-29b-C9C10C11) abolished the rapid decay of miR-29b (Figure 3B). It is questionable whether the change of observed decay rate is simply attributed to the alteration of miRNA GC content. Mutation into adenosine (miR-29b-A9A10A11) was examined and the result demonstrated a significant decrease in the decay rate (Figure 3B). Furthermore, to test whether position at nucleotides 9–11 is critical for the effect of uracil stretch, the nucleotides at position 9–11 were exchanged with that at position 12–14 (miR-29b-U12U13U14, see Figure 3A). Northern blotting showed that the change in position stabilized the wild-type miR-29b (Figure 3B). To test whether our observation would hold generally, the lifespan of miR-29b and miR-29b-C9C10C11 was chased in HEK293T cells. Indeed, mutation of centric uracils impeded the rapid degradation of miR-29b (Supplementary Figure S2). Our observation therefore demonstrated that uracil at the middle region of miR-29b (from 9 to 11 nt) was required for its rapid turnover.
Figure 3.
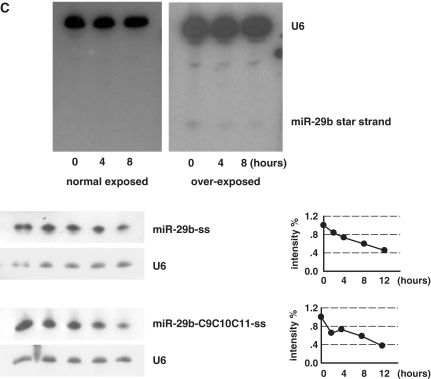
Uracils at nucleotides from 9 to 11 are required for the rapid decay of miR-29b. (A) Sequence of miR-29a/b and mutants. Nucleotides under investigation are underlined and uracils are marked as lowercase. (B) Northern blotting of indicated miRNAs in pulse–chase assay. The northern blot image was showed in the left panel and the quantification of bands in northern blotting images was displayed in the right panel. Intensity of miRNAs was normalized to U6. MiR-29a is much stable than miR-29b. Substitution of uracil at position 10 with cytosine for miR-29b (miR-29b-C10) has a detectable effect on its decay rate, while substitutions of nucleotides at both 9–10, 10–11 and from 9 to 11 (miR-29b-C9C10, -C10C11, -C9C10C11) significantly enhance the miRNA stability. Substitution with adenosine (miR-29b-A9A10A11) also delayed the decay. Movement of uracils from positions 9 to 11 to most near position (miR-29b-U12U13U14) stabilized the miRNA. (C) Upper: miR-29b mimicker (duplex) was transfected into HeLa cells for 4 h and the star strand of the mimicker was detected at the indicated time points. The star strand could not be detected unless film was over-exposed. Lower: Single strand of miR-29b and miR-29b mutant (miR-29b-C9C10C11) was transfected and chased by northern blotting. Both were at a rapid decay rate. Three independent experiments for each miRNA were performed and the typical one was showed here.
Due to nucleotides in positions 9 and 10 of miRNAs influence their sorting in Drosophila (28,29), we examined the star strand of miR-29b duplex. Northern blotting could hardly detect the existence of the star strand (Figure 3C, upper). To further explore the mechanism of the uracil-dependent decay, single strand but not duplex of miR-29b and miR-29b-C9C10C11 were chased. The result showed that the single strand of both miR-29b and miR-29b-C9C10C11 were rapidly decayed (Figure 3C, lower). This observation suggests that the uracil element function during the loading of miRNA into RISC or the unwinding of miR/miR* duplex.
A subset of miRNAs with uracils at nucleotide positions 9–11 possesses rapid turnover
Due to the above observation that uracils in positions 9–11 (U-rich sequence) was involved in miR-29b decay, the next question we asked was whether it is a general rule for the U-rich sequence to play a role as a cis-acting element in regulation of miRNA stability. MiRNAs are thought to contribute to the complexity of organism, thus majority of miRNAs emerge with cellular differentiation and development (30). We deduced that if U-rich sequence is truly involved in the acceleration of miRNA turnover, the expression of miRNAs with U-rich sequence will tend to be down-regulated during development. To this end, we listed all miRNAs with U-rich sequence and examined their expression profile during development in the published data which was summarized in Supplementary Table S1 (31–33). Factors other than internal stability of miRNAs can regulate their abundance, such as transcriptional regulation. To decrease this complication, the interval of developmental stage in expression data was restricted to no more than 24 h. There are 17 miRNAs with the U-rich sequence whose expression has been characterized (Table 1). As expected, 10 out of the 17 miRNAs exhibit the rapid decrease during development, while the majority of other miRNAs accumulates (Table 2), with a different expression dynamics (P = 0.004).
Table 1.
Developmental expression profiles of miRNAs containing U-rich sequence
| Species | miRNA | Sequence | profile | References |
|---|---|---|---|---|
| Worm | miR-43 | UAUCACAGUUUACUUGCUGUCGC | DR | 33 |
| miR-59 | UCGAAUCGUUUAUCAGGAUGAUG | NDR | ||
| miR-254 | UGCAAAUCUUUCGCGACUGUAGG | NDR | ||
| miR-787 | UAAGCUCGUUUUAGUAUCUUUCG | DR | ||
| miR-2208b-3p | AUGCAGAUUUUGGUACACUUCA | NDR | ||
| Fruit fly | bantam | UGAGAUCAUUUUGAAAGCUGAUU | DR | 34 |
| miR-14 | UCAGUCUUUUUCUCUCUCCUA | DR | ||
| miR-313 | UAUUGCACUUUUCACAGCCCGA | DR | ||
| miR-375 | UUUGUUCGUUUGGCUUAAGUUA | NDR | ||
| miR-983 | AUAAUACGUUUCGAACUAAUGA | NDR | ||
| miR-992 | AGUACACGUUUCUGGUACUAAG | DR | ||
| miR-996 | UGACUAGAUUUCAUGCUCGUCU | DR | ||
| Zebrafish | miR-29a | UAGCACCAUUUGAAAUCGGUUA | NDR | 35 |
| miR-29b | UAGCACCAUUUGAAAUCAGUGU | NDR | ||
| miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | DRa | ||
| miR-135b | UAUGGCUUUUUAUUCCUAUCUG | DR | ||
| miR-203a | GUGAAAUGUUUAGGACCACUUG | DR |
U-rich sequence is shown in bold. DR, down-regulated; NDR, not down-regulated.
aNot annotated as a/b.
Table 2.
Distinct expression profiles for miRNAs with and without U-rich sequence during development
| Down-regulated | Not down-regulated | |
|---|---|---|
| miR-UUU | 10 | 7 |
| Not miR-UUU | 106 | 311 |
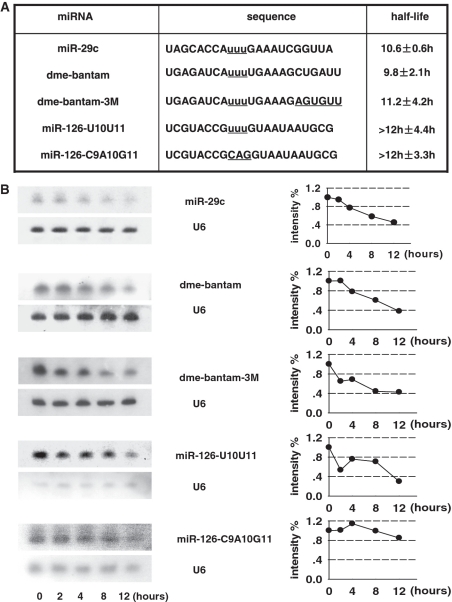
The decrease in expression level of miRNA does not necessarily attribute to miRNA degradation, thus we examined the stability of miRNAs in the above list. MiR-29c is another member of miR-29 family, and our assay showed it was much more unstable than miR-29a, with a half life of 10.6 h (Figure 4). The fruit fly miRNA bantam was sharply down-regulated during the development (34). The abundance of bantam decreased with a relative rapid rate in HeLa cell (Figure 4B).
Figure 4.
(A) Sequence of miRNAs with uracil-rich element and their mutants. Nucleotides under investigation are underlined and centric uracils are marked as lowercase. (B) A subset of miRNAs containing U-rich sequences has an accelerated turnover rate. MiR-29c and fly miRNA bantam show a relatively short lifespan, and replacement of bantam 3′-end sequence with that of miR-29b does not alter the decay dynamics. For two miR-126 mutants, both are long-lived although the one with CAG in positions 9–11 is more stable than one with UUU.
Further, we test whether the U-rich sequence functions dependent or independent of sequence context. First, bantam 3′-end sequence was replaced with that of miR-29b (Dbantam-3 M), and northern blot demonstrated that the sequence alternation did not visibly accelerate the decay of the hybrid (Figure 4B). To place the U-rich sequence in a random sequence background, we constructed two mutants of miR-126 by removing the nucleotides in positions 9–11 with UUU and CAG, respectively. A U-rich element accelerates the decay of miR-126 compared to element with no uracil (Figure 4B). However, the lifespan of U-rich miRNA mutant is still beyond the duration limit of our assay (>12 h). This indicates that the U-rich sequence confers the rapid turnover rate in a context dependent manner.
DISCUSSION
Despite the birth of functional miRNAs have been thoroughly studied, the disappearance of mature miRNAs has not yet fully understood. In the present work, we employed the pulse–chase assay to measure the lifespan of a subset of selected miRNAs. In HeLa cells, these miRNAs were relatively stable. The decay manner, however, is rather inhomogeneous. Mutation analysis of one of selected miRNAs, miR-29b, revealed that uracils at nucleotide positions 9–11 were required for its fast degradation. Two miRNAs that contain uracils in positions 9–11, miR-29c and bantam, also possessed a rapid turnover rate in our assay, and the effect of the U-rich sequence was dependent of sequence context.
In our study, we used the exogenous miRNAs. This strategy is aimed to prevent the continuous production of endogenous microRNA and allow the long-term chase. To assure the physiological relevance, we observed the silencing effect of exogenous miRNAs, which indicated that these miRNA mimickers could incorporate into RISC. Further, we examined the correlation of miRNAs between the transfection dose and cellular abundance. The good linearity suggested that there was no overload into RISC within the range of our assay. However, for an accurate assessment of miRNA metabolism, it should be considered to combine the interpretation from both endogenous and exogenous data.
On one hand, miRNA are actively involved in the regulation of target mRNA, they should be actively modulated to adapt the dynamic change of cellular network. On the other hand, miRNAs seemed to be tightly packaged by RISC which may make them resistant to the destruction (35,36). To discriminate these two possibilities, we measured the lifespan of synthetic miRNA mimickers in cell culture. The result partially supported the idea that majority of miRNAs are stable, with a half life as long as 12 h or more. By analogy, human mRNAs have a median lifespan of 10 h and transcription-related transcripts possess a marked decrease of half life (37). Compared with mRNA, miRNAs as a class of post-transcription regulators are considerably stable. Recently, Bail and colleagues carried out a genome-wide examination on the miRNA lifespan by microarray and revealed that up to observed 95% of miRNAs do not decrease after transcriptional halt (17). Their observation indicates that miRNA turnover is even slower than that demonstrated in our assay. This discrepancy is slight, which may be attributed to the overestimation of miRNA stability in their assay (17), or derived from that the flood of miRNA mimickers into the cell culture during our transfection might influence the metabolism of miRNAs. Notice that no signal could be detected for negative control RNA, the deviation is minimal to attribute to the transient transfection of exogenous mimickers.
Considering that miRNAs are as short as ∼22 nt, it is of apparent interest in the specificity of miRNA turnover. Apart from the seed sequence at 5′-end of miRNAs, two elements located at the 3′-end have been found. One element directs miRNAs to import from cytoplasm into nucleus (13), the other determines miRNA degradation (17). Here we reported that the middle region of miR-29b is required for its turnover. Together, all of these findings suggest that miRNAs contain abundant sequence information though they are short. Specific regulation of miRNA turnover is biologically relevant, and particular interest will concern the situation that miRNAs rapidly vanish in response to stimulation. Indeed, miR-125 in spleen is individually down-regulated in 1 h upon the LPS treatment (38), miR-183/96/182 in retinas are rapidly decayed during dark adaption and miR-29b destabilizes in HeLa cells once the cells enter the cell cycle (13,16).
Apart from the regulatory mechanism of miRNAs, our finding has some implication in siRNA design. The U-rich sequence as a trigger for degradation of miRNAs is probably true for siRNA. If so, avoiding the U-rich sequence at middle region of siRNA might prolong their lifespan and therefore be beneficial for enhancement of their silencing effect.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology (2007CB947002, 2010CB945600, 2011CB811304); National Natural Science Foundation of China (30770457, 30828006, 30971231); Chinese Academy of Sciences (KSCX2-YW-R-096, KSCX2-YW-R-233); Shanghai Pujiang Program (05PJ14105).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wen-Qing Li and An Zeng for helpful discussions.
REFERENCES
- 1.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 6.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 11.Smalheiser NR. Regulation of mammalian microRNA processing and function by cellular signaling and subcellular localization. Biochim. Biophys. Acta. 2008;1779:678–681. doi: 10.1016/j.bbagrm.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 13.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 14.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci. Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 16.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 17.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Yu Z, Hecht NB. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J. Androl. 2008;29:572–579. doi: 10.2164/jandrol.108.005090. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 22.Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 27.Hwang HW, Wentzel EA, Mendell JT. Cell–cell contact globally activates microRNA biogenesis. Proc. Natl Acad. Sci. USA. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr. Opin. Genet Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang F, Hajkova P, O’Carroll D, Lee C, Tarakhovsky A, Lao K, Surani MA. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem. Biophys. Res. Commun. 2008;372:24–29. doi: 10.1016/j.bbrc.2008.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.