Abstract
Heterochromatin Protein 1 (HP1) is a transcriptional repressor that directly binds to the methylated lysine 9 residue of histone H3 (H3K9me), which is a hallmark histone modification for transcriptionally silenced heterochromatin. Studies of homologs in different organisms have provided significant insight into the function of HP1 and the role of H3K9me. Initially discovered to be a major constituent of heterochromatin important for gene silencing, HP1 is now known to be a dynamic protein that also functions in transcriptional elongation, centromeric sister chromatid cohesion, telomere maintenance and DNA repair. Furthermore, recent studies have begun to uncover functional differences between HP1 variants and their H3K9me-independent mode of action. As our understanding of HP1 expands, however, conflicting data has also been reported that requires further reconciliation. Here we focus on some of the recent findings and controversies concerning HP1 functions in mammalian cells in comparison to studies in other organisms.
Keywords: HP1, HP1γ, cohesin, DNA repair, transcription, heterochromatin, D4Z4, FSHD, H3K9me3
HP1 is a conserved chromatin binding protein originally identified in Drosophila as a non-histone chromosomal protein functioning in heterochromatin-mediated gene silencing.1 Homologs and variants exist in organisms ranging from S. pombe to humans (Fig. 1). HP1 directly binds to the methylated K9 residue of histone H3 (H3K9me), an established marker of the transcriptionally repressed state of chromatin, and is critical for its maintenance.2–4 It has been demonstrated in several organisms, including humans, that artificial recruitment of HP1 to a gene promoter region results in gene repression, establishing the role of HP1 in gene silencing.5,6
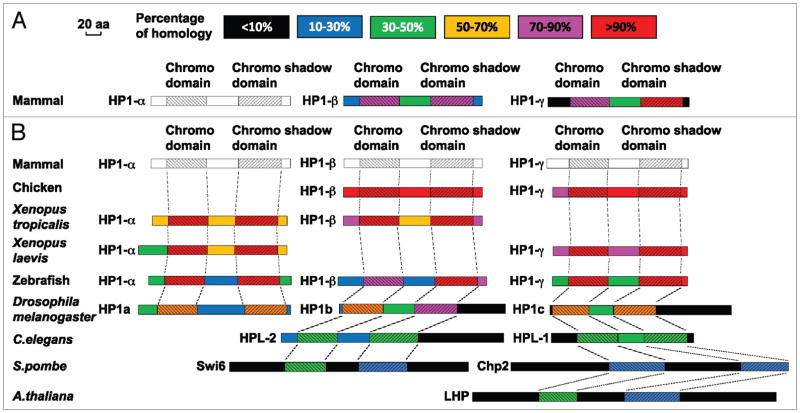
Figure 1.
Alignment of HP1 amino acid sequences. All available HP1 amino acid sequences are from the NCBI protein sequence database. In the alignment results, besides the chromo domain (CD) and the chromo shadow domain (CSD), the N-terminus adjacent to the CD, the C-terminus adjacent to the CSD, and the middle fragment between the two domains (i.e., the linker/hinge domain) are compared between the different proteins. Therefore, each protein diagram is separated into five fragments and in each fragment, the degree of homology is indicated with different colors. The length of each diagram is proportional to the protein size. (A) Alignment of mammalian HP1 amino acid sequences. The sequences of mammalian HP1β and HP1γ are compared with HP1α. The percentage of homology is based on comparison to HP1α (indicated in white/gray). (B) Alignment of HP1 amino acid sequences between different species. The HP1 amino acid sequences from several non-mammalian species are compared to the corresponding mammalian HP1 variant and their homology with mammalian HP1 is indicated in the color diagram. For C. elegans, S. pombe and A. thaliana, the homology between their HP1 proteins and each individual mammalian HP1 variant is low. Therefore, in the diagram, their sequences are compared to the mammalian HP1 variant with the highest alignment score. The alignment of the chromo and chromo shadow domains is delineated with dashed lines. Since all three mammalian HP1 sequences are used as standards for the individual alignments, they are all shown in white/gray.
HP1 proteins are characterized by two conserved domains: the chromodomain (CD) at the N-terminus, and the chromo shadow domain (CSD) at the C-terminus (Fig. 1). The CD was shown to directly bind to H3K9me, while the CSD is implicated in various protein:protein interactions and dimerization. A linker domain is involved in DNA and RNA binding (reviewed in refs. 7–9). While specific binding of HP1 to H3K9me is well described, recent studies have revealed its H3K9me-independent chromatin binding and have identified intriguing new dimensions of HP1 function in heterochromatin regulation, mitosis and DNA repair. Thus, in this review, we will briefly summarize recent findings and highlight the questions and controversies raised by these studies to encourage further investigation of HP1 and HP1-mediated chromatin organization in the cell.
HP1 Binds to the Methylated K9 Residue of Histone H3
Different histone modification patterns are associated with distinct transcriptional states of chromatin.10,11 The effect of histone methylation depends on the location of the modified amino acid residue and the degree of methylation.12–14 Individual silenced genes, as well as larger chromosomal domains that are transcriptionally repressed (so called “heterochromatin”), are often associated with histone H3 lysine 27 methylation (H3K27me) and H3K9me, which are critical markers of epigenetic silencing. Telomeres and centromeres are constitutively heterochromatic while other regions become heterochromatic or are transcriptionally silenced in a cell type- and differentiation-specific manner. H3K27me is part of the polycombmediated silencing pathway.15,16 H3K9me in mammals is effected by multiple histone methyltransferases (HMTases), such as SUV39 h (including both SUV39 h1 and SUV39 h2), SETDB1, and G9a, which have distinct targeting specificities and degrees of methylation (e.g., mono-, di- or tri-methylation).5,17–20 H3K9me at constitutive heterochromatin is maintained primarily by SUV39 h1 and SUV39 h2. The H3K9me-marked heterochromatin assembly pathway was best characterized in S. pombe, and involves non-coding RNA (ncRNA) transcribed from the region and RNA interference (RNAi) machinery which recruit Clr4 (SUV39 h homolog).21–23 H3K9me is then recognized by HP1, which mediates gene silencing. Although both the H3K9me-HP1 pathway and the H3K27me-polycomb pathway are involved in silencing and are often found in overlapping chromatin regions, the functional crosstalk between the two pathways is not well understood.
HP1 Variants
In S. pombe, the HP1 homolog Swi6 was extensively studied initially (Fig. 1), but recent studies of the second HP1 homolog Chp2 revealed that the two homologs are involved in gene silencing and heterochromatin organization in different ways by interacting with distinct sets of proteins.24–26 While Chp2 interacts with the SHREC complex containing ATP-dependent chromatin remodeling and HDAC activities, Swi6 interacts with many different types of nuclear proteins including those involved in chromatin remodeling and DNA replication as well as with centromere binding proteins.24,25 Thus, it is clear that despite the structural conservation and similar H3K9me binding, differences in primary amino acid sequence contribute to differential protein interactions and functions in the cell.
The HP1 variants in mammalian cells are HP1α, HP1β and HP1γ (Fig. 1). Based on the sequences, Swi6 is most closely related to HP1β, and Chp2 to HP1γ. Interestingly, however, HP1 homologs are smaller and lose unique sequences outside of the CD and CSD in higher eukaryotes compared to S. pombe, possibly reflective of species-specific functions and regulation (Fig. 1). In mammalian cells, the cytological distribution of the three HP1 variants was reported to be distinct. In general, HP1α and HP1β are found at constitutive heterochromatin such as centromeres and telomeres, while HP1γ is distributed in both heterochromatic and euchromatic regions.27,28 Consistent with this notion, HP1γ, but not HP1α or HP1β, was found to associate with actively transcribed gene regions (still in a H3K9me-dependent manner) and plays a role in efficient transcriptional elongation.29,30 Interestingly, knockout of HP1β, but not HP1α, was associated with genome instability in mice while depletion of HP1γ, but not HP1α or HP1β, led to mitotic defects in human tissue culture cells.31,32 It remains to be determined whether these observations reflect species-specific or inherent differences in variant functions and/or the underlying mechanisms. Nonetheless, the results further suggest the functional specificities of HP1 variants in the cell.
HP1’s Role in Cohesin Recruitment to Centromeres
In S. pombe, Swi6 not only functions in pericentromeric heterochromatin silencing, but also recruits cohesin, which is important for centromeric sister chromatid cohesion and kinetochore biorientation (Fig. 2).33,34 Cohesin is a conserved and essential protein complex required for sister chromatid cohesion (reviewed in ref. 35 and 36). In chicken DT40 cells, depletion of Dicer, a nuclease important for RNA-dependent heterochromatin assembly, was also shown to disrupt HP1/cohesin localization at centromeres, resulting in premature sister chromatid separation, closely paralleling the observations in S. pombe.37
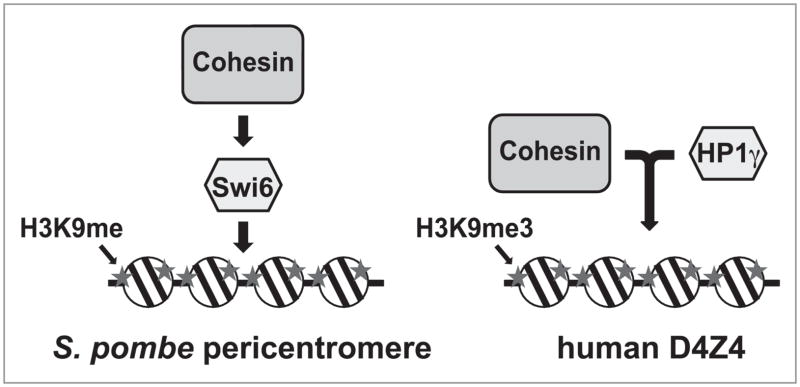
Figure 2.
Relationship between HP1 and cohesin. The sequential recruitment of Swi6/HP1 and cohesin to S. pombe pericentromeric heterochromatin regions (left) and the interdependent recruitment of HP1γ and cohesin to human D4Z4 repeats (right) are depicted.
Cohesin is composed of the two SMC family proteins SMC1 and SMC3 and the two non-SMC subunits Rad21 (also known as Scc1 or Mcd1) and SA (also known as Scc3). By both coimmunoprecipitation after crosslinking and yeast two-hybrid assay, Swi6 was shown to interact with Psc3 (an SA homolog) in S. pombe.34 Several reports in S. pombe and human cells indicate, however, that the cohesin loading factor Scc2 (also known as NIPBL), rather than cohesin, interacts with HP1.24,32,38,39 Human HP1 and S. pombe Swi6 were also found to interact with Sgo1. This interaction is important for Sgo1 localization at centromeres in early mitosis, where it plays a role in cohesin retention and sister chromatid cohesion.40 The interactions with Scc2 and Sgo1 are both through the CSD of HP1.38,40 Thus, HP1 interacts with both the cohesin loader and cohesin retainer in an evolutionarily conserved manner to collectively ensure the enrichment of cohesin at centromeres.
Surprisingly, decreased H3K9 tri-methylation (H3K9me3) and HP1 did not appear to affect cohesin association at centromeres in SUV39 h1 and SUV39 h2 double-knockout mouse embryonic fibroblasts.41 Furthermore, another study reported that HP1 depletion by siRNA in human cells failed to affect cohesin recruitment.32 These two studies suggested that the H3K9me-HP1 pathway is not important for cohesin recruitment to centromeres in mitosis. In the case of the SUV39 h1/h2 knockout MEFs, however, it is possible that redundant HMTases complemented SUV39 h1/h2 function. Thus, although reduced, residual H3K9me and HP1 might have been sufficient to retain cohesin. Another potential challenge is the use of tagged recombinant cohesin components for detection of centromeric localization of cohesin (in the presence of the endogenous proteins) in mammalian cells. Due to the difficulty of visualizing endogenous cohesin at centromeres (possibly because of an antibody accessibility problem), tagged recombinant proteins such as myc-tagged Rad21 (Scc1-myc) or hSMC1-GFP expressed in stable cell lines have been used.41–43 However, unlike in yeast in which the endogenous gene can be replaced by the tagged version to confirm its functionality, the recombinant fusions in mammalian cells are expressed in the presence of the endogenous proteins. Although the tagged cohesin components were shown to be incorporated into the cohesin complex, there is some uncertainty as to whether the expressed fusion proteins have the same functionality and sensitivity for factor requirements as the endogenous species. For siRNA experiments, incomplete depletion of targeted factors is a serious concern. Indeed, the effect of Dicer depletion using the tet-off system on cohesin recruitment to centromeres can only be observed after ~120 h in chicken DT40 cells whose normal cell cycle spans 8 h.37 Finally, it is formally possible that in mammalian cells the presence of other proteins in the centromere-kinetochore complex may offer multiple pathways to ensure cohesin localization at centromeres, thus compensating for the loss of one factor.
HP1’s Role in Cohesin Recruitment to Non-Centromeric Heterochromatin
Despite the controversy surrounding the H3K9me-HP1-cohesin pathway at centromeres, our recent study demonstrated the clear dependence of cohesin on H3K9me3 and HP1 at a specific non-centromeric heterochromatic region in human cells.39 The 3.3 kb D4Z4 repeat is a type of macrosatellite repeat sequence.44 Using chromatin immunoprecipitation (ChIP) and PCR with specific primers, we found that D4Z4 contains both euchromatic (H3K4me2 and H3Ac) and heterochromatic (H3K9me3 and H3K27me3) domains and binding of HP1γ and cohesin.39 SiRNA against SUV39 h1 abolished H3K9me3 as well as HP1 and cohesin binding at D4Z4, but not at other repeats, indicating that HP1γ and cohesin are recruited to D4Z4 in a H3K9me3-dependent manner.44
Interestingly, only HP1γ, not HP1α or HP1β, is detected at D4Z4, and its depletion was sufficient to remove cohesin from this site, indicating the unique involvement of HP1γ at D4Z4.39 Furthermore, loss of cohesin by either depleting the cohesin component Rad21 or the cohesin loading factor Scc2 led to the loss of HP1γ, indicating that binding of cohesin and HP1γ to D4Z4 is inter-dependent. This is different from the relationship between Swi6 and cohesin at the pericentromeric hetero-chromatin in S. pombe, in which Swi6 does not require cohesin for its recruitment to H3K9me3-bearing heterochromatin (Fig. 2).34 Scc2 and cohesin depletion had no effect on HP1γ binding at other repeat sequences, suggesting that this relationship is site-specific. We also found that cohesin and HP1γ binding to D4Z4 bearing H3K9me3 is cell type-specific. Despite the abundant H3K9me3 at D4Z4, HP1γ and cohesin binding to D4Z4 was minimal in lymphoblasts, implying that additional cell type- and repeat sequence-specific histone modification mark(s) and/or binding protein(s) may contribute to the restricted recruitment of HP1γ and exclusion of HP1α and HP1β at D4Z4.
Our findings indicated that cohesin and HP1 function together at a certain type of non-centromeric heterochromatin. Interestingly, H3K9me3 and HP1γ-cohesin are specifically lost from D4Z4 in facioscapulohumeral muscular dystrophy (FSHD), suggesting that they may play a role in gene regulation important for muscle function. This notion is particularly attractive given recent reports of cohesin’s involvement in gene regulation through long-distance chromatin interactions.45–47 Although these studies are presently limited to cohesin at CTCF binding sites that are not in the context of heterochromatin, it is equally plausible to imagine that cohesin recruited to heterochromatin by H3K9me3 and HP1 also mediates silenced chromatin interactions. In fact, HP1 has also been implicated in heterochromatin bundling in Drosophila, and was shown to interact with the lamin B receptor to possibly link chromatin to the nuclear envelope.6,48 Thus, it is conceivable that in a specific context, cohesin and HP1 act together to mediate such chromatin interaction and organization in the nucleus.
H3K9me-Independent Recruitment of HP1 in DNA Repair
HP1 attracted much attention recently for its involvement in DNA repair. As summarized previously,49 HP1 appears to have two opposing functions in DNA repair in the cell. First, depletion of HP1 was found to alleviate the requirement for ATM for efficient repair of heterochromatic regions, suggesting its suppressive role in repair of heterochromatic DNA regions.50 It was also found that casein kinase 2 (CK2)-mediated phosphorylation of HP1 induced in response to DNA damage decreased its CD-dependent affinity for H3K9me leading to the transient displacement of HP1 from DNA damage sites.51 Based on the idea that HP1 has a negative effect on DNA repair in the context of heterochromatin, it was thought that the transient displacement of HP1, especially from heterochromatin, may be part of the necessary cellular response to DNA damage, suggesting an inhibitory role for HP1 in DNA repair. Interestingly, however, HP1 was also found to accumulate at the damage sites, indicating a more active involvement in DNA repair.52 Importantly, this recruitment is mediated by the CSD, not the CD, and therefore is H3K9me-independent. It was proposed that HP1 changes the way it associates with chromatin in response to DNA damage (i.e., from CD-dependent H3K9me binding to CSD-dependent H3K9me-independent binding).49 This model has been further elaborated in two recent reviews.53,54 Supporting this contention is evidence that HP1 is rapidly recruited to oxidative DNA damage, but continues to be actively exchanged at the damage sites.55
CD-dependent and -independent binding of HP1 is analogous to the differential mechanisms of HP1 association with centromeres in interphase and mitosis in human cells (i.e., CD- and H3K9me-dependent association in inter-phase and CD-independent association in mitosis).56 In Drosophila, HP1 association with telomeres critical for telomere capping was shown to be CD- and H3K9me-independent while HP1 binding important for telomere silencing and elongation is through H3K9me.57 Thus, HP1 appears to dynamically utilize different mechanisms to associate with specific sites on chromatin. It is currently unclear whether different modes of chromatin binding result in, or are reflective of, distinct functions of HP1.
Mechanistically, dissociation of HP1 from H3K9me is well-explained by CK2-dependent phosphorylation within the CD, which decreases its affinity for methylated H3K9 in vitro.51 It is unclear, however, as to what recruits HP1 to DNA damage sites. According to findings using different wavelength lasers (i.e., UVC, UVA with Hoechst, green with ethidium bromide) and charged particle irradiation systems, HP1 appears to be recruited to several types of DNA damage, including UV crosslinks, double-strand and single-strand breaks (DSBs and SSBs), and base damage.52,55 Furthermore, HP1 recruitment does not seem to depend on DNA damage recognition factors for each pathway (e.g., DDB2 and XPC for nucleotide exchange repair (NER), and Ku for DSBs).52 So what is the common denominator present in these different types of DNA damage that is responsible for HP1 recruitment? Does HP1 directly recognize damaged DNA or indirectly react to a downstream signal(s)? As damaging systems often introduce a mixture of lesions,58 it is possible that HP1 recognizes one form of damage or chromatin state that is commonly induced by the different irradiation systems. It is critical, therefore, to confirm the recruitment of HP1, for example, to a defined DSB site induced by endonucleases using ChIP.59–61
Finally, depletion or mutation of each of the HP1 variants in C. elegans exhibited different damage-related phenotypes, which contrasts with the findings in mammalian cells where no variant-specific recruitment was detected.52 Puzzlingly, depletion of all three variants had no effect on γ ray-induced DSB damage sensitivity.50 Thus, the functional significance and specificity of HP1 variant recruitment to DNA damage sites remains an interesting question to be addressed in future work.
With the inherent complexity associated with studying cellular pathways in higher eukaryotes, differing results and/or interpretations will likely continue to emerge. While careful evaluation of negative results is vital, it remains an essential aspect of healthy scientific endeavor to publish quality work even if it generates controversy. The efforts to resolve this controversy will frame new questions and studies that will advance the HP1 field. Studies of homologs and variants in multiple species and in various fields (e.g., chromatin, transcription, DNA repair and telomere biology) will continue to provide us with valuable opportunities to further our insight into the ever evolving role of HP1 in the cell.
Abbreviations
- HP1
heterochromatin protein 1
- SMC
structural maintenance of chromosomes
- FSHD
facioscapulohumeral muscular dystrophy
- H3K9me
H3 lysine 9 methylation
- H3K9me3
H3 lysine 9 tri-methylation
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/11683
References
- 1.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SCR. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophiola melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sims R, Jr, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–39. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 4.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 5.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–24. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- 7.Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–74. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 10.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–76. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Brit J Cancer. 2004;90:761–9. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Cur Opin Cell Biol. 2001;13:263–73. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat REv Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 15.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 17.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 19.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher IFJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 21.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–80. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by non-overlapping mechanisms. Mol Cell. 2008;32:778–90. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–88. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–34. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 28.Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–84. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 29.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–15. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 30.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Aucott R, Bullwinkel J, Yu Y, Shi W, Billur M, Brown JP, et al. HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J Cell Biol. 2008;183:597–606. doi: 10.1083/jcb.200804041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano A, Rodríguez-Corsino M, Losada A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLoS One. 2009;4:5118. doi: 10.1371/journal.pone.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard P, Maure J-F, Partridge JF, Genier S, Javerzat J-P, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–42. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 34.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SS, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 35.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–87. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 36.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–8. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 37.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–91. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 38.Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher F., Jr The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun. 2005;331:929–37. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Zeng W, de Greef JC, Chen Y-Y, Chien R, Kong X, Gregson HC, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–5. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 41.Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39 h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- 42.Hou F, Chu CW, Kong X, Yokomori K, Zou H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J Cell Biol. 2007;177:587–97. doi: 10.1083/jcb.200701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 44.Chadwick BP. Macrosatellite epigenetics: the two faces of DXZ4 and D4Z4. Chromosoma. 2009;118:675–81. doi: 10.1007/s00412-009-0233-5. [DOI] [PubMed] [Google Scholar]
- 45.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–45. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–6. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 49.Ball ARJ, Yokomori K. Revisiting the role of hetero-chromatin protein 1 in DNA repair. J Cell Biol. 2009;185:573–5. doi: 10.1083/jcb.200904033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with hetero-chromatin. Mol Cell. 2008;31:167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 51.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 52.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2945–50. [PubMed] [Google Scholar]
- 54.Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol. 2009;29:6335–40. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry. 2009;75:619–25. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- 56.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–38. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 57.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, et al. HP1 controls telomere capping, telomere elongation and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–76. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 58.Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, et al. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nuc Acids Res. 2009;37:68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–70. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 60.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 61.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–29. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]