Abstract
Small-molecule compounds approved for use as drugs may be “repurposed” for new indications and studied to determine the mechanisms of their beneficial and adverse effects. A comprehensive collection of all small-molecule drugs approved for human use would be invaluable for systematic repurposing across human diseases, particularly for rare and neglected diseases, for which the cost and time required for development of a new chemical entity are often prohibitive. Previous efforts to build such a comprehensive collection have been limited by the complexities, redundancies, and semantic inconsistencies of drug naming within and among regulatory agencies worldwide; a lack of clear conceptualization of what constitutes a drug; and a lack of access to physical samples. We report here the creation of a definitive, complete, and nonredundant list of all approved molecular entities as a freely available electronic resource and a physical collection of small molecules amenable to high-throughput screening.
Introduction
The sequencing of the human genome and subsequent translational efforts have brought about unprecedented opportunities for the rapid application of new biological knowledge to improve human health. While diagnostic applications of genomic information have been relatively straightforward to develop, advances in therapy have been slower, due in part to the time (10–15 years) and expense (~$1B) of new drug development (1).
New chemical entities (NCEs) are the focus of most drug development efforts, in part because of the need for novel composition of matter intellectual property to recoup the cost of drug research and development. However, the propensity of drugs to act on more than one target, or to act on their intended target in an unanticipated system, has long been noted to occur with regularity in clinical medicine, manifesting as either additional therapeutic uses for a drug, or adverse events. With the recent difficulties of the biopharmaceutical industry in developing NCEs, and the focus on drug safety, more attention has been placed on drugs already approved for clinical use. Nowhere has this attention been greater than in rare and neglected diseases, where the expected return on investment makes NCE development particularly challenging.
Rare diseases are defined by the U.S. Orphan Drug Act as those with <200,000 prevalence in the U.S.; while rare diseases are frequently neglected in drug development due to their low prevalence, the term “neglected” diseases generally refers to tropical diseases that may be highly prevalent but occur in developing nations unable to afford treatments (2). There are over 6000 rare and neglected diseases, of which fewer than 300 have any therapy currently available (http://rarediseases.info.nih.gov/). As a result, particular interest has arisen in finding drugs for rare and neglected diseases in the current pharmacopeia, by finding new therapeutic indications for already approved drugs – a process frequently referred to as “repurposing”.
A drug must be demonstrated to be reasonably safe and effective in the treatment of a disease or condition in order to receive regulatory approval (http://www.fda.gov/Drugs/; http://www.ema.europa.eu/). However, when used in larger populations, many drugs are subsequently discovered to have clinical utility or toxicity not appreciated at the time of approval. This can result in expansion of a drug’s clinical use to new indications (pregabalin) (3–5) or withdrawal (fenfluramine) (6, 7) of marketing authorization. Extension of the clinical use of a drug to a new indication has historically occurred via serendipitous clinical observation (e.g., sildenafil for erectile dysfunction), but more recently has occurred via logical connection of a disease’s pathophysiology to a drug’s target (e.g., losartan for Marfan syndrome, thalidomide for multiple myeloma) (8–10). These effects may be the result of the interaction of the drug on its intended target in a different organ (e.g., sildenafil) (11) or to the action of the drug on a different target (so-called “off-target” effects), and may be beneficial (e.g., imatinib action on c-kit) (12) or harmful (e.g., cisapride action on hERG) (13). Thalidomide (9, 10) was originally introduced in the late 1950s as a sedative drug and an effective antiemetic used widely to treat morning sickness. It was withdrawn later on due to teratogenicity and neuropathy. Recently there has been growing interests in thalidomide because it has been found effective against leprosy and multiple myeloma through inhibition of tumor necrosis factor alpha and angiogenesis. The US FDA approved the use of thalidomide for the treatment of lesions associated with Erythema Nodosum Leprosum (ENL) in 1998 and granted accelerated approval in 2006 for thalidomide in combination with dexamethasone for the treatment of newly diagnosed multiple myeloma (MM) patients. Studies are currently underway to determine the effect of thalidomide on arachnoiditis and several other types of cancers.
An alternative approach to repurposing, which does not require a priori knowledge of disease or drug mechanism, is screening drugs for activity in cell-based models of disease (e.g., ceftriaxone in ALS (14), astemizole in malaria (15)). These anecdotal successes raise the possibility that a substantial percentage of rare and neglected diseases (RND) might be treatable with drugs in the current pharmacopeia. Biopharmaceutical companies understandably have been less enthusiastic about testing their drugs for these indications, since if they are still covered by patents any adverse events in RND patients could adversely affect revenue, and if they are generic, the new RND indication would provide little financial return. Academic investigators or disease foundations may be interested in pursuing this approach, but frequently lack the infrastructure and expertise to do so, and even when successful the data are generally not aggregated with other assays or made public, preventing use of the data by others or cross-assay comparisons that would likely reveal important relationships among diseases and targets.
The NIH Chemical Genomics Center (NCGC) is a national resource for translation of the genome into biological insights and new therapeutics, particularly for rare and neglected diseases (16, 17). The NCGC collaborates with disease and target experts worldwide to develop chemical probes for novel biological and therapeutic systems, utilizing its assay development, quantitative high-throughput screening, informatics, and medicinal chemistry platforms (17). As part of its chemical genomics program aimed at understanding the features of small molecules that are important for biological activity, the NCGC has assembled a large and diverse collection of bioactive compounds. While most such compounds have been shown to be active only in cell-free, cell-based, or animal model systems, a small percentage have been tested in, or approved for, use in humans and animals. These latter compounds, for which we reserve the termed “drugs”, make up the class of bioactive small molecules useful for repurposing applications. A comprehensive understanding of the activities of all drugs in the pharmacopeia would facilitate the greatest possible application of known drugs across the full spectrum of human diseases, and help explain or predict their toxicities.
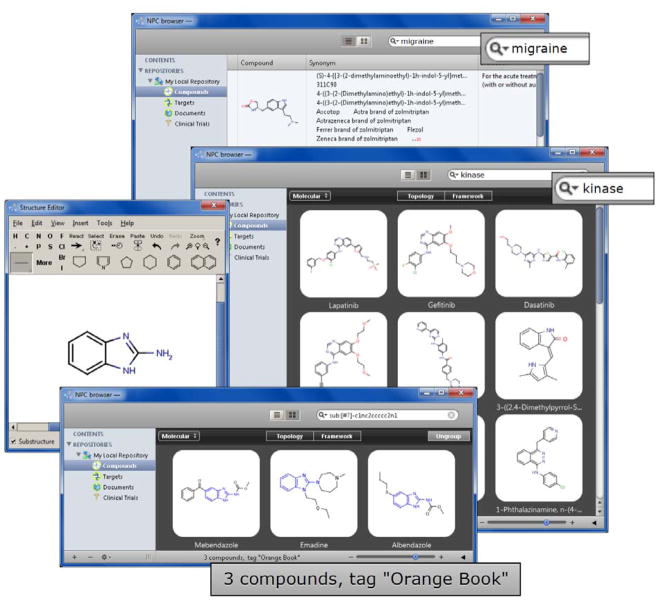
In order to enable this systematic drug mechanism and repurposing effort, we desired to identify and then acquire the complete, non-redundant collection of small molecule drugs approved for human or veterinary use by regulatory agencies worldwide. However, upon initiating this effort, it rapidly became evident that neither such a complete non-redundant list of drugs, nor such a physical collection of them, existed. Several attempts have been made to do so (18–22), but upon scrutiny all turned out to be incomplete and/or mixed approved drugs with bioactive compounds not approved for use in humans. We therefore resolved to assemble a definitive collection, and since this need only be done once and would be an enormous resource for the research and clinical communities, we resolved to make the information available through our website, and the collection available to collaborators who wish to bring their projects to the NCGC. This report describes our success in creating the NCGC Pharmaceutical Collection (NPC), a definitive collection of drugs registered or approved for use in humans or animals, as both an Informatics and a Screening Resource, and delineates our approach to profiling the activity of this collection across a broad array of human pathways and diseases. This ongoing project will benefit the medical, systems biology, and toxicology communities, via the NPC Informatics Resource browser (Figure 1) we describe here, available at http://tripod.nih.gov/npc/, and via the NCGC’s collaborative screening programs. Data on the activities of these drugs generated through screening of the NPC Screening Resource will be made publicly available through PubChem (23), with links provided on the NPC Informatics Resource browser.
Figure 1.
The NPC database browser (http://tripod.nih.gov/npc/) provides users with a graphical interface to explore drugs by a number of attributes including, but not limited to, name, structure, approval status, indication and target information. Complex queries implementing concurrent searches by multiple attributes are also available.
What is a “drug”?
Approval of a drug for human use is assumed to be an unambiguous event, so when we embarked on this comprehensive drug collection project, we were surprised to find that estimates of the number of approved drugs in the literature vary widely (15, 19, 24). The US Food and Drug Administration (FDA) does not maintain a single list of drugs that they have approved; instead, there are several such listings, each with its own particular legal origin, intent, and limitations. The United States Pharmacopeia-National Formulary (USP-NF), which is published by the non-governmental organization United States Pharmacopeia, is cited by the Federal Food, Drug, and Cosmetic Act of 1938 as the official compendium of the United States, but is incomplete (25). The FDA publishes several official lists of its own including: the Orange Book, the National Drug Code (NDC), the Drugs@FDA webpage, the Over-the-Counter (OTC) listings from the Office of Nonprescription Products, and its Substance Registration System’s Unique Ingredient Identifier (UNII) (26). An August 2006 Department of Health and Human Services, Office of Inspector General audit of the NDC found that over 14,000 drug products were missing from official government registries and that over 34,000 listed products were no longer being marketed (27). FDA has acknowledged these informatics shortcomings and has recently introduced an initiative to correct this issue (28, 29), but that work has not yet been completed. Several others have also attempted to generate a precise listing of FDA approved drugs (30–35). Comparing these resources, we discovered that none of these lists fully concur with one another, likely due to varying definitions of the term “drug”, the complexity of the regulatory process, and the complexity of FDA’s own publications. Research, medical, marketing, and lay communities use the word “drug” quite differently, and it is only in precisely defining the term did the number and identity of all substances to be included in our comprehensive collection become clear.
For the purpose of pharmacologic, mechanistic, and repurposing studies, the term “drug” refers to a molecular entity (ME)(36) that interacts with one or more molecular targets and effects a change in biological state. These may be small molecules, proteins, antibodies, or other substances such as siRNAs or aptamers. This term unambiguously denotes a unique molecule with a known structure.
The term “Active Pharmaceutical Ingredient” (API) refers to the molecule in physical form, and is more specific since different esters or salt forms of the same ME are designated as different APIs (e.g., paroxetine mesylate, paroxetine hydrochloride). Though different APIs of the same ME may exhibit distinct ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties in vivo, this distinction is rarely relevant for in vitro and in silico studies, so the NCGC Pharmaceutical Collection contains only one API for each ME.
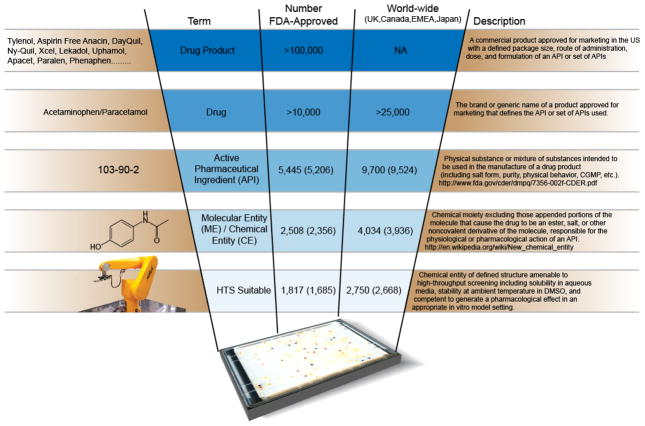
Next on the scale of lexical specificity (Figure 2) is the term ”Drug” as it is used by FDA, which is a name approved for marketing that defines an API, or set of APIs, used. Thus, a given API may be found in multiple drugs (e.g., ibuprofen, Motrin, Advil, Nuprin). This term includes drugs requiring a prescription and those that do not (OTC, or “over-the-counter”), and those covered by current market exclusivity (“brand”) and those that are not (“generic”), and may be small molecules, proteins, antibodies, or other substances or groups of substances. Also included in the category FDA (and other regulatory agencies) terms “Drugs” are substances or extracts without a defined molecular structure and products that are used for supportive or diagnostic purposes but are not intended to be used as specific modifiers of a particular disease or condition. These include thousands of allergenic extracts, commonly used intravenous fluids such as lactated Ringer’s solution and D5W (5% Dextrose in Water), oxygen, and purified air.
Figure 2.
Drug: What’s in a name? Definitions of the terms drug product, drug, API and ME and the numbers associated with each term. Record counts include veterinary drugs. Numbers of substances approved only for human use are shown in parentheses.
Finally, the most inclusive term for what FDA regulates is “Drug Product”. Most Drugs are marketed in a wide variety of dosages, forms (e.g., oral, intravenous, intramuscular), combinations, and packages, and each of these is referred to as a “Drug Product” by FDA.
These definitions make clear why there has been such confusion about what a “Drug” is and how many there are, and make possible the building of a truly comprehensive and non-redundant list and collection. What the lay public refers to as a “Drug” is more properly termed a “Drug Product”. In mechanistic research parlance, however, “Drug” refers to a Molecular Entity, or an API when the physical substance is being considered.
The example of Tylenol® illustrates the importance of these distinctions to properly classifying and counting drugs. N-(4-hydroxyphenyl)ethanamide, known as “acetaminophen” in the U.S., is the active pharmaceutical ingredient (API) in the common analgesic and antipyretic medicine Tylenol®. However, this API is also marketed under many other brand names, each registered as a separate Drug by the FDA. These include Aceta, Actimin, Anacin-3, Apacet, Aspirin Free Anacin, Atasol, Banesin, Crocin, Dapa, Dolo, Datril Extra-Strength, DayQuil, Depon & Depon Maximum, Feverall, Few Drops, Fibi, Fibi plus, Genapap, Genebs, Lekadol, Liquiprin, Lupocet, Neopap, Ny-Quil, Oraphen-PD, Panado, Panadol, Paralen, Phenaphen, Plicet, Redutemp, Snaplets-FR, Suppap, Tamen, Tapanol, Tempra, Valorin and Xcel. Each brand name also comes in different forms, such as tablet, capsule, liquid suspension, suppository, intravenous, and intramuscular form, and doses, and each of these is listed as a separate “Drug Product”. Outside the U.S., this API is known not as acetaminophen, but as paracetamol, and each paracetamol-containing product is also listed separately in regulatory databases. Thus worldwide, there are hundreds of different “Drugs” and “Drug Products” that appear separately in drug databases, but all are composed of or contain the same ME/API, N-(4-hydroxyphenyl)ethanamide. This circumstance exists for many APIs, essentially none of which is marketed under only one name in only one dose, form, or combination.
How many drugs are there? - Compiling complete and non-redundant lists of Drug Products, Drugs, APIs, and Molecular Entities
Assembling a comprehensive enumeration
Using the definitions above, each category was enumerated using data first from the FDA, then from regulatory agencies outside the US. In this step, inclusion and completeness was the goal, with redundancy to be eliminated later (see following section). Lists of drug names approved for human use were obtained from the FDA official publications including the Orange Book, the National Drug Code (NDC), the Drugs@FDA webpage, the Over-the-Counter (OTC) listings from the Office of Nonprescription Products, and its Substance Registration System’s Unique Ingredient Identifier (UNII). After assigning structures and Chemical Abstracts Service (CAS) numbers to these names wherever applicable and removing duplicate entries, we found that FDA has over 100,000 Drug Products registered. These Drug Products have in them over 10,000 Drugs. However, this latter number, though formally correct, is misleading since the majority of these 10,000 are different brands of the same API, different APIs of the same ME, or chemically undefined substances (e.g. allergenic extracts) (Supplementary Table 1).
We considered the possibility that additional Drugs might exist that are not listed by the FDA, if they were in use prior to the relevant statutes took effect in 1938 (37). We could find no evidence of such additional drugs, and FDA considers such drugs unlikely (38). FDA has made efforts to evaluate drugs in use prior to 1938, and has taken proactive steps to exclude them from the market if the science was found lacking, as was the case with ethyl nitrite (“sweet spirit of nitre”) (39, 40).
Two categories of compounds not currently approved for human use were then added to this enumeration due to their potential for human application: veterinary products listed in the Green Book, and drugs previously approved for human use but subsequently withdrawn from the market. Ivermectin is a broad-spectrum antiparasitic that was first approved for veterinary use, and subsequently repurposed for treatment of human helminthic diseases, particularly onchocerciasis (41). Thalidomide is a prominent example of a previously withdrawn drug being repurposed for another indication and re-approved (10). Drug withdrawal may occur either because FDA or another regulatory agency withdraws marketing approval (42, 43), or because the manufacturer voluntarily ceases production (e.g., mesoridazine); in the latter case, the drug may remain listed in regulatory publications. Several resources that list and monitor drug withdrawals (44–49) were included in the NPC. However, the designation “withdrawn” is not unambiguous, since drugs are frequently withdrawn for one indication while remaining approved for others, or withdrawn in one country/market but while remaining on the market in others. The veterinary drugs and drugs withdrawn for certain indications are labeled as such in the NPC browser (http://tripod.nih.gov/npc/).
Drugs are frequently approved in other countries but not approved by the U.S. FDA; we wished to capture these Drugs for the NPC as well. We therefore performed analogous definition of terms, enumeration of categories, and definition of structures for Drugs approved by regulatory agencies in other countries (Table 1). We obtained listings from the Dictionary of Medicines and Devices published by the U.K. National Health Service Information Authority (NHS), Health Canada’s (HC) Drug Products Database, the European Medicines Agency (EMEA), and an English translation of the Japanese Pharmacopeia, Fourteenth Edition.
Table 1.
| a) Data sources for approved drugs
| ||||
|---|---|---|---|---|
| Data Source | URL | Country | Agency | Type |
| Approved Drug Products (Orange Book) | http://www.fda.gov/cder/ob/ | USA | FDA | Human |
| FDA Office of Nonprescription Products (OTC Ingredient List) | http://www.fda.gov/Cder/Offices/OTC/Ingredient_List_A-C.pdf | USA | FDA | Human |
| FDA National Drug Code Directory (NDC) | http://www.fda.gov/cder/ndc/ | USA | FDA | Human |
| Drugs@FDA | http://www.accessdata.fda.gov/scripts/cder/drugsatfda/ | USA | FDA | Human |
| Approved Animal Drug Products (Green Book) | http://www.fda.gov/cvm/greenbook.html | USA | FDA | Vet |
| Dictionary of Medicines and Devices | http://www.dmd.nhs.uk/ | UK | NHS | Human |
| EPARs for authorised medical products for human use | http://www.emea.europa.eu/htms/human/epar/a.htm | EU | EMEA | Human |
| Human medicines - Orphan medicinal products | http://www.emea.europa.eu/htms/human/orphans/opinions.htm | EU | EMEA | Human |
| EPARs for authorised medical products veterinary use | http://www.emea.europa.eu/htms/vet/epar/eparintro.htm | EU | EMEA | Vet |
| Therapeutic Products Directorate | http://www.hc-sc.gc.ca/dhp-mps/prodpharma/databasdon/ | Canada | HC | Human |
| The Japanese Pharmacopeia, Fourteenth Edition | http://jpdb.nihs.go.jp/jp14e/monographs2.html | Japan | NHI | Human |
| The Japanese Pharmacopeia, Supplement I | http://jpdb.nihs.go.jp/jp14supp1e/ | Japan | NHI | Human |
| The Japanese Pharmacopeia, Supplement II | http://jpdb.nihs.go.jp/jp14supp2e/ | Japan | NHI | Human |
| The Japanese Pharmacopeia, Name Database | http://moldb.nihs.go.jp/jp/index.html | Japan | NHI | Human |
| b) Sources for other bioactive compounds tested in human
| |||
|---|---|---|---|
| Data Source | URL | Country | Agency |
| DEA Drug Scheduling | http://www.usdoj.gov/dea/pubs/scheduling.html | USA | DEA |
| World Health Organization International Nonproprietary Names (INN) | http://www.who.int/medicines/publications/druginformation/innlists/en/index.html | WHO | |
| United States Adopted Names (USAN) | http://www.ama-assn.org/ama/pub/about-ama/our-people/coalitions-consortiums/united-states-adopted-names-council/adopted-names.shtml | USA | AMA/USP/APhA |
| Harmonized Tariff Schedule of the United States | http://hotdocs.usitc.gov/docs/tata/hts/bychapter/0600PHARMAPPX.pdf | USA | U.S. ITC |
| FDAMDD: FDA Maximum (Recommended) Daily Dose | http://www.epa.gov/ncct/dsstox/sdf_fdamdd.html | USA | |
| KEGG Drug | http://www.genome.jp/kegg/drug/ | Japan/USA/Europe | |
While currently approved, or previously approved, drugs may be those most amenable to repurposing, compounds that have been registered for human testing but not necessarily approved by any regulatory agency, represent potentially attractive starting points for further testing and/or medicinal chemistry optimization, so were also included in the NPC. This category includes unapproved Drugs registered by the US Drug Enforcement Agency (DEA), compounds listed in the World Health Organization (WHO) International Nonproprietary Names (INN) and United States Adopted Names (USAN) registries, and compounds listed on the US tariff schedule (Table 1(b)). These listings include Drugs which have an approved Investigational New Drug (IND) application with the FDA or analogous approval by regulatory agencies outside the U.S., and those that are being tested or have been tested in clinical trials in humans. Importantly, inclusion in any of these latter registries does not indicate that the drug has in fact been tested in humans, much less the stage of testing (e.g., Phase I, II, III). The latter information is impossible to determine systematically since it is generally not disclosed by the company doing the trials; ClinicalTrials.gov, for instance, requires prospective registration of the trial but does not standardize disclosure of the API(s) being tested. While not immediately useful in repurposing applications, these USAN/INN drugs may be considered partially developed drugs and therefore require less effort to achieve regulatory approval than compounds in preclinical stages of drug development. Detailed information on the number of drug records obtained from each source can be found in Supplementary Table S1.
Generating a non-redundant list of molecular entities
Having produced an aggregate enumeration that was complete, we then devised a process to eliminate redundancy to arrive at a list that was non-redundant – i.e., in which each ME was represented only once. There are multiple mechanisms by which a single molecular entity may be listed more than once in drug listings, including by simple duplication within or between countries, or by listing of MEs as distinct APIs, Drugs, or Drug Products. Since this redundancy is the source of much of the confusion in the literature about how many MEs exist, this process of redundancy elimination is described here in some detail.
Different regulatory agencies often assign different names for the same API (e.g., paracetamol/acetaminophen), do not adhere to standard ways of listing active ingredients (e.g., terazosin, terazosin hydrochloride, and terazosin hydrochloride anhydrous are all used to refer to the same API); idiosyncratic and inconsistent naming made synonym identification difficult. Several heuristics were created to reliably eliminate redundancy. Since the only completely unambiguous identifier was chemical structure, but the regulatory agencies did not supply structural information, APIs were matched to chemical structures via names, synonyms, and/or CAS numbers. Structures were primarily derived from ChemIDPlus’s PubChem deposited substances (50), Prous’ PubChem deposited substances (search for “Prous Science Drugs of the Future”[SourceName] under PubChem Substances) (23), FDAMDD (51), commercial supplier catalog structures, SciFinder searches, or ChemOffice’s name-to-structure utility (52) if an IUPAC name was available, and were manually checked against existing literature including drug labels (53). CAS identifiers are included in the Japanese Pharmacopeia, and were also obtained from ChemIDPlus via PubChem, and manually from SciFinder. We relied on SciFinder to find the correct mapping from name to CAS and to structure once an inconsistency was identified. As a structure source, commercial supplier catalogs (Supplementary Tables S3 and S4) were found to be less reliable (i.e., more error prone) than ChemIDPlus and FDAMDD. An initial scan identified 1770 inconsistencies (different structures having the same name) from 12,800 vendor record indicating an error rate of approximately 14%. Based on the incorrect structures identified during the manual curation process, ChemIDPlus, of which we curated 12,300 records linked to approved drugs, appeared to be a reliable source for structures with an estimated error rate of 1.3% (155 errors found) and FDAMDD, with 990 of its 1,217 structures curated, was of similar quality with an error rate of 2.0% (20 errors found).
Many compound structures included salt and solvent. Common salts were removed, and remaining mixtures separated into component MEs, utilizing automated software followed by manual curation to verify the results. As structure representations of heavy metal containing compounds are frequently problematic, a special set of heuristics were applied to these drugs, where any fragment without a carbon or nitrogen and with less than 6 atoms was removed, and the rest of the molecule treated as one molecular entity. Structures were then canonicalized to facilitate molecular entity matching using the IUPAC International Chemical Identifier or InChI hash key (54) and a NCGC software package for structure standardization (http://tripod.nih.gov/?p=61) (55). To accomplish this, API records were first merged by canonical molecular entities, then by CAS number, and finally by names and synonyms. Each unique Molecular Entity was assigned a unique ID. If more than one unique ID shared the same CAS numbers or name, an alert flag was added indicating a potential error in the structure, name or CAS. Manual curation was then performed to correct such mistakes (Supplementary Table 2). Since a unique ID was assigned to each unique molecular entity, mixtures (i.e., drugs made up of multiple molecular entities) had more than one unique ID assigned.
Comparison to previous estimates of drug numbers
The NPC is the most comprehensive and accurate exposition to date of molecular entities registered or approved for human or veterinary use worldwide. There have been many previous efforts at compiling drug lists, but upon examination it was evident that all have suffered from substantial overcounting, undercounting, and/or misclassification. Much of the confusion derives from different definitions of what a “drug” is, the redundancy of drug name listings, the often opaque nature of regulatory agency databases, and lack of connection of drug names to unambiguous chemical identifiers such as structures or SMILES. In addition, the term “approved” has previously been used in different ways. A compound may be “approved” for listing in a database (such as the INN), “approved” for use in experimental settings only (as indicated by IND approval by FDA), “approved” by one or more regulatory agencies for specific clinical uses and marketing, or may have been previously “approved” but subsequently withdrawn from the market. Compounds in all of these categories have been included in previous listings of “approved” drugs, though only a fraction can actually be used in medical practice. In the NPC, “approved” means that marketing and use in medical practice, for the prevention or treatment of one or more disease indications, is currently allowed by one or more regulatory agencies worldwide.
These ambiguities, and the lack of methodological detail in many previous estimates of drug listings, make comparison to our results difficult. Previous estimates of the number of “FDA-approved” drugs for screening have ranged from 1382 (56) to 6534 (31) and of approved drugs worldwide up to 9990 (19). Scrutiny of these databases revealed undercounting of approved drugs, misdesignation of tested drugs as approved, and inclusion of the same ME more than once due to naming ambiguities. Finally, some reports have incorrectly indicated that compound listing in the USAN or INN indicates either entry into Phase II clinical trials or approval for regulatory use outside the U.S., greatly inflating the reported number of “approved” drugs available for repurposing. USAN/INN listing in fact only indicates registration by a sponsor of intention to file for human use at some point, but not approval for any such use (57).
How many “drugs” are there?
This rigorous process of definition, enumeration, and redundancy elimination allowed us to arrive at definitive numbers of MEs, APIs, Drugs, and Drug Products, approved for human and/or veterinary use, in the US and/or other countries; these are summarized in Figure 2 and listed in detail in Supplementary Table S1. Since for scientific purposes the term “drug” is most properly used to describe MEs, the most accurate answer to the question, “How many drugs are there?” is in reality “How many approved molecular entities are there?” The answer to this question is that 2356 MEs are approved for human use by the US FDA, and 3936 MEs are approved for human use in major markets worldwide including the US. These will be the richest source for repurposing applications, since they are already approved for human use and thus approval of new indications will be most straightforward.
When molecular entities approved for veterinary use are included, the numbers of MEs increase to 2508 FDA, and 4034 worldwide. Finally, there are 4935 unique molecular entities included in the USAN, INN, US Tariff schedule, WHO, DEA, KEGG drugs and FDAMDD database listings of compounds registered for experimental human use but not approved by any regulatory agency for marketing (Supplementary Table 1). While not immediately useable for human applications, these MEs would provide a “jump start” to new drug approvals, so are of interest and importance as well. Taken together, the 4034 unique worldwide approved MEs and 4935 unique worldwide registered MEs sum to a total unique 8969 molecular entities which are represent the universe of compounds for repurposing, advanced drug development, and chemical genomics.
The NPC Informatics Resource
From these listings, we created the NPC Informatics Resource, available at http://tripod.nih.gov/npc/. The NPC Informatics Resource lists all drug MEs and APIs, whether they are suitable for laboratory-based screening or not, and enables informatics-based rational repurposing and chemical genomics applications. All APIs corresponding to these MEs are also included. Currently, the NPC Informatics Resource numbers 2508 FDA approved MEs and 4034 worldwide approved MEs. Including the 4935 unique MEs from the USAN and INN registries, the NPC Informatics Resource contains 8969 unique MEs. Information will be periodically updated as curation proceeds, new MEs are added as they are registered or approved, and errors are found. This process will benefit enormously from community feedback, and we users to utilize the error report mechanism on the site (http://tripod.nih.gov/npc/). Database updates with new records and error fixes will be released periodically with distinct database version numbers.
The NPC Screening Resource
Laboratory-based high-throughput screening (HTS) places certain constraints on the types of molecules that can be tested; therefore, a subset of the NPC Informatics Resource, termed the NPC Screening Resource, was created for HTS applications. The NPC Screening Resource excludes large molecules (e.g., proteins, antibodies, >1500 MW), as well as small molecule that are insoluble in DMSO, unstable at room temperature, have less than 16 atoms, or have no carbon or nitrogen atoms. Since different salt forms of the same ME behave similarly in in vitro assays, only one API corresponding to each ME was included. The APIs suitable and not suitable for HTS are labeled accordingly in the NPC Informatics Resource (http://tripod.nih.gov/npc/). The NPC Screening Resource listing numbers 2750 worldwide approved MEs (including 1817 FDA approved MEs) and 4881 USAN/INN MEs, for a total of 7631 MEs (see http://tripod.nih.gov/npc/ and Supplementary Table S1).
Acquisition of all approved drugs
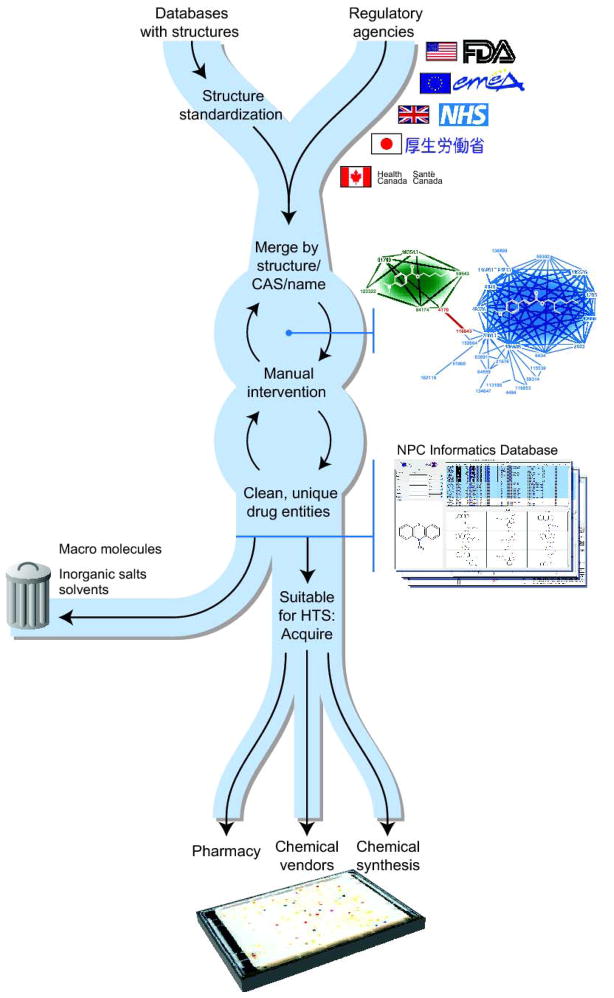
Acquisition of physical samples of the 2750 worldwide approved ME/APIs on the NPC Screening Resource list was surprisingly challenging, principally because chemical vendors generally list their inventory by structure, IUPAC name, or CAS number, and none of this information is routinely available from the regulatory agencies. Therefore, we used intermediary data sources to individually connect ME/APIs with vendor entries (Figure 3). In addition, different vendors frequently represented a given ME with different structures, so software was written to detect discrepancies and resolve them automatically whenever possible. When resolving ambiguities, ChemIDPlus (http://chem.sis.nlm.nih.gov/chemidplus) was particularly accurate and useful.
Figure 3.
Workflow for the NPC library construction process. This process is convoluted by the high prevalence of mistakes in the data sources. Octylmethoxycinnamate is shown as a semantic web diagram as an example. In the diagram, each node represents one entry from a data source. Lines represent identity relationships between source nodes, matching either on 1) name 2) CAS or 3) chemical structure. The line in red represents a spurious synonym linkage (Escalol 506) between padimate A and octinoxate.
Compound acquisition was prioritized by approval status, ease of acquisition, and cost. Currently approved drugs were assigned a higher priority for acquisition than investigational drugs, and drugs registered in the U.S. were assigned a higher priority than drugs registered in other countries. Drugs were procured from commercial bioactive compound collections (e.g. LOPAC) and bulk chemical suppliers (e.g. Sigma) first, from which large numbers of compounds were available at relatively low cost, with structures provided for all compounds. Procurement aggregators such as ChemNavigator and specialty chemical vendors (Supplementary Table S4) that generally supply compounds at higher costs were utilized next. If no commercial supplier could be found for a drug, the drug product was obtained from pharmacies and the API purified. For compounds not available commercially, drugs were custom synthesized either by NCGC chemists or via outsourcing; cost for custom synthesis depended on structural complexity and number of synthetic steps, and ranged from $1,000 to $40,000 for 100 mg of compound.
The current acquisition status of the NPC Screening Resource is summarized in Table 2. The majority (64%) of the NPC Screening Resource approved drugs, totaling 1767 compounds, were obtained from major suppliers, including Sigma-Aldrich (St. Louis, MO), Tocris Bioscience (Ellisville, Missouri), MicroSource Discovery Systems (Gaylordsville, CT), Enzo Life Sciences International, Inc. (Formerly BIOMOL International, L.P., Plymouth Meeting, PA), Prestwick Chemical (Illkirch, France), the United States Pharmacopeia (USP), the National Institute on Drug Abuse (NIDA), and and the National Cancer Institute (NCI) (Supplementary Table 3). Controlled substances were mainly procured from the NIDA and Sigma, after licensing of the NCGC by the U.S. Drug Enforcement Administration (DEA). These suppliers were willing to provide compounds in 96-well plate or 96-tube rack formats, making them also the easiest to prepare for screening. Approximately 15% of the collection (404 compounds) were sourced as individual compounds from over 70 smaller chemical suppliers (Supplementary Table 4), either directly from the supplier or through procurement aggregators such as ChemNavigator (San Diego, CA). This was a time-intensive process, requiring the iterative manual compilation of a master list of compound names and structures. Continual changes in vendor offerings led us to create a custom structure comparison tool (MolOverlap v1.0), which compares SD files from all vendors to a master list of structures, and outputs a text file of matching structures to be obtained; this tool is freely available at http://tripod.nih.gov/moloverlap/. This allowed us to rapidly, accurately, and ongoingly extract updated catalog items and procure them for the collection. APIs not commercially available were sourced as drugs from pharmacies and purified. Approximately 21% of the collection (579 compounds) were not available from any vendor, so required custom synthesis, either by NCGC chemists or via contract synthesis. As of this publication, syntheses have completed for 220 of these compounds; synthesis of the remaining 359 will be completed over the next six months.
Table 2.
Procurement status of NPC.
| Drug Source | In current collection | Procurement/synthesis in process | Total |
|---|---|---|---|
| US FDA* | 1635 | 182 | 1817 |
| UK/EU/Canada/Japan* | 756 | 177 | 933 |
| Total Approved* | 2391 | 359 | 2750 |
| Testing Only | 928 | 3953 | 4881 |
| Total | 3319 | 4312 | 7631 |
These counts include approved veterinary drugs.
Acquisition of compounds registered but not approved for human use
Of the 4881 MEs identified as appropriate for inclusion in the NPC Screening Resource that are registered only by the World Health Organization (WHO) International Nonproprietary Names (INN), and the United States Adopted Names (USAN), or compounds listed on the US tariff schedule, only a small proportion are available commercially. Currently 928 (19%) of these compounds have been procured or synthesized. Approximately 20% of the remaining MEs are obtainable from chemical vendors, and the remaining 60%, totaling nearly 3000 compounds, will require synthesis at the NCGC or via contract synthesis (Table 2). Given the cost and time required for custom synthesis, we expect the expansion of the NPC Screening Resource to include all registered MEs will be a long-term effort, but a critically important one. When considering starting points for chemical optimization for a new drug, these 3000 compounds may be considered advanced leads with likely attractive activity, physicochemical, and ADME properties, which may therefore allow considerable time saving compared to leads generated from conventional HTS of diversity collections.
Recently, we have begun actively procuring or synthesizing drugs approved in countries other than the US, UK, Europe, Canada and Japan, and active metabolites of approved drugs. The building of the NPC Informatics and Screening resources will be ongoing, and as new compounds are added to regulatory databases, and physical samples are obtained, the Informatics and Screening Resource pages will be updated on our website. For immediate use by the community, we have listed all compounds in the NPC Informatics and Screening Resources by regulatory agency and supplier in Figure 4 and Table S4, and at the NPC website, http://tripod.nih.gov/npc/.
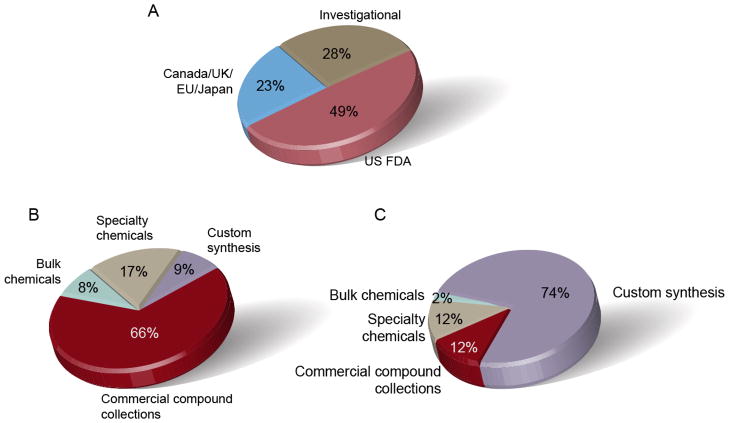
Figure 4.
The composition of NPC by a) regulatory agency, b) supplier type and c) sample cost. If a drug is listed by more than one regulatory agency, it is counted only once following the approval status priority rank: 1. US FDA, 2. UK/Canada/EU/Japan, 3. investigational.
QC of the Screening Resource
Ensuring the correct identity and purity of compounds in screening collections is a critical aspect of drawing reliable conclusions from HTS data (58). This is particularly critical for the NPC, since the data generated on these compounds may be used to advance new clinical applications and draw conclusions about the universe of targets affected by clinically approved drugs, and will be made publically available. Therefore though we have received suppliers’ Certificates of Analysis, each sample has also been subjected to independent QC at the NCGC to ensure identity and >90% purity by LC/MS. Three types of detectors are used for the analysis. The primary analytical technique for assessing compound identity is mass spectrometry. Identification is based on the expected nominal mass being detected. The primary technique for assessing analytical purity is an Evaporative Light Scattering Detection (ELSD). The secondary technique for accessing compound purity is UV absorbance at a wavelength of 220 nanometers. UV detection becomes important for samples which give a poor response to the ELSD (58).
Applications
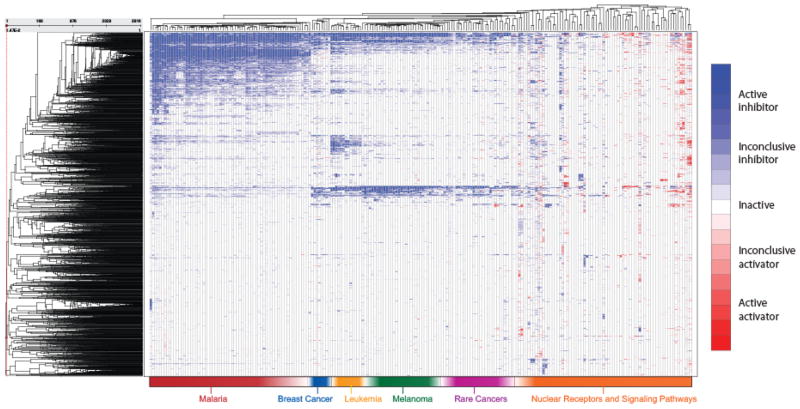
The NCGC Pharmaceutical Collection Screening Resource has thus far been screened against over 200 assays of targets, pathways, and cellular phenotypes (Figure 5). All screening of the NPC is done using the NCGC’s Quantitative High Throughput Screening (qHTS) paradigm, wherein every drug is screened at 6 or more concentrations over 4–5 orders of magnitude in the primary screen (59). The percentage of NPC compounds with activity in the assays screened so far averages 4.2% (hits classified according to (59)), above the average rate of 1.8% in assays across the NCGC’s larger screening collection (principally the Molecular Libraries Small Molecule Repository (MLSMR)), consistent with the notion that “bioactive” chemical structures frequently have multiple activities that may not be predictable a priori. Importantly, the assays against which the NPC has been screened have a wide diversity of formats and readouts, eliminating the possibility of this high hit rate being due to an assay platform artifact (60, 61). Though beyond the purview of this paper, the individual and aggregate assay screening data generated using the NPC will be of great interest for repurposing and chemical genomics, and will be published (e.g., (62) and (63)) and made publicly available via the NPC Browser (http://tripod.nih.gov/npc/) and PubChem (23).
Figure 5.
Activities of the NPC screened against approximately 200 assays of targets, pathways, and cellular phenotypes. The heat map, where each row represents a drug and each column an assay, is colored by the drug activity observed (type of concentration response curve produced) such that activation is colored red, inhibition blue, inactive white, and missing data is colored grey. A darker shade of red or blue indicates more conclusive activity (significant concentration response).
The NPC is currently being utilized for three principal purposes: drug repurposing for the treatment of rare and neglected diseases; defining the universe of pathway activities of known drugs for improved toxicological understanding, modeling and prediction; and defining characteristics of small molecule compounds that confer biological activity.
Repurposing
Expansion of a drug’s use to diseases other than that for which it was originally intended is commonly referred to as “repurposing”. While many individual examples exist of successful repurposing, only recently has the concept of large-scale, even comprehensive, examination of the disease applications of clinically used drugs been considered (19, 20). In order to be maximally reliable and useful, the collection being screened must be truly comprehensive, the screening paradigm must minimize false positives and false negatives, confirmatory testing should be done, and the data should be made publically available. The substantial infrastructure and diverse disease expertise required for such an effort has until now prevented comprehensive repurposing from being implemented. The NPC, in the context of the collaborative mission of the NCGC and the NIH Therapeutics for Rare and Neglected Diseases (TRND) program (64) makes this comprehensive approach to drug repurposing feasible, and the enormous unmet medical need – over 6000 rare and neglected diseases currently have no treatment – makes it urgent. Such repurposing will not only provide the possibility of rapid therapeutic advances, but also obviate the need for NME development, a long and expensive process. Ultimately, application of the NPC to a large number of diseases will help determine the proportion of human diseases are amelioratable by a drug in the current pharmacopeia; this question has both theoretical and practical importance, informing questions of common disease mechanisms and helping determine the scope of the problem of therapeutic development of the thousands of diseases currently without treatment.
Virtual screening of the NPC Informatics Resource can be performed by any investigator worldwide with an internet connection, and we encourage researchers worldwide to do so. To enable the research community and build the knowledge base of drug activities, we encourage researchers to inform us of their successes (and failures) using the Resource, and contribute their results to PubChem. Laboratory-based screening of the NPC Screening Resource is done at the NCGC via collaboration with any researcher who has a disease-relevant assay. The screening requirements for the NPC are much less demanding than a typical HTS campaign given the small number of compounds (3,500, compared to >350,000 for a typical HTS); in our experience the assays that produce results most directly applicable to clinical applications utilize primary patient cells. We encourage any researcher to contact us with their interest. Given the expense of building and maintaining the NPC Screening Resource, we cannot send copies of the collection to collaborators (100 screens can be performed at the NCGC with the amount of compound required to send to one collaborator), but we routinely have collaborators bring their assays to the NCGC, or send them to us, for collaborative screening. A solicitation for development and screening of rare/neglected disease assays for repurposing applications will be released by the TRND program shortly (http://trnd.nih.gov). For those researchers who prefer to reproduce all or part of the NPC Screening Resource in their own laboratories, information on suppliers of all compounds can be found in Table S4 and on our website at http://tripod.nih.gov/npc/.
Toxicology
While unanticipated biological activities of known drugs may be therapeutically beneficial for repurposing, those activities may also be responsible for unanticipated toxicological effects of drugs. Drug toxicity is one the major reason for failure of new drug development programs (65), and approved drugs are regularly removed from the market because of adverse effects; frequently, the mechanism by which the toxicity occurs is not known. To improve the reliability and mechanistic understanding of toxicity of chemicals, the NPC will be screened across a very broad range of pathways and cellular phenotypes relevant to toxicity as part of the Tox21 program, a collaboration between the NCGC, the National Toxicology Program, the US Environmental Protection Agency, and the US Food and Drug Administration (66).
Chemical Genomics
Ultimately, improvements in the efficiency of drug development and application to disease will rely on improved understanding, and therefore predictability, of the general principles by which small molecules interact with their biological targets. This long-term goal will be greatly advanced by the broad and rigorous profiling to which the NPC will be subjected. All data generated by the NPC will be placed into the NCGC’s publically available relational browser (NPC browser v2.0.0) (67) that will allow relationships between targets, pathways, diseases, and drugs to be queried in a user-defined fashion. This browser is currently available at http://tripod.nih.gov/npc/, and improvements will be made regularly in data richness and analysis capabilities.
A National Collaborative Resource
The creation of the NCGC Pharmaceutical Collection as a definitive informatics and screening resource is an important milestone, but is only the first step in its utilization for repurposing and chemical genomics. We hope that the enumeration of all drugs registered and/or approved for human and veterinary use, and the creation of laboratory resource for their screening, will allow effort to turn to the more important questions of the Resources’ scientific and medical applications. The NPC will only achieve its potential as a community resource, since the scientific and medical problems to which it can be applied will require the full breadth of target, pathway, and disease expertise. The NPC is intended as a collaborative instrument, and we encourage researchers to utilize the NPC interactively via our website, and via screening projects. All NCGC programs are partnerships, and the Center currently has over 200 collaborations with investigators worldwide.
Having provided what to our knowledge is a definitive listing of drugs intended or approved for human use, we hope that the chemical genomics and drug development communities will utilize the NPC to realize the full potential of these drugs for human health, addressing the many devastating and untreatable diseases for which therapeutics are so urgently needed.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Program of the National Human Genome Research Institute, National Institutes of Health. We thank in particular Paul Loebach at FDA, Colonel Colin Ohrt at Walter Reed Army Medical Center, Hari Singh at NIDA, Jill Heemskerk at NINDS, David Sullivan at Johns Hopkins University, Stephen White at NCI, Doug Livingston at Galapagos, Gopal Potti and Robert DeChristoforo at the NIH Clinical Center pharmacy for help with source lists of approved drugs and discussions on procurement, Mike Philippi and Peggy McClelland for help with procurement, Bill Leister for QC, Craig Thomas for compound synthesis consultation and Darryl Leja for illustration.
Footnotes
Author contributions
R.H. and N.S. coordinated the project, sourced and compiled drug lists to construct the NPC, helped to build the NPC database and browser, helped with the NPC procurement, and wrote the manuscript; P.S. and A.Y. helped to find drug sources, procured compounds for the NPC, and helped to write the manuscript; Y.W. built the NPC database and browser; D.-T.N. helped to build the NPC database and browser; C.P.A. conceived and directed the project, and wrote the manuscript.
Reference and Notes
- 1.DiMasi JA, Grabowski HG. The Cost of Biopharmaceutical R&D: Is Biotech Different? Manage and Decis Econ. 2007;28:469. [Google Scholar]
- 2.Feasey N, Wansbrough-Jones M, Mabey DCW, Solomon AW. Neglected tropical diseases. Br Med Bull. 2010;93:179. doi: 10.1093/bmb/ldp046. [DOI] [PubMed] [Google Scholar]
- 3.Blommel ML, Blommel AL. Pregabalin: an antiepileptic agent useful for neuropathic pain. American Journal of Health-System Pharmacy. 2007;64:1475. doi: 10.2146/ajhp060371. [DOI] [PubMed] [Google Scholar]
- 4.Mease PJ, et al. A randomized, double-blind, placebo-controlled, phase III trial of pregabalin in the treatment of patients with fibromyalgia. Journal of Rheumatology. 2008;35:502. [PubMed] [Google Scholar]
- 5.Spaeth M. Is pregabalin a safe and effective treatment for patients with fibromyalgia? Nature Clinical Practice Rheumatology. 2008;4:514. doi: 10.1038/ncprheum0906. [DOI] [PubMed] [Google Scholar]
- 6.Curzon G, Gibson EL. The serotonergic appetite suppressant fenfluramine: Reappraisal and rejection. Advances in Experimental Medicine and Biology. 1999;467:95. doi: 10.1007/978-1-4615-4709-9_13. [DOI] [PubMed] [Google Scholar]
- 7.Ioannides-Demos LL, Proietto J, Tonkin AM, McNeil JJ. Safety of drug therapies used for weight loss and treatment of obesity. Drug Safety. 2006;29:277. doi: 10.2165/00002018-200629040-00001. [DOI] [PubMed] [Google Scholar]
- 8.Lacro RV, et al. Rationale and design of a randomized clinical trial of b-blocker therapy (atenolol) versus angiotensin II receptor blocker therapy (losartan) in individuals with Marfan syndrome. American Heart Journal. 2007;154:624. doi: 10.1016/j.ahj.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito T, et al. Identification of a Primary Target of Thalidomide Teratogenicity. Science (Washington, DC, United States) 2010;327:1345. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson JM. Thalidomide: a remarkable comeback. Expert Opinion on Pharmacotherapy. 2000;1:849. doi: 10.1517/14656566.1.4.849. [DOI] [PubMed] [Google Scholar]
- 11.Ghofrani HA, Osterloh IH, Grimminger F. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nature Reviews Drug Discovery. 2006;5:689. doi: 10.1038/nrd2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves Patrick M, et al. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med. 2005;11:731. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- 13.Griffin JP. Prepulsid withdrawn from UK & US markets. Adverse drug reactions and toxicological reviews. 2000;19:177. [PubMed] [Google Scholar]
- 14.Rothstein JD, et al. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005 Jan 6;433:73. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 15.Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ., Jr A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006 Aug;2:415. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 16.Austin CP. The impact of the completed human genome sequence on the development of novel therapeutics for human disease. Annu Rev Med. 2004;55:1. doi: 10.1146/annurev.med.55.091902.104426. [DOI] [PubMed] [Google Scholar]
- 17.Thomas CJ, et al. The pilot phase of the NIH Chemical Genomics Center. Curr Top Med Chem. 2009;9:1181. doi: 10.2174/156802609789753644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spectrum, The Spectrum Collectionhttp://www.msdiscovery.com/spectrum.html
- 19.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007 Aug 9;448:645. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 20.Heemskerk J. Screening existing drugs for neurodegeneration: The National Institute of Neurologic Disorders and Stroke (NINDS) model. Retina. 2005 Dec;25:S56. doi: 10.1097/00006982-200512001-00025. [DOI] [PubMed] [Google Scholar]
- 21.NCC, NIH Clinical Collectionhttp://www.nihclinicalcollection.com/(2008)
- 22.LOPAC1280, Library of Pharmacologically Active Compoundshttp://www.sigmaaldrich.com/chemistry/drug-discovery/validation-libraries/lopac1280-navigator.html
- 23.PubChem. 2009 http://www.ncbi.nlm.nih.gov/sites/entrez.
- 24.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009 Nov 12;462:175. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya L, et al. The value of USP public standards for therapeutic products. Pharm Res. 2004 Oct;21:1725. doi: 10.1023/b:pham.0000045222.01170.4c. [DOI] [PubMed] [Google Scholar]
- 26.FDA, The Orange Book is composed of approved prescription drugs, approved OTC drug products for those drugs that may not be marketed without New Drug Applications (NDAs) or Abbreviated New Drug Application (ANDAs) because they are not covered under existing OTC monographs, drug products with approval under Section 505 of the Act administered by the Center for Biologics Evaluation and Research, and finally an accumulative list of approved products that have never been marketed, are for exportation or military use, have been discontinued from marketing, or have had their approvals withdrawn for reasons other than safety or efficacy subsequent to being discontinued from marketing. The National Drug Code (NDC) lists all drugs manufactured, prepared, propagated, compounded, or processed for commercial distribution. Currently, properly listed marketed prescription drug products and insulin are published into the NDC Directory. Drugs@FDA contains prescription and over-the-counter human drugs and therapeutic biologicals currently approved for sale in the United States, and also includes discontinued drugs and new indication approvalshttp://www.accessdata.fda.gov/Scripts/cder/DrugsatFDA/).Drugs@FDA does not include OTC products approved for marketing through a process other than submission of a NDA or Biologic License Application. The OTC Ingredient List contains a listing of the ingredients considered in the OTC Drug Review that were classified and published in various Federal Register publications. This list includes over 2,700 ingredients (records) that provide a classification history throughout the rulemaking process. The UNII is a non- proprietary, free, unique, unambiguous, non semantic, alphanumeric identifier based on a substance’s molecular structure and/or descriptive information used in the generation of structured product labels, among other things. The Facts@FDA site provides up-to-date labeling information using the XML-based Structured Product Labeling format. While there was hope that labeling for all approved prescription drugs would be available by the end of 2006, labels on many older medications are still not available (e.g. terfenadine). The FDA’s substance registry system Unique Ingredient Identifier is integrated into the Structured Product Labeling process, and currently lists over 11,000 entries. Unfortunately, the service has multiple entries for the same molecular entity (for example terazosin has three UNIIs for terazosin, terazosin hydrochloride, and terazosin hydrochloride anhydrous even though terazosin hydrochloride is the only approved formulation of this compound) and contains inactive ingredients (microcrystalline cellulose) as well as ingredients not yet approved by the FDA (zosuquidar)
- 27.Levinson DR. O o I G Health and Human Services. OEI-06-05-00060 2006. [Google Scholar]
- 28.Kim S, Linehan J. Science Board Subcommittee Review of Information Technology. 2009 August; http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/ScienceBoardtotheFoodandDrugAdministration/UCM200667.pdf.
- 29.FDA. Substance Registration System - Unique Ingredient Identifier (UNII) http://www.fda.gov/ForIndustry/DataStandards/SubstanceRegistrationSystem-UniqueIngredientIdentifierUNII/default.htm.
- 30.Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36:D901. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leadscope, Leadscope Known Drugs Database. http://www.leadscope.com/db_kd_product.php.
- 32.GVKBIO, GVKBIO Drug Database. http://www.gvkbio.com/database_pdf/Drug.pdf.
- 33.Drugs.com. http://www.drugs.com/
- 34.NLM, Drug Information Portal. http://druginfo.nlm.nih.gov/drugportal/drugportal.jsp.
- 35.RxNorm. http://www.nlm.nih.gov/research/umls/rxnorm/
- 36.FDA 21 C.F.R. § 314.108 (2010) defines an Active moiety as the molecule or ion, excluding those appended portions of the molecule that cause the drug to be an ester, salt (including a salt with hydrogen or coordination bonds), or other noncovalent derivative (such as a complex, chelate, or clathrate) of the molecule, responsible for the physiological or pharmacological action of the drug substance. In the same section, a new chemical entity is defined as a drug that contains a new active moiety. However, new molecular entity is defined on the Drugs@FDA website (http://www.fda.gov/Drugs/informationondrugs/ucm079436.htm) as an active ingredient that has never before been marketed in the United States in any form – which is ambiguous with respect to whether different forms of the same active moiety are considered the same active ingredient. In the present manuscript, we have opted to interpret a ME as equivalent to active moiety as stated in the federal code.
- 37.USC, The Federal Food, Drug, and Cosmetic (FDC) Act of 1938, Kefauver-Harris Drug Amendments to the FDC Act of 1962.
- 38.FDA. CPG Sec. 440.100 Marketed New Drugs Without Approved NDAs and ANDAs. http://www.fda.gov/ICECI/ComplianceManuals/CompliancePolicyGuidanceManual/ucm074382.htm.
- 39.FDA. Rulemaking History for OTC Sweet Spirits of Nitre Drug Products. http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/Over-the-CounterOTCDrugs/StatusofOTCRulemakings/ucm072140.htm.
- 40.NRC. The Drug Efficacy Study of the National Research Council’s Division of Medical Sciences, 1966–1969. http://www7.nationalacademies.org/archives/drugefficacy.html.
- 41.Omura S. Ivermectin: 25 years and still going strong. Int J Antimicrob Agents. 2008 Feb;31:91. doi: 10.1016/j.ijantimicag.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 42.FDA. List of Drug Products That Have Been Withdrawn or Removed From the Market for Reasons of Safety or Effectiveness. http://www.fda.gov/ohrms/dockets/98fr/100898b.txt.
- 43.USC, 21 C.F.R. PART 216.24. Drug products withdrawn or removed from the market for reasons of safety or effectiveness. http://law.justia.com/us/cfr/title21/21-4.0.1.1.11.html
- 44.Wikipedia. Category:Withdrawn drugs. http://en.wikipedia.org/wiki/Category:Withdrawn_drugs.
- 45.Wikipedia. List of withdrawn drugs. http://en.wikipedia.org/wiki/List_of_withdrawn_drugs.
- 46.Coleman V. Betrayal of Trust. http://www.vernoncoleman.com/downloads/betrayal.htm.
- 47.Lexchin J. Drug withdrawals from the Canadian market for safety reasons, 1963–2004. CMAJ. 2005 Mar 15;172:765. doi: 10.1503/cmaj.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.EMEA. Withdrawals and Refusals. http://www.ema.europa.eu/withdraw.htm.
- 49.Wysowski DK, Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969–2002: the importance of reporting suspected reactions. Arch Intern Med. 2005 Jun 27;165:1363. doi: 10.1001/archinte.165.12.1363. [DOI] [PubMed] [Google Scholar]
- 50.ChemIDplus. 2006 http://www.ncbi.nlm.nih.gov/sites/entrez?db=pcsubstance&term=chemidplus.
- 51.FDAMDD. FDA Maximum (Recommended) Daily Dose. 2006 http://www.epa.gov/ncct/dsstox/sdf_fdamdd.html.
- 52.ChemOffice. 2007 http://www.cambridgesoft.com/software/ChemDraw/
- 53.DailyMed. 2009 http://dailymed.nlm.nih.gov/dailymed/about.cfm.
- 54.InChi. 2009 http://www.iupac.org/inchi/release102.html.
- 55.ChemAxon. 2009 http://www.chemaxon.com/
- 56.DrugBank. DrugBank. http://www.drugbank.ca/
- 57.WHO. Guidance on INN. http://www.who.int/medicines/services/inn/innquidance/en/index.html.
- 58.MacArthur R, et al. Monitoring compound integrity with cytochrome P450 assays and qHTS. J Biomol Screen. 2009 Jun;14:538. doi: 10.1177/1087057109336954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006 Aug 1;103:11473. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auld DS, et al. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008 Apr 24;51:2372. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 61.Simeonov A, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008 Apr 24;51:2363. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 62.Miller SC, et al. Identification of known drugs that act as inhibitors of NF-kappaB signaling and their mechanism of action. Biochem Pharmacol. 2010 May 1;79:1272. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shukla SJ, et al. Identification of clinically used drugs that activate pregnane X receptors. Drug Metab Dispos. 2010 Jan;39:151. doi: 10.1124/dmd.110.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.TRND, Therapeutics for Rare and Neglected Diseases. http://rarediseases.info.nih.gov/TRND/
- 65.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004 Aug;3:711. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 66.Kavlock RJ, Austin CP, Tice RR. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009 Apr;29:485. doi: 10.1111/j.1539-6924.2008.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ngyuen D. NCGC Pharmaceutical Collection. 2011 http://tripod.nih.gov/npc/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.