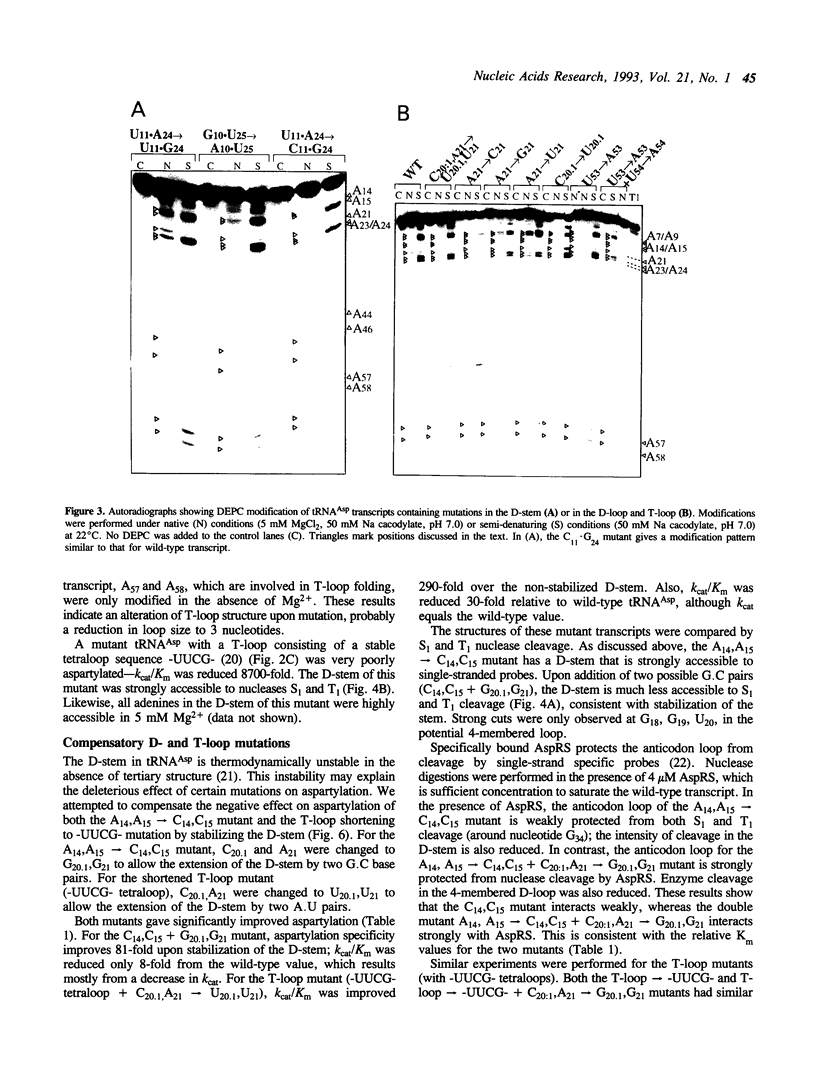
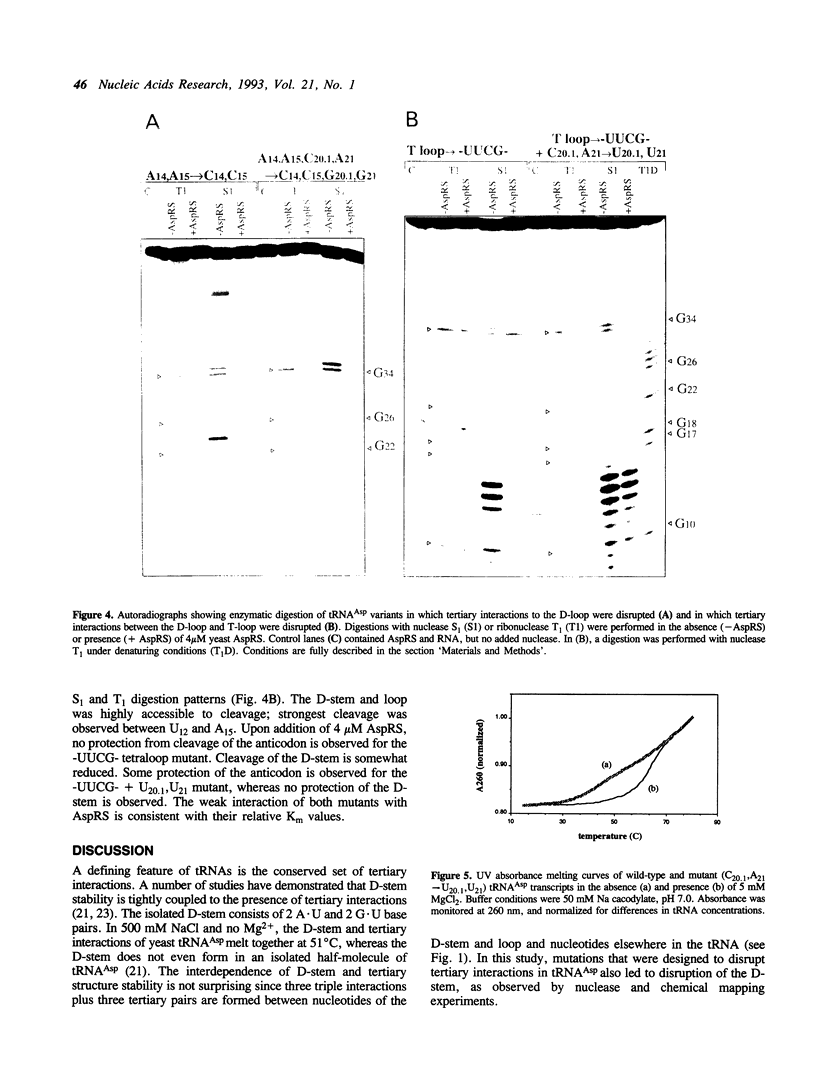
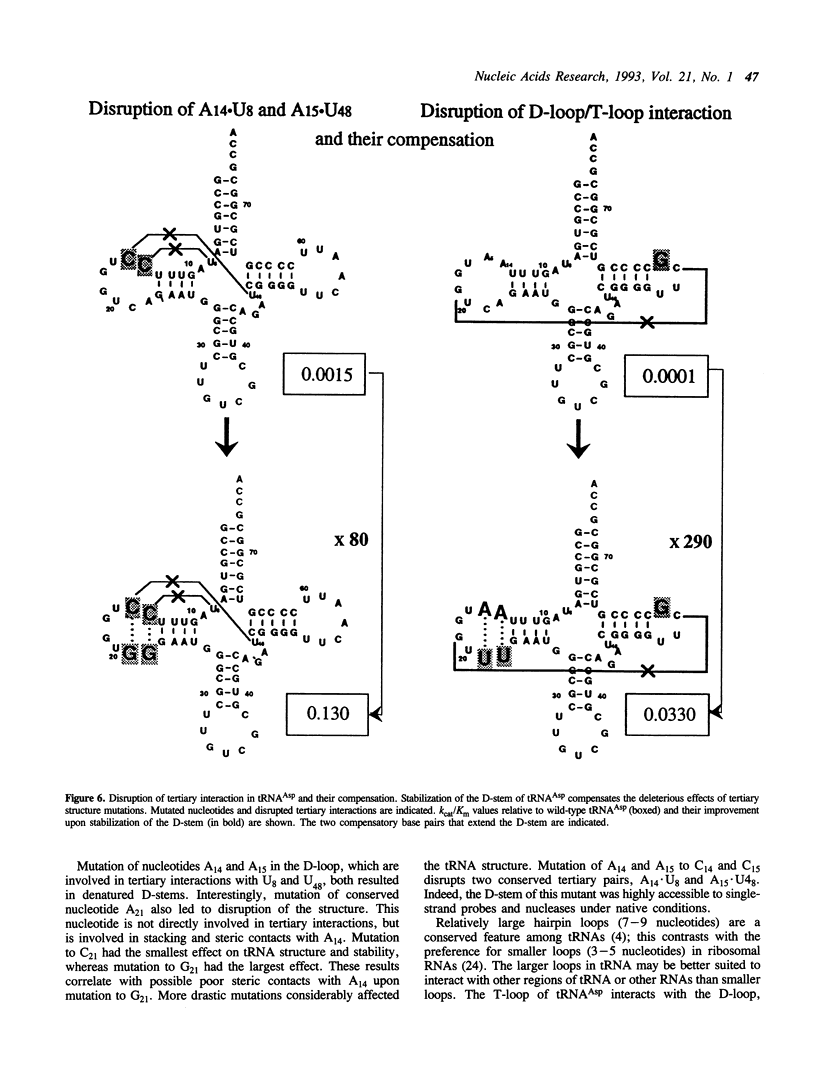
Abstract
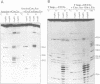
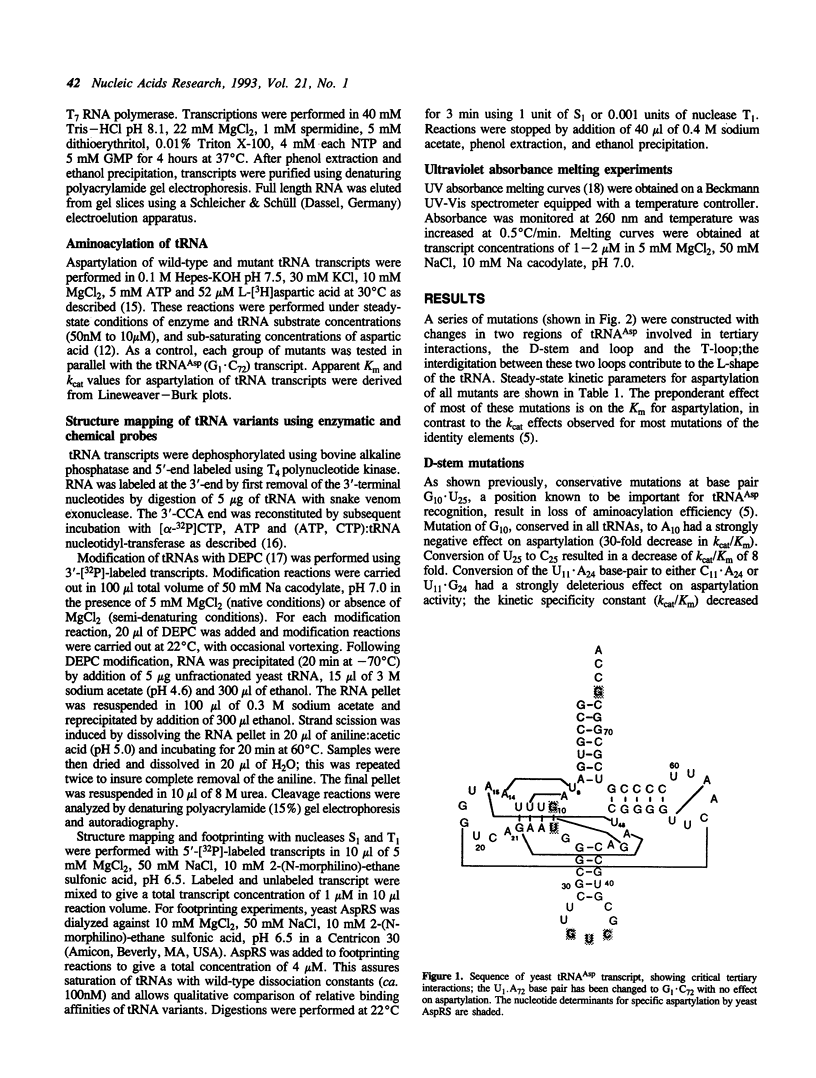
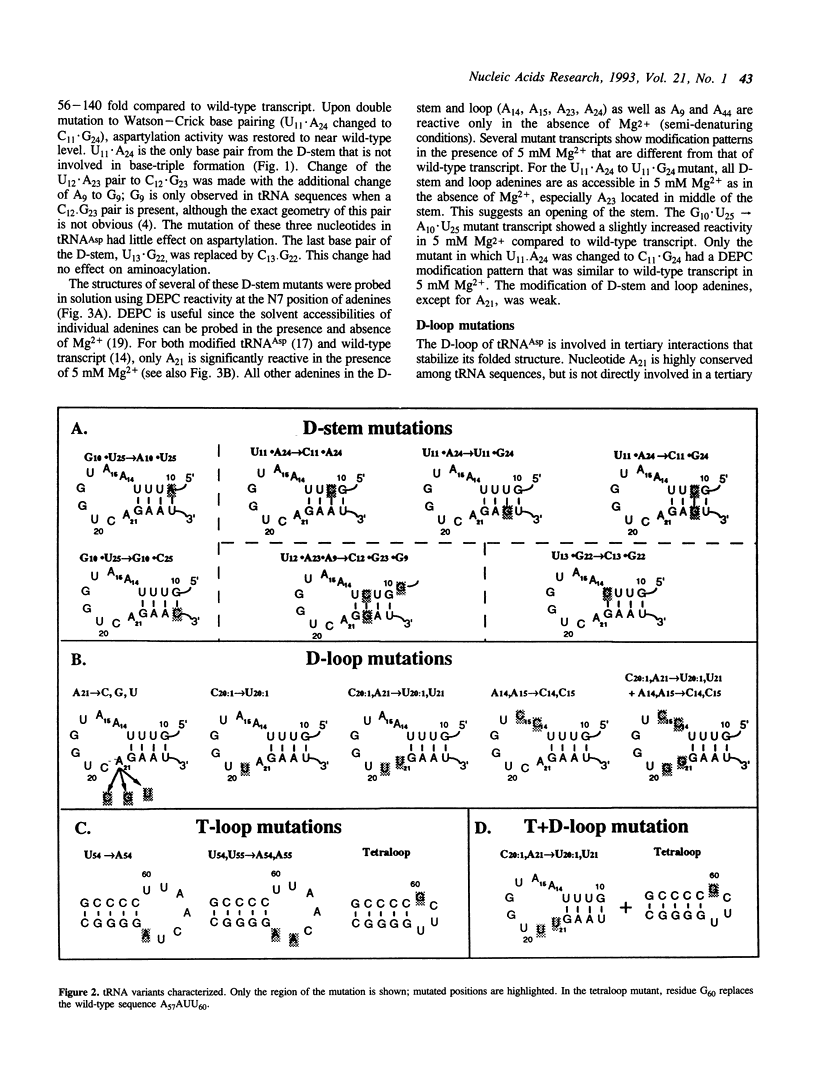
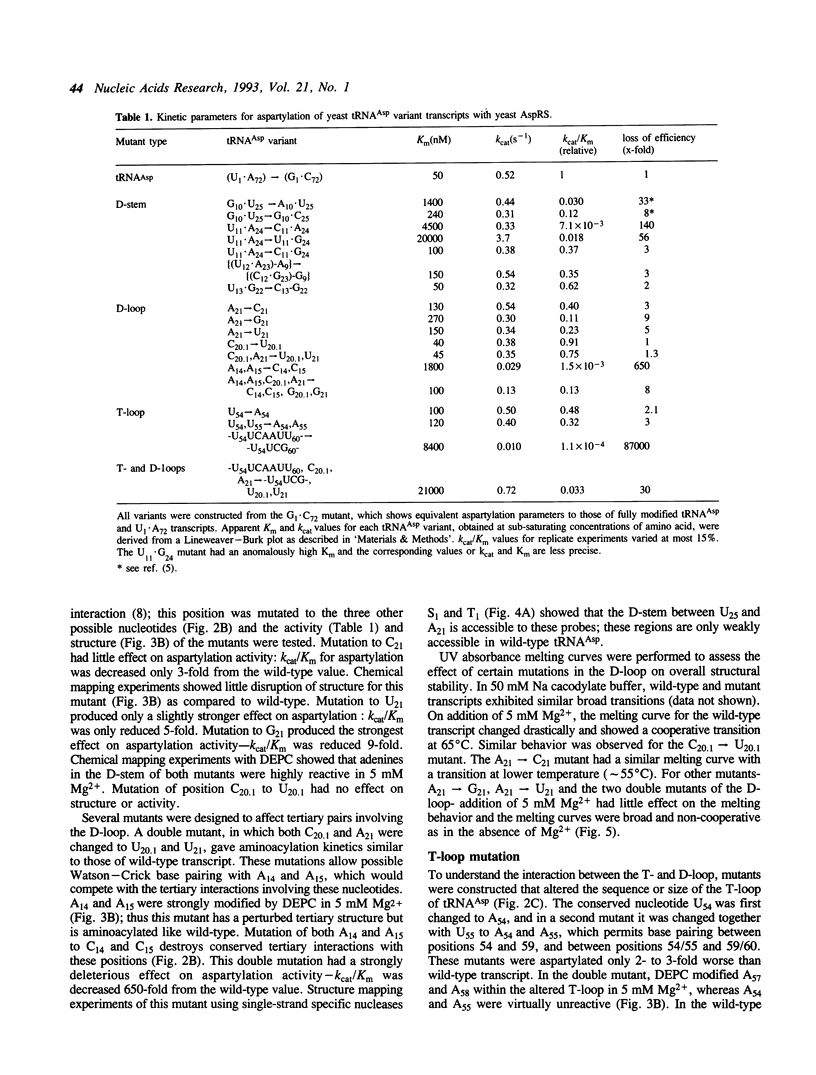
Mutations have been designed that disrupt the tertiary structure of yeast tRNA(Asp). The effects of these mutations on both tRNA structure and specific aspartylation by yeast aspartyl-tRNA synthetase were assayed. Mutations that disrupt tertiary interactions involving the D-stem or D-loop result in destabilization of the base-pairing in the D-stem, as monitored by nuclease digestion and chemical modification studies. These mutations also decrease the specificity constant (kcat/Km) for aspartylation by aspartyl-tRNA synthetase up to 10(3)-10(4) fold. The size of the T-loop also influences tRNA(Asp) structure and function; change of its T-loop to a tetraloop (-UUCG-) sequence results in a denatured D-stem and an almost 10(4) fold decrease of kcat/Km for aspartylation. The negative effects of these mutations on aspartylation activity are significantly alleviated by additional mutations that stabilize the D-stem. These results indicate that a critical role of tertiary structure in tRNA(Asp) for aspartylation is the maintenance of a base-paired D-stem.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coutts S. M., Gangloff J., Dirheimer G. Conformational transitions in tRNA Asp (brewer's yeast). Thermodynamic, kinetic, and enzymatic measurements on oligonucleotide fragments and the intact molecule. Biochemistry. 1974 Sep 10;13(19):3938–3948. doi: 10.1021/bi00716a019. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y., Yokogawa T., Hasegawa E., Miura K., Watanabe K. The aminoacylation of structurally variant phenylalanine tRNAs from mitochondria and various nonmitochondrial sources by bovine mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 1989 Aug 5;264(22):13005–13011. [PubMed] [Google Scholar]
- Lorber B., Kern D., Dietrich A., Gangloff J., Ebel J. P., Giegé R. Large scale purification and structural properties of yeast aspartyl-tRNA synthetase. Biochem Biophys Res Commun. 1983 Nov 30;117(1):259–267. doi: 10.1016/0006-291x(83)91569-3. [DOI] [PubMed] [Google Scholar]
- Mans R. M., Pleij C. W., Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991 Oct 15;201(2):303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- Mohr G., Lambowitz A. M. Integration of a group I intron into a ribosomal RNA sequence promoted by a tyrosyl-tRNA synthetase. Nature. 1991 Nov 14;354(6349):164–167. doi: 10.1038/354164a0. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Okimoto R., Wolstenholme D. R. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm: towards a minimal tRNA adaptor. EMBO J. 1990 Oct;9(10):3405–3411. doi: 10.1002/j.1460-2075.1990.tb07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret V., Florentz C., Puglisi J. D., Giegé R. Effect of conformational features on the aminoacylation of tRNAs and consequences on the permutation of tRNA specificities. J Mol Biol. 1992 Jul 20;226(2):323–333. doi: 10.1016/0022-2836(92)90950-o. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Romby P., Moine H., Lesage P., Graffe M., Dondon J., Ebel J. P., Grunberg-Manago M., Ehresmann B., Ehresmann C., Springer M. The relation between catalytic activity and gene regulation in the case of E coli threonyl-tRNA synthetase. Biochimie. 1990 Jun-Jul;72(6-7):485–494. doi: 10.1016/0300-9084(90)90072-o. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Dumas P., Ebel J. P., Giegé R. Comparison of the tertiary structure of yeast tRNA(Asp) and tRNA(Phe) in solution. Chemical modification study of the bases. J Mol Biol. 1987 May 5;195(1):193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Rudinger J., Puglisi J. D., Pütz J., Schatz D., Eckstein F., Florentz C., Giegé R. Determinant nucleotides of yeast tRNA(Asp) interact directly with aspartyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5882–5886. doi: 10.1073/pnas.89.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry. 1990 Mar 13;29(10):2523–2532. doi: 10.1021/bi00462a014. [DOI] [PubMed] [Google Scholar]
- Stein A., Crothers D. M. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry. 1976 Jan 13;15(1):160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Hydration of transfer RNA molecules: a crystallographic study. Biochimie. 1988 Feb;70(2):145–165. doi: 10.1016/0300-9084(88)90056-9. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr A. 1988 Mar 1;44(Pt 2):112–123. [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]
- de Bruijn M. H., Klug A. A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire 'dihydrouridine' loop and stem. EMBO J. 1983;2(8):1309–1321. doi: 10.1002/j.1460-2075.1983.tb01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]