Abstract
Background
Germline mutations and deletions of SMARCB1/INI1 in chromosome band 22q11.2 predispose patients to rhabdoid tumor and schwannomatosis. Previous estimates suggested that 15–20% of rhabdoid tumors were caused by an underlying germline abnormality of SMARCB1. However, these studies were limited by case selection and an inability to detect intragenic deletions and duplications.
Procedure
One hundred matched tumor and blood samples from patients with rhabdoid tumors of the brain, kidney, or soft tissues were analyzed for mutations and deletions of SMARCB1 by FISH, multiplex ligation-dependent probe amplification (MLPA), sequence analysis and high resolution Illumina 610K SNP based oligonucleotide array studies.
Results
Thirty-five of 100 patients were found to have a germline SMARCB1 abnormality. These abnormalities included point and frameshift mutations, intragenic deletions and duplications, and larger deletions including regions both proximal and distal to SMARCB1. There were 9 cases that demonstrated parent to child transmission of a mutated copy of SMARCB1. In 8 of the 9 cases, one or more family members were also diagnosed with rhabdoid tumor or schwannoma, and 2 of the 8 families presented with multiple affected children in a manner consistent with gonadal mosaicism.
Conclusions
Approximately one third of newly diagnosed patients with rhabdoid tumor have an underlying genetic predisposition to tumors due to a germline SMARCB1 alteration. Families may demonstrate incomplete penetrance and gonadal mosaicism, which must be considered when counseling families of patients with rhabdoid tumor.
Keywords: SMARCB1, INI1, rhabdoid, schwannomatosis
INTRODUCTION
Rhabdoid tumors are particularly aggressive pediatric malignancies that primarily develop in infancy and early childhood. They are referred to as atypical teratoid/ rhabdoid tumor (AT/RT) when they arise in the central nervous system and malignant rhabdoid tumor (MRT) when they are found in renal or extra-renal sites. The vast majority of tumors (both AT/RT and MRT) are characterized by loss of function of the SMARCB1/INI1/SNF5/BAF47 gene in chromosome band 22q11.2 [1,2]. SMARCB1 is an invariant member of the SWI/SNF chromatin-remodeling complex of proteins and thus functions in controlling gene transcription. The role of SMARCB1 and the SWI/SNF complex in cancer has recently been reviewed [3,4].
Consistent with its role as a tumor suppressor gene, tumors arise following inactivation of both alleles of SMARCB1. While the majority of tumors are histologically classified as rhabdoid tumors, loss of expression of SMARCB1 has also been observed in other tumors including epithelioid sarcoma, renal medullary carcinoma, undifferentiated pediatric sarcomas, and a subset of hepatoblastomas [5–9]. It has been previously estimated that 15–20% of all rhabdoid tumors are caused by germline mutations of SMARCB1. In these cases predisposing germline deletions or mutations constitute the so-called “first hit”, and can be de novo or inherited from a parent. However, these studies may have been biased by case selection, and did not include intragenic deletions and duplications that are now detectable with improvements in molecular genetic screening methods, particularly MLPA [10]. A recent study of 50 rhabdoid patients reported germline deletions or mutations in 20% of patients, but abnormalities were not identified in all of the cases [11].
Retinoblastoma provides a useful paradigm in considering the role of germline mutations in rhabdoid tumor. Retinoblastoma can arise unilaterally or bilaterally. Forty percent of retinoblastomas are due to germline mutations in RB1, most of which are de novo [12]. If a germline mutation in RB1 exists, there is a ninety-five percent chance of a second mutation developing in the normal allele in at least one retinal cell [12]. Germline mutations, therefore, overwhelmingly cause bilateral disease. Indeed, only fifteen percent of unilateral retinoblastomas are caused by a germline mutation [12]. Furthermore, the de novo mutations in retinoblastoma show a bias towards originating on the paternal allele [13]. Studies show that unaffected carriers of RB1 mutations are more likely to show a maternal mutant allele, whereas the mutant allele is more likely to be of paternal origin in patients who develop retinoblastoma [14]. The parental origin of mutations or deletions of SMARCB1 have not been extensively studied in rhabdoid tumor.
As with bilateral retinoblastoma, two distinct rhabdoid tumors developing in different sites, such as in the brain and the kidney, are a strong indication of an underlying germline alteration in SMARCB1. While it has been argued that a tumor in a second location could be a metastasis, the identification of distinct second “hits”, either mutations or deletions, provides conclusive support for their independent origin. Patients who present with multiple primary tumors are inevitably found to have a germline alteration, however, children with apparently sporadic tumors have also been reported to have germline mutations or deletions in SMARCB1 [2]. The a priori risk of developing a rhabdoid tumor, or another related tumor, in the setting of a germline mutation is not yet known. Whether this risk could also be affected by the parental origin of the mutated or deleted allele is also not known.
Although specific genotype-phenotype correlations are still in their infancy, we have previously reported that germline deletions of chromosome 22 that include the SMARCB1 gene increase the risk of a variety of phenotypic findings, as well as rhabdoid tumor [10]. These may include heart defects, developmental delay and other congenital abnormalities, depending on the size of the deletion and the specific genes involved. Recent studies have also shown that constitutional mutations in SMARCB1 predispose to schwannomatosis [15–17]. Only one family has been reported with both rhabdoid tumor and schwannomatosis due to an inherited exon 6 duplication in SMARCB1 [17]. The goal of the present study was to obtain a more accurate estimate of germline SMARCB1 abnormalities in rhabdoid tumor patients and build a knowledge base that will eventually allow for estimates of disease risk based on mutation status.
METHODS
Case selection
Studies were performed in accordance with an Institutional Review Board approved protocol. All families gave consent for germline SMARCB1 testing as part of this research protocol, although they may have also been enrolled in a clinical therapeutic trial. Studies were performed in a laboratory accredited by the College of American Pathologists and according to Clinical Laboratory Improvement Amendments so that results could be provided back to physicians for patient management and counseling.
Patients were ascertained by clinical report of a rhabdoid tumor, which was confirmed to demonstrate loss of SMARCB1 protein expression by immunohistochemistry. Cases were selected for inclusion in this study based on the availability of matched tumor and normal tissue, and the identification of both inactivating events in the SMARCB1 gene in the tumor. A total of 100 cases met the selection criteria. Tumor and blood samples were obtained from 100 patients from a total of 57 institutions. Parental blood samples, or samples from siblings, were available for 42 of the 100 cases.
Tumor DNA was isolated using a Qiagen Gentra PureGene kit (Germantown, MD), and screened for a mutation or deletion according to previously reported methods that included PCR amplification and sequencing, MLPA, FISH and/or high resolution SNP array analysis using an Illumina 610K Beadchip [18]. Matched blood DNA samples were then screened for the abnormalities detected in the tumor. If a germline mutation or deletion was identified, both parental blood samples were tested to determine if the alteration was de novo or inherited. When possible, an analysis of genotype calls in Illumina's Beadstudio software or chromosome 22q specific microsatellite assays were used to identify the parental origin of the deleted chromosome.
RESULTS
The 100 cases included 65 patients with CNS AT/RT, 12 with MRT of the kidney, 17 with extra-renal tumors, and 6 patients with multiple primary tumors (Tables I, II, and III). There were 49 males and 51 females. As shown in Table I, Table III and Figure 1, 35 of 100 patients had a germline deletion or mutation. Patients with a germline abnormality ranged from birth to 5 years with a median age at diagnosis of 5 months. The age at diagnosis for patients with sporadic tumors (only somatic mutations/deletions detected) ranged from birth to 17 years with a median age of 18 months. Five of the 65 patients with sporadic tumors were diagnosed over 6 years of age, and two of these were teenagers.
TABLE I.
SMARcb1 Alterations in 35 Patients With Germline Abnormalities Predisposing to Rhabdoid Tumor
| Sample ID | Sex | Dx age (month) | Anatomic site | Germline abnormality | Acquired abnormality | Inheritance |
|---|---|---|---|---|---|---|
| 98-074 | F | 6 | CNS | c.152G>A | Deletion | Unknown |
| 99-084 | M | 3 | Renal,1 CNS | c.192G>A | Deletion | Unknown |
| 00-189 | F | 24 | Renal | Deletion | c.346de6c | Unknown |
| 00-241 | F | 1 | Renal,1 CNS | Deletion E4–5 | CN LOH | De novo (maternal) |
| 01-135 | M | 5 | Extra-renal | Deletion E1–2 | CN LOH | De novo (maternal) |
| 01-247 | M | 6 | CNS | Deletion | c.1125_1130dup6 | Unknown |
| 02-0662, 3 | F | 48 | CNS | c.859G>T | c.847_848delAT | Maternal |
| 04-0642, 3 | M | 48 | CNS | c.986+1G>C | CN LOH | Paternal |
| 04-273 [30]2, 3 | M | 17 | CNS | c.373_376dup4bp | Deletion | Maternal |
| 05-132[10] | M | 56 | Renal | Deletion | c.667_668delTG | Unknown |
| 05-136 | M | 1 | Renal | c.592C>T | Deletion | Unknown |
| 05-208[10] | F | 13 | CNS | Deletion | c.605_612dup8bp | Unknown |
| 06-147[10] | M | 7 | Renal,1 CNS | Deletion | Deletion | De novo (paternal) |
| 06-165 [17]2, 3 | F | 20 | CNS | Deletion E6 | c.578_581dup8bp | Paternal |
| 06-175 [10] | M | 1 | Extra-renal,1 renal | Deletion | Deletion | De novo (paternal) |
| 06-216 | M | 5 | CNS | Deletion | Deletion | Unknown |
| 06-249 | M | 21 | CNS | c.853_854delGA | Deletion | De novo |
| 07-0052 | M | 3 weeks | CNS | Duplication E4–5 | CN LOH | Gonadal mosaicism |
| 07-158 | M | 5 | Extra-renal1 (multiple) | Deletion | c.842G>A | De novo (paternal) |
| 07-313 | F | 10 | CNS | Deletion E1–5 | CN LOH | De novo |
| 07-3673 | F | 2 | CNS | c.472C>T | Deletion | Paternal |
| 08-092 | M | Birth | Extra-renal | Deletion | c.548_566dup19 | De novo (paternal) |
| 08-1433, 3 | F | 11 | CNS | c.367C>T | CN LOH | Maternal |
| 08-187 | M | 11 | CNS | c.472C>T | Deletion | De novo |
| 08-192 | M | 14 | CNS | c.168_169delTG | CN LOH | De novo |
| 08-304 | M | 5 | Extra-renal | Deletion E-–5 | c.146_147ins25 | De novo |
| 09-0132 | F | 2 | CNS | c.20_43delinsT | Deletion | Gonadal mosaicism |
| 09-044 | F | 9 | CNS1, renal | c.141C>A | CN LOH | De novo (paternal) |
| 09-0453 | M | 5 | CNS | Deletion E7 | Deletion | Maternal |
| 09-131 | M | 3 | CNS | c.93G>C | Deletion | Unknown |
| 09-223 | F | 1 | CNS | c.601C>T | CN LOH | De novo (paternal) |
| 09-261 | F | 5 years | CNS | Duplication E6 | Deletion | Unknown |
| 09-289 | M | 2 weeks | CNS | Deletion E4–6 | Deletion | Unknown |
| 09-312 | F | 4 | CNS | Deletion E6 (mosaic) | CN LOH | Unknown |
| 09-359 | F | 2 | CNS | Deletion exon 1 | Deletion | Unknown |
TABLE II.
SMARCB1 Alterations in 65 Patients With Rhabdoid Tumor Caused by Somatic Abnormalities
| Sample ID | Sex | Dx age (month) | Anatomic site | Abnormality 1 | Abnormality 2 |
|---|---|---|---|---|---|
| 00-008 | F | 2 | CNS | Deletion | Deletion |
| 00-163 | M | 22 | CNS | c.252insA | Deletion |
| 00-315 [18] | F | 13 years | Extra-renal | Deletion | Deletion |
| 01-090 [18] | M | 36 | CNS | c.1143delG | Deletion |
| 01-154 | F | 4 | Extra-renal | Deletion | Deletion |
| 01-179 | M | 6 | CNS | Deletion | Deletion |
| 01-323 [18] | F | 18 | CNS | c.118C>T | CN LOH |
| 02-040 [18] | M | 24 | CNS | Deletion | Deletion |
| 02-171 | M | 53 | CNS | Deletion E6–7 | CN LOH |
| 02-203 [18] | F | 22 | CNS | c.1143delG | Deletion |
| 02-237 [18] | M | 48 | CNS | c.1143delG | Deletion |
| 02-291 [18] | F | 36 | CNS | c.601C>T | CN LOH |
| 03-085 | M | 12 | CNS | Deletion | Deletion |
| 03-102 | F | 24 | Renal | Deletion | Deletion |
| 03-105 | M | Birth | Extra-renal | c.472C>T | c.550_556dup7bp |
| 03-119 | F | 18 | CNS | c.1145delC | Deletion |
| 03-128 [18] | M | 12 | CNS | c.1143delG | Deletion |
| 03-287 [18] | F | 3 | CNS | c.157C>T | CN LOH |
| 04-007 [18] | F | 14 | Renal | c.601C>T | CN LOH |
| 04-053 [18] | F | 6 | CNS | c.1145delC | Deletion |
| 04-092 | F | 24 | CNS | c.1143delG | Deletion |
| 04-119 | M | 11 years | CNS | c.1143delG | Deletion |
| 05-024 [18] | F | 14 | CNS | Deletion | Deletion |
| 05-036 [18] | M | 5 | CNS | Deletion | Deletion |
| 05-109 | M | 19 | CNS | c.472C>T | Deletion |
| 05-187 | F | 36 | Extra-renal | Deletion | Deletion |
| 06-148 | F | 11 | CNS | c.601C>T | Deletion |
| 06-315 [18] | M | 6 | Extra-renal | Deletion | Deletion |
| 07-050 | M | 8 years | Extra-renal | Deletion | Deletion |
| 07-071 | F | 2 weeks | CNS | Deletion | Deletion |
| 07-104 | M | 17 years | CNS | c.323delA | Deletion |
| 07-167 | M | 12 | Renal | Deletion E1–5 | Deletion |
| 07-221 [18] | F | 3 | CNS | Deletion E7–9 | CN LOH |
| 07-352 | F | 5 years | CNS | c.1143delG | Deletion |
| 07-371 | F | 9 | Extra-renal | Deletion | Deletion |
| 07-386 | M | 15 | Extra-renal | Deletion | Deletion |
| 08-071 | M | 36 | Extra-renal | Deletion | Deletion |
| 08-114 | M | 18 | CNS | Deletion | Deletion |
| 08-145 | M | 9 | CNS | c.1145delC | Deletion |
| 08-158 | F | 6 years | Extra-renal | c.762_774dup13bp | Deletion |
| 08-172 | F | 6 | CNS | Deletion E6–9 | Deletion |
| 08-186 | M | 24 | CNS | c.1145delC | Deletion |
| 08-197 | M | 2 | CNS | Deletion | Deletion |
| 08-215 | F | 12 | CNS | c.1145delC | Deletion |
| 08-230 | M | 12 | CNS | Deletion | Deletion |
| 08-237 | M | 18 | CNS | c.553_565dup13 | Deletion |
| 08-251 | M | 24 | Renal | Deletion | Deletion |
| 08-255 | F | 9 | Renal | Deletion | Deletion |
| 08-311 | M | 11 | Renal | c.617G>A | CN LOH |
| 09-008 | F | 25 | Renal | Deletion | Deletion |
| 09-019 | F | 22 | Renal | c.501-1G>C | CN LOH |
| 09-027 | F | Birth | Extra-renal | Deletion | Deletion |
| 09-028 | F | 24 | CNS | c.1145delC | Deletion |
| 09-034 | F | 24 | CNS | c.1143delG | c.1145delC |
| 09-046A | F | 24 | CNS | c.472C>T | Deletion |
| 09-125 | F | 26 | CNS | c.1145delC | Deletion |
| 09-130 | M | 7 | Extra-renal | Deletion E4–5 | CN LOH |
| 09-153 | F | 16 | CNS | c.1143delG | Deletion |
| 09-170 | F | 34 | Renal | c.157C>T | CN LOH |
| 09-182 | F | 5 years | CNS | Deletion | Deletion |
| 09-196 | F | 19 | CNS | c.601C>T | CN LOH |
| 09-230 | F | 30 | CNS | Deletion | Deletion |
| 09-236 | F | 27 | Extra-renal | Deletion E1–6 | Deletion |
| 09-252 | M | 9 years | Extra-renal | Deletion E4–5 | Deletion |
| 09-317 | M | 18 | CNS | Deletion | Deletion |
Ref. [18]: previously reported cases.
TABLE III.
Germline Alterations in Rhabdoid Tumors by Anatomic Site
| Anatomic site | Total | Number (%) |
|---|---|---|
| CNS | 65 | 23 (35) |
| Kidney | 12 | 3 (25) |
| Multiple primaries | 6 | 6 (100) |
| Extra-renal | 17 | 3 (18) |
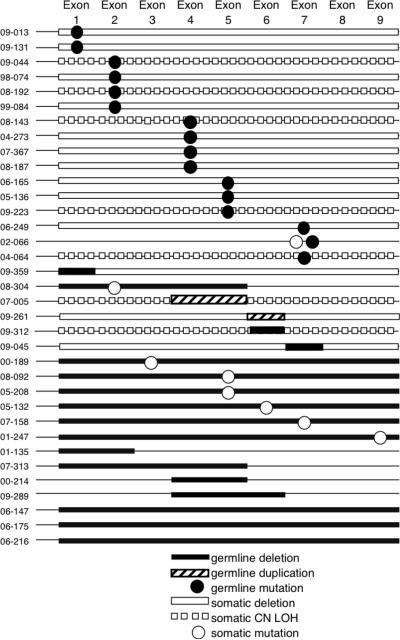
Fig 1.
Germline alterations were found in 35 cases and included mutations, duplications and deletions (shown in black). The “second hit,” found only in tumor cells, is shown in white. The last seven patients (01-135, 07-315, 00-214, 09-289, 06-147, 06-175, and 06-216) have homozygous deletions in their tumors that are the same size as germline heterozygous deletions, so they appear only as black bars.
As was expected, patients with multiple tumors were most likely to have a germline mutation. Six of 6 patients with two primary tumors, as assessed by clinical presentation and/or distinct molecular alterations as the second inactivating event, had a germline mutation. Twenty-three of 65 patients with CNS tumors had a germline mutation, compared with 3 of 12 patients with renal tumors and 3 of 17 patients with soft tissue tumors (Table III). In contrast, there was a high frequency (9 of 17 cases) of somatic homozygous whole gene deletions in tumors from extra-renal sites, as previously reported [19].
The most common germline abnormalities detected were point or frameshift mutations resulting in premature truncation of the protein (14 of 35 cases). Other germline abnormalities included heterozygous loss of SMARCB1 (9 of 35 cases), and duplications (3 of 35 cases) or deletions (8 of 35 cases) of one or more exons within SMARCB1 (Fig. 1). All of the germline deletions of 22q11.2 that included the entire SMARCB1 locus appeared to be de novo. There was one family with a splice site mutation (04–064). A mutation in exon 4 (c.472C>T) was demonstrated in two cases. All of these abnormalities were also found in patients with somatic alterations of SMARCB1. The second inactivating event in the tumors from these patients was a deletion (16), copy number neutral loss of heterozygosity (CN LOH) (10 tumors), or a frameshift mutation (8). There was only one tumor with a point mutation as the second hit (case 07–158).
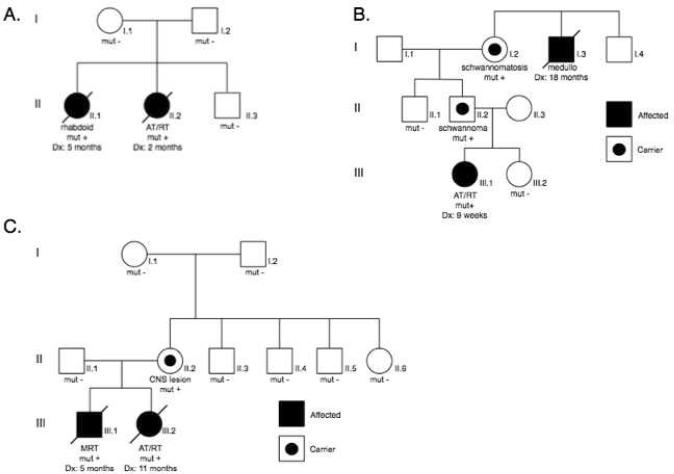
Both parents' blood samples were available for testing in 22 of the 35 cases with germline mutations. In 7 of these 22 cases a parent was a carrier of the germline abnormality, four of which were maternally inherited and 3 of which were paternally inherited (Table I). Two of the carrier fathers (cases 06–165 and 07–367) developed schwannomas (Fig. 3B). The mother of patient 08–143 was diagnosed with a benign CNS lesion (Fig. 3C). The other 4 carrier parents were unaffected. In two additional cases (07-005 and 09–013) the parents' blood samples were normal, but there were two affected siblings with the same mutation, consistent with gonadal mosaicism (Fig. 3A).
Fig. 3.
Pedigree of family 09-013. Two affected sisters had the same SMARCB1 exon 1 mutation (c.20_43delinsT). Neither parent had the mutation, consistent with gonadal mosaicism (A). Pedigree of family 07-367. The proband (III.1) was found to have a germline mutation in SMARCB1 exon 4 (c.472C>T), inherited from her paternal grandmother (B). Pedigree of family 08-143. The SMARCB1 exon 4 mutation (c.367C>T) was identified in two affected siblings as well as their mother. She had a de novo mutation and was subsequently diagnosed with a benign CNS lesion (C).
The deletions and duplications within SMARCB1 encompassed anywhere from 1 exon to all 9 exons (Fig. 1). These small changes in copy number preferentially included exons 4 and 5, which seem to be a particular hot spot for both germline and somatic abnormalities. Duplications and deletions proximal and distal to SMARCB1 were identified by both MLPA and SNP array analyses (data not shown). The largest germline deletions detected by MLPA all spanned from 22q11.21–q11.23 and included MAPK1, PRAME and SMARCB1 (6 of 9 cases). Two of these children presented with phenotypic abnormalities, and were analyzed by SNP array. All genomic positions were based upon hg18 (March, 2006) from the UCSC Genome Browser (http://genome.ucsc.edu). Case 07–158 demonstrated a de novo deletion from 20,247,190 to 22,973,609. He presented with several phenotypic defects including ptosis, small cupped auricular deformity, and pectus excavatum. He also had a small ventricular septal defect. Case 08–092 showed a de novo deletion spanning from 20,128,907 to 23,011,579. He was noted to have a patent foramen ovale, a slightly upslanting palpebral fissure and his ears were low set with posterior rotation. We previously reported similar phenotypic findings in rhabdoid patients with germline deletions proximal to, and including, SMARCB1 [10].
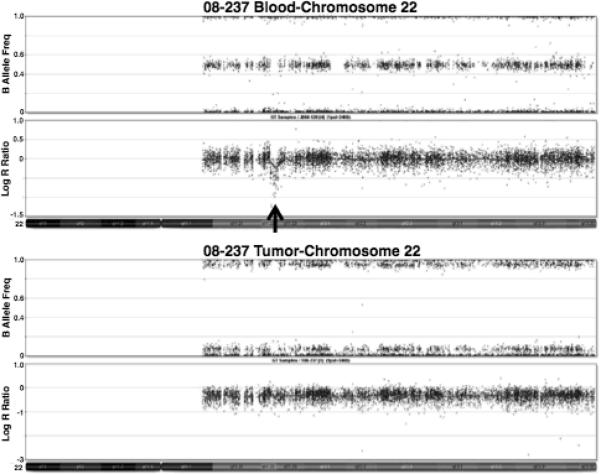
In one case (08–237) there was a germline deletion of GNAZ in chromosome band 22q11.22, proximal to SMARCB1 (Fig. 2). The tumor demonstrated loss of the deleted chromosome, as well as a mutation in the remaining copy of SMARCB1. As the GNAZ deletion was also found in the proband's unaffected brother and mother, it was not considered to be the predisposing rhabdoid tumor deletion. However, there may be some underlying genetic instability in these individuals that resulted in multiple alterations of chromosome 22 in the affected child.
Fig. 2.
Illumina Beadstudio depiction of chromosome 22 showing a heterozygous deletion in the blood of patient 08-237. A decrease in the Log R ratio corresponding to the absence of heterozygous SNPs indicates a deletion, the location of which is demonstrated by the black arrow. This deletion includes GNAZ, but not SMARCB1. The tumor shows a whole chromosome 22 deletion.
In two cases, mutations were found in the splice sites of exons. In one family (case 04–064) three of four children were diagnosed with AT/RT and showed an intron 7 point mutation (c.986+1G>C) in the donor splice site (Fig. 1). Sequence analysis of a RT-PCR product spanning exons 5 to 9 demonstrated that the splice site mutation resulted in the exclusion of exon 7. A similar mutation in the donor splice site of exon 7 was previously reported in a family with a history of posterior fossa brain tumors in infancy [20]. Both parents were screened and the father was found to carry the same mutation as the affected children. The paternal grandmother and aunts were also tested and found to be normal. Clinical details of this case have been published separately [21]. The second case (09–019) had a somatic mutation in the acceptor splice site of exon 5 (c.501−1G>C) and CN LOH of the other allele that spanned from 22q11.21 to the terminus.
There were seven patients in the present study that had at least one affected sibling. The pedigree shown in Figure 3A, case 09–013, represents a family with two affected daughters. The first daughter was diagnosed at 5 months with a bladder mass, reported as a poorly differentiated sarcoma. The second daughter developed a brain tumor at 2 months of age. This tumor was diagnosed as an AT/RT, and SMARCB1 mutation screening was performed. Sequence analysis revealed a mutation in exon 1 in the blood and tumor (c.20_43delinsT) (Fig. 1). MLPA analysis was consistent with a deletion of the second allele. Formalin fixed tissue preserved from the older daughter's tumor was then tested and found to have the same mutation. MLPA analysis of both of their tumors showed a deletion of probe E1b, which overlaps the mutated region of exon 1. The sequencing and MLPA studies were normal in the peripheral blood DNA from both parents. Results from the SNP array studies confirmed the paternity and there were no significant abnormalities found in either parent. Gonadal mosaicism is therefore the most likely explanation for the inheritance of the germline mutation seen in these two siblings.
A second family (case 07-005) in our study population showed a similar pattern of inheritance. This family presented with two children with rhabdoid tumors, a male and female. Both siblings showed a duplication of SMARCB1 exons 4–5 by MLPA (Fig. 1). The MLPA results from the parents' blood DNA indicated normal SMARCB1 copy number. These results were confirmed with a TaqMan assay designed specifically to reveal copy number of SMARCB1 exons 4–5 (data not shown). The SNP array confirmed paternity and did not reveal any significant abnormalities. Details of this family have been published separately [21].
The pedigree shown in Figure 3B represents the second family to be reported in which a germline mutation predisposed to both schwannomatosis and CNS AT/RT. The proband (III.1) was diagnosed with an AT/RT. Testing of peripheral blood DNA was prompted due to the identification of an exon 4 (c.472C>T) mutation in the tumor, as this frequently presents as a germline mutation (Fig. 1). The paternal grandmother also had a history of multiple schwannomas. The father and grandmother were both found to be carriers. Although the grandmother's brother (I.3) was reported to have died from medulloblastoma, a diagnosis of AT/RT is more likely, given that there is a SMARCB1 mutation segregating in the family. The patient's father (II.2) was subsequently found to have a schwannoma. Prenatal testing for III.2 was performed, and there was no mutation identified.
The third pedigree (Fig. 3C) depicts a family in which the son was initially reported to have an epithelioid malignant peripheral nerve sheath tumor. His sister was subsequently diagnosed with a CNS AT/RT, prompting genetic testing and pathologic review of the son's tumor. The tumor from III.1 had the same mutation (c.367C>T) found in the tumor and blood from his sister (III.2) and was ultimately re-classified as MRT. The mother was found to carry the same exon 4 mutation. Extensive testing of her parents and siblings suggested that this was a de novo mutation. She was later found to have an ill-defined CNS lesion, not suggestive of AT/RT, with retained SMARCB1.
There were 65 of the 100 cases in which both inactivating SMARCB1 events were somatic in origin (Table II). These abnormalities included 89 deletions in 26 patients, 7 intragenic deletions in 7 patients, 22 frameshift mutations in 20 patients, and 12 point mutations in 12 patients. The most common finding was a homozygous deletion (26 of 65 tumors). The deletions were approximately the same size as the deletions in the germline associated cases. The most common mutation was a frameshift (16 tumors) in exon 9, either c.1143delG or c.1145delC, both of which are predicted to cause an elongation of the protein by 104 amino acids. The protein in these tumors is not expressed, since immunohistochemical analysis always demonstrates loss of expression in tumor nuclei [22]. These single base deletions are the most common mutations observed in CNS AT/RTs and yet have never been seen in the germline. To date, they have not been reported in renal or extra-renal malignancies. Whether this could be due to some dominant negative effect in the germline is not known. As in the germline cases, an exon 4 mutation, c.472C>T, was seen multiple times (3 of 12 point mutations). An exon 5 mutation, c.601C>T, was the most common point mutation observed (4 of 12 point mutations).
A total of 23 trios (patients and parents) were analyzed to determine the parental origins of chromosomes harboring the SMARCB1 abnormalities (11 sporadic tumors and 12 germline associated tumors). Overall, of 15 mutations, 7 were found on the paternal allele and 8 were found on the maternal allele. Of 4 intragenic deletions, 1 was on the paternal allele, and 3 were on the maternal allele. Among 27 deletions, 15 were on the paternal allele and 12 were on the maternal allele. These data do not seem to show a bias in the parental origin of the abnormality; however, further studies are required in order to draw any significant conclusions.
DISCUSSION
The goal of the present study was to ascertain a more accurate estimate of the frequency of germline abnormalities in patients with rhabdoid tumor and related entities. We now estimate that as many as 35% of rhabdoid tumors may be associated with a germline deletion or mutation of SMARCB1, significantly higher than previous estimates. These studies depend on a comprehensive strategy using MLPA, direct sequencing, and SNP array analysis to identify all of the mutations, copy number changes and LOH events. The mutations and deletions predispose to rhabdoid tumors in all sites, but may show variable expressivity and reduced penetrance. Several unaffected carriers were identified in the course of these studies, who are thus at risk for development of both benign and malignant tumors, including schwannoma and rhabdoid tumor.
Alterations of SMARCB1 are not specific for rhabdoid tumor, which may make risk assessment with regard to benign vs. malignant tumors difficult. Cases of choroid plexus carcinoma, epithelioid sarcoma, peripheral primitive neuroectodermal tumor, undiffererentiated sarcoma and epithelioid malignant peripheral nerve sheath tumor have all been reported to demonstrate loss of expression of SMARCB1, although the number of cases with inactivating gene deletions or mutations in SMARCB1 is limited [5–8,19,22–24]. The histology of these cases seems to rule out rhabdoid tumor, despite the molecular evidence to the contrary. Any report of a SMARCB1 abnormality, even if another tumor type is suspected, warrants further genetic testing to rule out a germline mutation.
A history of schwannomatosis is also an indication for genetic testing, due to the high likelihood of carrying a germline SMARCB1 mutation, and the associated risk of developing a rhabdoid tumor [15–17]. The family shown in Figure 3B had no history of CNS tumors, but had a history of schwannomatosis and reported medulloblastoma. Slides were not available for review of the medulloblastoma, however, it is likely that the actual diagnosis was AT/RT, given that medulloblastomas do not have alterations in SMARCB1 [22]. There are several families reported in the literature that show similar histories of schwannomatosis and SMARCB1 mutation. We previously reported a family in which two sisters developed MRT [17]. Molecular screening indicated a duplication of exon 6 by high-resolution chromosome 22 specific array analysis, as well as MLPA, in both sisters and their unaffected brother. A father, paternal uncle and paternal grandmother also carry the duplication and had a history of schwannomatosis. All such family members should thus be tested for a SMARCB1 mutation, and counseled regarding the risk of rhabdoid tumors.
The present report describes two new families with findings consistent with gonadal mosaicism predisposing to rhabdoid tumor. Sévenet et al. first reported gonadal mosaicism and rhabdoid tumor in two families with multiple affected siblings and normal parents [24]. A case of gonadal mosaicism and SMARCB1 mutation has recently been reported in a family with schwannomatosis [25]. A mutation in the donor splice site of exon 8 was found in two affected siblings, but was not present in either parent. Haplotype analysis revealed a maternal origin of the mutation [25]. Gonadal mosaicism, although rare, has been observed in other cancers. These include colorectal cancer, associated with familial adenomatous polyposis and mutation in the adenomatous polyposis coli (APC) gene; retinoblastoma and neurofibromatosis type 1 [26–28].
In the present study, cases were specifically selected based on histology and loss of SMARCB1 expression by immunohistochemistry. In fact, 98% of rhabdoid tumors demonstrate SMARCB1 loss and concomitant deletions or mutations of the SMARCB1 gene [18]. A recent report of a SMARCA4/BRG1 mutation in a family with rhabdoid tumor suggests that there is at least one other rhabdoid tumor locus, which may account for a small number of SMARCB1 positive rhabdoid tumors [29]. It is likely that additional loci will be identified in association with rhabdoid tumor, and it will be of interest to determine the incidence of germline vs. somatic mutations associated with these tumor associated genes as well.
Due to the varied treatment strategies employed over the last ten years, reports of early treatment failures in children who are carriers of germline mutations are mostly limited to small series of patients. The present study was not designed to assess clinical outcome. Clinical correlative studies are in progress to determine if these patients have an inherent difference in response to radiation and/or chemotherapy that impacts survival, or have a different prognosis than patients with sporadic tumors because of age, or presence of multiple tumors. In fact, the median age of children with rhabdoid tumor and a germline mutation or deletion is younger (5 months) than children with apparently sporadic disease (18 months). In the future, treatment approaches may differ based on the presence of a germline SMARCB1 alteration, as these patients are younger, and already predisposed to cancer.
At present, it is important for clinicians to facilitate genetic testing and refer families for genetic counseling. Knowing that a germline SMARCB1 mutation increases the risk for rhabdoid tumor and schwannomatosis can be taken into consideration when planning to have more children. Discussions regarding the risk of gonadal mosaicism are complex, and are best initiated by a genetics professional. Prenatal testing can be performed in situations where a specific SMARCB1 mutation or deletion has been documented in the family. Screening protocols for carriers who are identified before the occurrence of a rhabdoid tumor, or after successful treatment for a tumor, are clearly indicated. While specific guidelines for screening have not yet been established, baseline CNS MRIs and renal ultrasounds are standard practice and further monitoring can be implemented as needed.
Acknowledgements
This work was supported by a grant from the NIH (CA46274 to J.A.B).
The technical assistance of Casey Belcher-Timme and Amalie Chen is greatly appreciated.
We thank the following individuals for contributing cases to this study: Brad Angle, Ronald Barr, Jean Belasco, Anne Bendel, David Biddle, J. Bourgeois, Daniel C. Bowers, Carole Brathwaite, Carol Bruggers, Paulette Bryant, Beth Ann Burt, Laura Campbell, Gabriel Chamyan, W. Bruce Cherny, Kudakwashe Chikwava, Katrina Conard, Linda Cooley, Odile David, Dennis Davidson, Daphne E. deMello, Steven Diamond, John Donahue, Michael S. Edwards, Anat Erdreich-Epstein, Linda Ernst, Michael M. Etzl, Lauren Evans, Jonathan Finlay, Michael Fisher, Nicholas Foreman, Ralph Franciosi, Jeffrey Goldstein, Eric Gratias, Paul Grundy, Marta Guttenberg, Sridharan Gururangan, Caroline Hastings, Patrick Healey, Brian Jeffrey, Michael A. Jones, Sarah Johnson-Welch, J. Martin Johnson, Marjolijn Jongmans, Mark Kayton, Tammy Kang, Portia Kreiger, Yves Lacassie, Martin Lammens, Michael P. Laquaglia, Amy Lowichik, David Malkin, Christopher Maloney, Arthur E. Marlin, Mark R. Matthews, Michael D. Medlock, Christopher Moertel, Hector Monforte, Jaume Mora, Robert Newbury, Sarah M. Nikkel, Dmitriy Niyazov, Sonia Partap, Kathleen Patterson, Laszlo Perlaky, Peter Phillips, Sharon Plon, Ted Pysher, R. H. Rhodes, Lucy Rorke-Adams, Marc K. Rosenblum, Lei Shao, Hiroyuki Shimada, Richard E. Slavin, Stacie L. Stapleton, Duncan Stearns, Phillip Storm, Louise C. Strong, Leslie Sutton, Benjamin Warf, Bradley E. Weprin, Cynthia Wetmore, Jennifer A. Wright, David Zagzag, Holly Zhou, and Craig Zuppan
References
- 1.Versteege I, Sévenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 2.Biegel JA, Zhou J, Rorke LB, et al. Germ-line and acquired mutations of INI1 in atypical teratoid rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 3.Roberts CWM, Biegel JA. The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer Biol Ther. 2009;8:412–416. doi: 10.4161/cbt.8.5.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 5.Hornick JL, Dal Cin P, Fletcher CDM. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am J Surg Pathol. 2009;33:542–550. doi: 10.1097/PAS.0b013e3181882c54. [DOI] [PubMed] [Google Scholar]
- 6.Modena P, Lualdi E, Facchinetti F, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 2005;65:4012–4019. doi: 10.1158/0008-5472.CAN-04-3050. [DOI] [PubMed] [Google Scholar]
- 7.Cheng JX, Tretiakova M, Gong C, Mandal S, et al. Renal medullary carcinoma: rhabdoid features and the absence of INI1 expression as markers of aggressive behavior. Mod Pathol. 2008;21:647–652. doi: 10.1038/modpathol.2008.44. [DOI] [PubMed] [Google Scholar]
- 8.Kreiger PA, Judkins AR, Russo PA, et al. Loss of INI1 expression defines a unique subset of pediatric undifferentiated soft tissue sarcomas. Mod Pathol. 2009;22:142–150. doi: 10.1038/modpathol.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trobaugh-Lotrario AD, Tomlinson GE, Finegold MJ, et al. Small cell undifferentiated variant of hepatoblastoma: adverse clinical and molecular features similar to rhabdoid tumors. Pediatr Blood Cancer. 2009;52:328–334. doi: 10.1002/pbc.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson EM, Shaikh TH, Gururangan S, et al. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet. 2007;122:117–127. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- 11.Kordes U, Gesk S, Fruhwald MC, et al. Clinical and molecular features in patients with atypical teratoid rhabdoid tumor or malignant rhabdoid tumor. Genes Chromosomes Cancer. 2010;49:176–181. doi: 10.1002/gcc.20729. [DOI] [PubMed] [Google Scholar]
- 12.Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer. 2007;46:617–634. doi: 10.1002/gcc.20457. [DOI] [PubMed] [Google Scholar]
- 13.Kato MV, Ishizaki K, Shimizu T, et al. Parental origin of germ-line and somatic mutations in the retinoblastoma gene. Hum Genet. 1994;94:31–38. doi: 10.1007/BF02272838. [DOI] [PubMed] [Google Scholar]
- 14.Klutz M, Brockmann D, Lohmann DR. A parent-of-origin effect in two families with retinoblastoma is associated with a distinct splice mutation in the RB1 gene. Am J Hum Genet. 2002;71:174–179. doi: 10.1086/341284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulsebos TJM, Plomp AS, Wolterman RA, et al. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–810. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd C, Smith MJ, Kluwe, et al. Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clin Genet. 2008;74:358–366. doi: 10.1111/j.1399-0004.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 17.Swensen JJ, Keyser J, Coffin CM, et al. Familial occurrence of schwannomas and malignant rhabdoid tumour associated with a duplication in SMARCB1. J Med Genet. 2009;46:68–72. doi: 10.1136/jmg.2008.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson EM, Sievert AJ, Gai X, et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15:1923–1930. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sévenet N, Lellouch-Tubiana A, Schifield D, et al. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Molc Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- 20.Taylor MD, Gokgoz N, Andrulis IL, et al. Familial posterior fossa brain tumors of infancy secondary to germline mutation of the hSNF5 gene. Am J Hum Genet. 2000;66:1403–1406. doi: 10.1086/302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruggers CS, Bleyl SB, Pysher T, et al. Clinicopathologic comparison of familial versus sporadic atypical teratoid/rhabdoid tumors (AT/RT) of the central nervous system. Pediatr Blood Cancer. doi: 10.1002/pbc.22757. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judkins AR, Mauger J, Ht A, et al. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol. 2004;28:644–650. doi: 10.1097/00000478-200405000-00013. [DOI] [PubMed] [Google Scholar]
- 23.Bourdeaut F, Freneaus P, Thuille B, et al. hSNF5/INI1-deficient tumours and rhabdoid tumours are convergent but not fully overlapping entities. J Pathol. 2007;211:323–330. doi: 10.1002/path.2103. [DOI] [PubMed] [Google Scholar]
- 24.Sévenet N, Sheridan E, Amram D, et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulsebos TJM, Kenter SB, Jakobs MF, et al. SMARCB1/INI1 maternal germ line mosaicism in schwannomatosis. Clin Genet. 2010;77:86–91. doi: 10.1111/j.1399-0004.2009.01249.x. [DOI] [PubMed] [Google Scholar]
- 26.Schwab AL, Tuohy TMF, Condie M, et al. Gonadal mosaicism and familiar adenomatous polyposis. Fam Cancer. 2008;7:173–177. doi: 10.1007/s10689-007-9169-1. [DOI] [PubMed] [Google Scholar]
- 27.Mohammed AM, Kamel AK, Hammad SA, et al. Constitutional retinoblastoma gene deletion in Egyptian patients. World J Pediatr. 2009;5:222–225. doi: 10.1007/s12519-009-0042-1. [DOI] [PubMed] [Google Scholar]
- 28.Lazaro C, Ravella A, Gaona A, et al. Neurofibromatosis type 1 due to germ-line mosaicism in a clinically normal father. NEJM. 1994;331:1403–1407. doi: 10.1056/NEJM199411243312102. [DOI] [PubMed] [Google Scholar]
- 29.Schneppenheim R, Fruhwald MC, Gesk S, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janson K, Nedzi LA, David O, et al. Predisposition to atypical teratoid/ rhabdoid tumor due to an inherited INI1 mutation. Pediatr Blood Cancer. 2006;47:279–284. doi: 10.1002/pbc.20622. [DOI] [PubMed] [Google Scholar]