Abstract
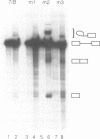
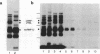
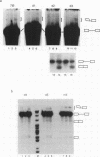
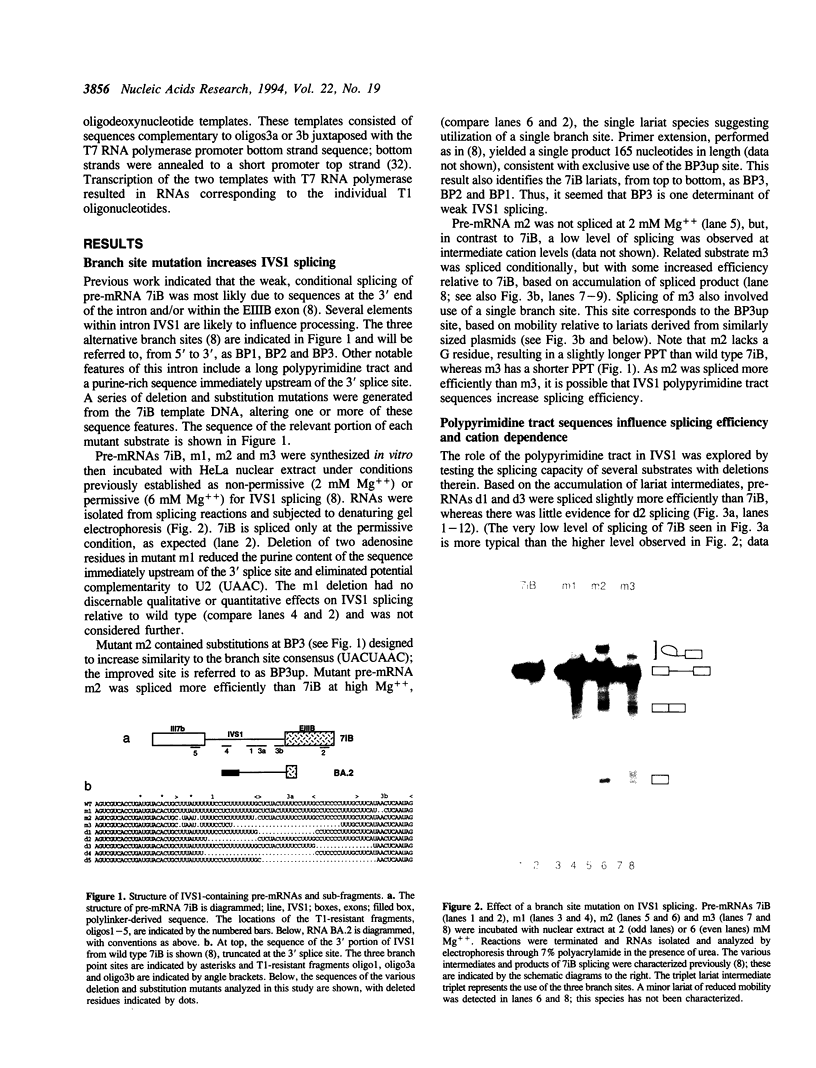
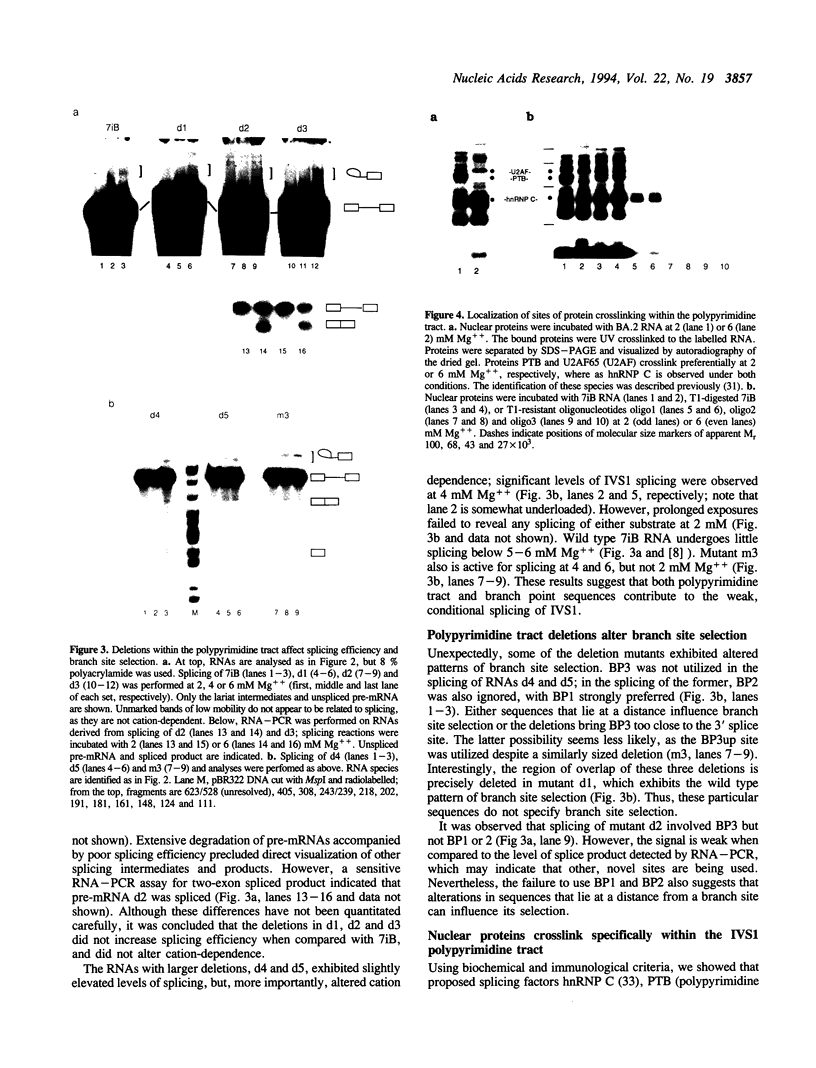
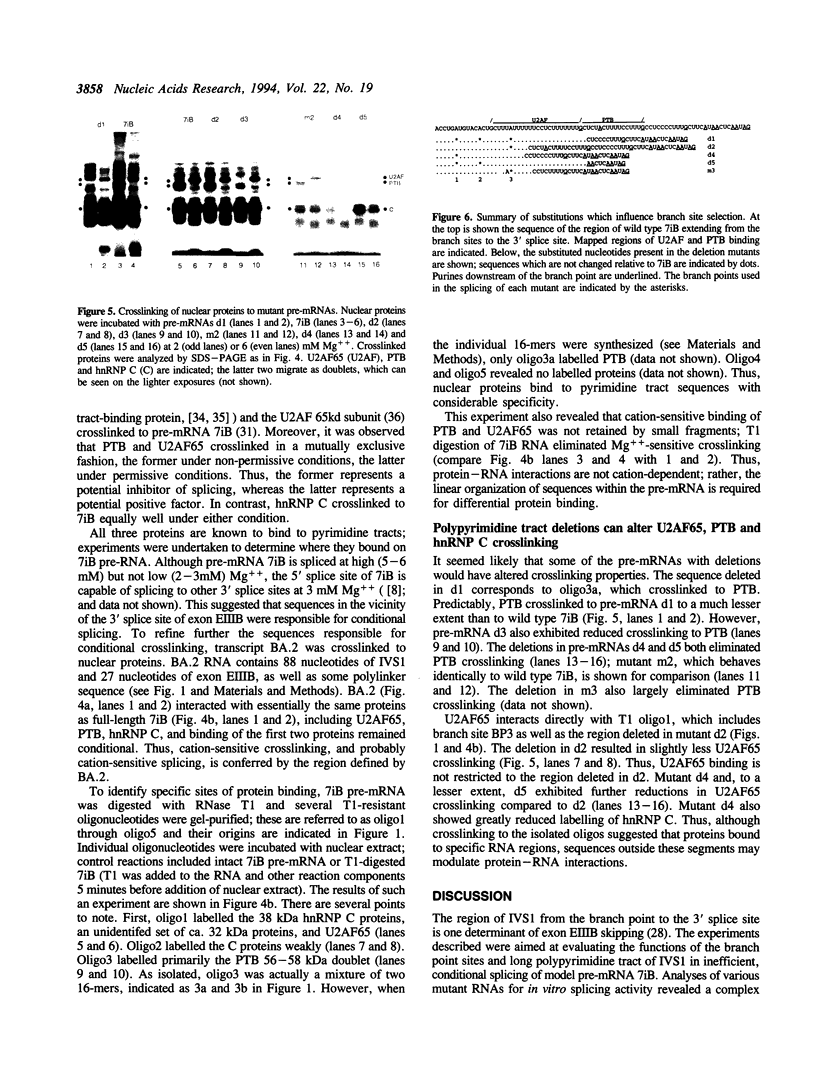
IVS1, an intron derived from the rat fibronectin gene, is spliced inefficiently in vitro, involving the use of three alternative branch sites. Mutation of one branch point site, BP3, so as to increase complementarity to U2 snRNA resulted in exclusive use of that site and improved splicing efficiency, indicating that the wild type BP3 site is one determinant of poor IVS1 splicing. Deletions within the polypyrimidine tract had a variable effect on splicing efficiency and altered the pattern of branch site usage. Selection of each branch site was influenced negatively by purine substitutions ca. 20 nucleotides downstream. It is proposed that all three IVS1 branch sites are pyrimidine tract-dependent. Pyrimidine tract deletions also influenced the crosslinking of PTB (the polypyrimidine tract-binding protein), hnRNP C, and splicing factor U2AF65. All three proteins bound preferentially to distinct regions within the polypyrimidine tract and thus are candidates for mediating pyrimidine tract-dependent branch site selection. The findings indicate the complexity of the IVS1 polypyrimidine tract and suggest a crucial role for this region in modulating branch site selection and IVS1 splicing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., O'Brien J. P., Hurwitz J. Alterations in the polypyrimidine sequence affect the in vitro splicing reactions catalyzed by HeLa cell-free preparations. J Biol Chem. 1989 Sep 5;264(25):14631–14637. [PubMed] [Google Scholar]
- Fu X. D., Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3' splice site. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Gattoni R., Schmitt P., Stevenin J. In vitro splicing of adenovirus E1A transcripts: characterization of novel reactions and of multiple branch points abnormally far from the 3' splice site. Nucleic Acids Res. 1988 Mar 25;16(6):2389–2409. doi: 10.1093/nar/16.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I. R., Hamshere M., Eperon I. C. Alternative splicing of a human alpha-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992 Sep;12(9):3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Huh G. S., Hynes R. O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993 Sep;13(9):5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison S. F., Crow A., Garcia-Blanco M. A. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol. 1992 Oct;12(10):4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991 Jun;10(6):1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- McKeown M. Alternative mRNA splicing. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Kakegawa T., Kaspar R. L., White M. W. Polypyrimidine tracts and their binding proteins: regulatory sites for posttranscriptional modulation of gene expression. Biochemistry. 1993 Mar 30;32(12):2931–2937. doi: 10.1021/bi00063a001. [DOI] [PubMed] [Google Scholar]
- Mulligan G. J., Guo W., Wormsley S., Helfman D. M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992 Dec 15;267(35):25480–25487. [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Norton P. A. Alternative pre-mRNA splicing: factors involved in splice site selection. J Cell Sci. 1994 Jan;107(Pt 1):1–7. doi: 10.1242/jcs.107.1.1. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Hynes R. O. Characterization of HeLa nuclear factors which interact with a conditionally processed rat fibronectin pre-mRNA. Biochem Biophys Res Commun. 1993 Aug 31;195(1):215–221. doi: 10.1006/bbrc.1993.2032. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Hynes R. O. In vitro splicing of fibronectin pre-mRNAs. Nucleic Acids Res. 1990 Jul 25;18(14):4089–4097. doi: 10.1093/nar/18.14.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton P. A., McInnis R. Selective enzymatic depletion during RNA-PCR: improved detection of rare RNA forms. Biotechniques. 1993 Jun;14(6):954–957. [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988 Nov;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Patton J. G., Mayer S. A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991 Jul;5(7):1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Patton J. G., Porro E. B., Galceran J., Tempst P., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993 Mar;7(3):393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Roscigno R. F., Weiner M., Garcia-Blanco M. A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem. 1993 May 25;268(15):11222–11229. [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Valcárcel J., Singh R., Zamore P. D., Green M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993 Mar 11;362(6416):171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Patton J. G., Green M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992 Feb 13;355(6361):609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]