Abstract
Recent advances in sequencing technology make it possible to comprehensively catalogue genetic variation in population samples, creating a foundation for understanding human disease, ancestry and evolution. The amounts of raw data produced are prodigious and many computational steps are required to translate this output into high-quality variant calls. We present a unified analytic framework to discover and genotype variation among multiple samples simultaneously that achieves sensitive and specific results across five sequencing technologies and three distinct, canonical experimental designs. Our process includes (1) initial read mapping; (2) local realignment around indels; (3) base quality score recalibration; (4) SNP discovery and genotyping to find all potential variants; and (5) machine learning to separate true segregating variation from machine artifacts common to next-generation sequencing technologies. We discuss the application of these tools, instantiated in the Genome Analysis Toolkit (GATK), to deep whole-genome, whole-exome capture, and multi-sample low-pass (~4×) 1000 Genomes Project datasets.
Introduction
Recent advances in NGS technology now provide the first cost-effective approach to large-scale resequencing of human samples for medical and population genetics. Projects such as the 1000 Genomes 1, The Cancer Genome Atlas and numerous large medically-focused exome sequencing projects 2 are underway in an attempt to elucidate the full spectrum of human genetic diversity 1 and the complete genetic architecture of human disease. The ability to examine the entire genome in an unbiased way will make possible comprehensive searches for standing variation in common disease; mutations underlying linkages in Mendelian disease 3; as well as spontaneously arising variation for which no gene-mapping shortcuts are available (e.g., somatic mutations in cancer 4–6 and de novo mutations 7,8 in autism and schizophrenia).
Many capabilities are required to obtain a complete and accurate record of the variation from NGS from sequencing data. Mapping reads to the reference genome9–12 is a first critical computational challenge whose cost necessitates each read be aligned independently, guaranteeing many reads spanning indels will be misaligned. The per-base quality scores, which convey the probability that the called base in the read is the true sequenced base 13, are quite inaccurate and co-vary with features like sequencing technology, machine cycle and sequence context 14–16. These misaligned reads and inaccurate quality scores propagate into single nucleotide polymorphism (SNP) discovery and genotyping, a general problem that becomes acute in projects with multiple sequencing technologies, generated by many centers using rapidly evolving experimental processing pipelines, such as the 1000 Genomes Project.
Given well mapped, aligned, and calibrated reads, resolving even simple SNPs, let alone more complex variation such as multi-nucleotide substitutions, insertions and deletions, inversions, rearrangements, and copy number variation requires sensitive and specific statistical models 9–12,16–24. Separating true variation from machine artifacts due to the high rate and context-specific nature of sequencing errors is the outstanding challenge in NGS analysis. Previous approaches have relied on filtering SNP calls that exhibit characteristics outside of their normal ranges, such as occurring at sites with too much coverage 18,20, or by requiring non-reference bases to occur on at least three reads in both synthesis orientations 21. Though effective, such hard filters are frustratingly difficult to develop, require parameterization for each new data set, and are necessarily either restrictive (high specificity, as in 1000 Genomes) or tolerant (high sensitivity, used in Mendelian disease studies, with concomitantly more false positives). Moreover, all of these challenges must be addressed within the context of a proliferation of sequencing technology platforms and study designs (e.g. whole genome shotgun, exome capture sequencing, multiple samples sequenced at shallow coverage), a point not tackled in previous work.
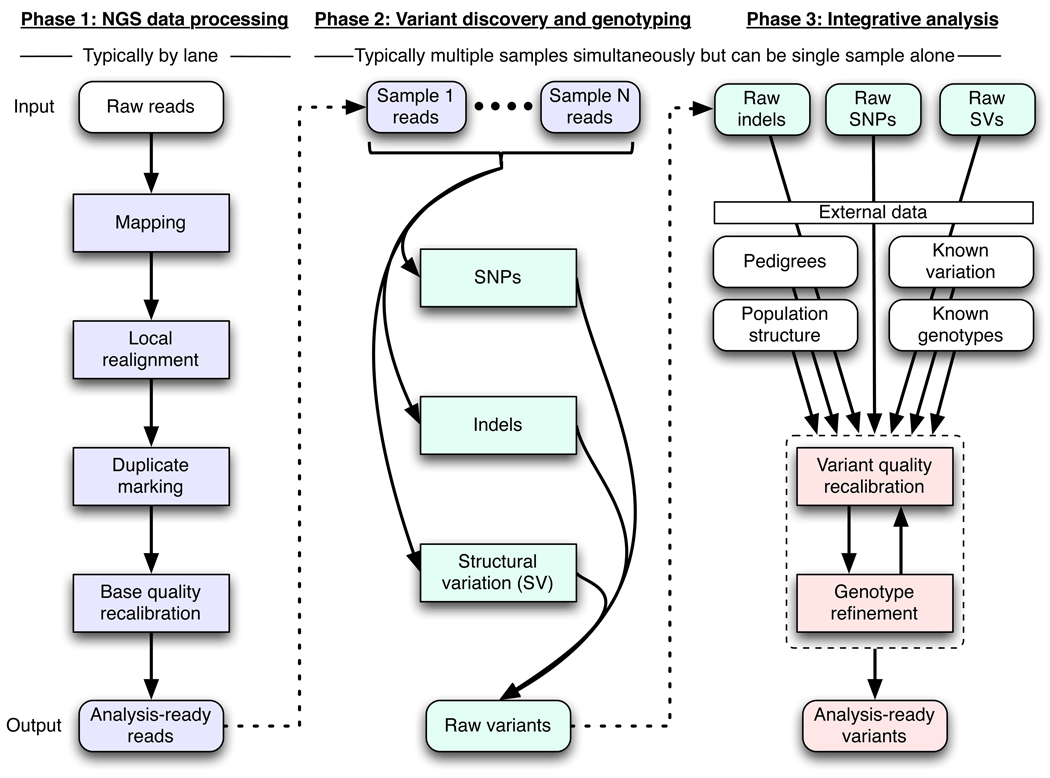
Here we present a single framework and associated tools capable of discovering high-quality variation and genotyping individual samples using diverse sequencing machines and experimental designs (Figure 1). We present several novel methods addressing the challenges listed above in local realignment, base quality recalibration, multi-sample SNP calling and adaptive error modeling, which we apply to three prototypical NGS data sets (Table 1). In each data set we include CEPH individual NA12878 to demonstrate the consistency of results for this individual across all three data sets.
Figure 1.
Framework for variation discovery and genotyping from next-generation DNA sequencing. See text for a detailed description.
Table 1.
Next-generation DNA sequencing data sets analyzed
| HiSeq | Exome | Low-pass | |
|---|---|---|---|
| Samples | NA12878 | NA12878 | NA12878 + 60 unrelated CEPH individuals |
| Sequencing technologies | Whole genome shotgun; Illumina HiSeq 2000 18 | Agilent exome hybrid capture 32,33; Illumina GenomeAnalyzer 18 | Whole genome shotgun; Illumina GenomeAnalyzer 18; Life/SOLiD 34; Roche/454 20 |
| Coverage per sample | ~60× | ~150×; 93% of bases at >20× coverage | ~4× |
| Read architecture | 101bp paired end | 76/101bp paired end | 25, 36, 51, 76, ~250 (454) bp single and paired ends |
| Targeted area | 2.85 Gb of autosomes and chrX | 28 Mb | 2.85 Gb of autosomes and chrX |
| Data set source | Novel, generated for this article | Novel, generated for this article | 1000 Genomes Project |
| Aligner(s) | BWA 11 | MAQ 10 | MAQ 10; Corona Lite; SSAHA 12 |
Results
Here we describe a three-part conceptual framework (Figure 1):
Phase 1: raw read data with platform-dependent biases is transformed into a single, generic representation with well-calibrated base error estimates, mapped to their correct genomic origin, and aligned consistently with respect to one another. Mapping algorithms place reads with an initial alignment on the reference genome, either generated in, or converted to, the technology-independent SAM/BAM reference file format 25. Next, molecular duplicates are eliminated (Suppl. Mats), initial alignments are refined by local realignment, and then an empirically accurate per-base error model is determined.
Phase 2: the analysis-ready SAM/BAM files are analyzed to discover all sites with statistical evidence for an alternate allele present, among the samples including SNPs, short indels, and CNVs. CNV discovery and genotyping methods, though part of this conceptual framework, are described elsewhere 26.
Phase 3: technical covariates, known sites of variation, genotypes for individuals, linkage disequilibrium, and family and population structure are integrated with the raw variant calls from phase 2 to separate true polymorphic sites from machine artifacts, and at these sites high-quality genotypes are determined for all samples.
All components after initial mapping and duplicate marking are instantiated in the Genome Analysis ToolKit (GATK) 27.
Applying the analysis pipeline to HiSeq data at ˜60× of NA12878
2.72B bases (~96%) of the 2.83B non-N bases in the autosomal regions and chromosome X of the human reference genome have sufficient coverage to call variants in the 101bp paired-ended HiSeq data (Table 1). Even though the HiSeq reads were aligned with the gap-enabled BWA, more than 15% of the reads that span known homozygous indels in NA12878 are misaligned (Supplemental Table 1). Realignment corrects 6.6M of 2.4B total reads in 950K regions covering 21Mb in the HiSeq data, eliminating 1.8M loci with significant accumulation of mismatching bases (Supplemental Table 2). The initial data processing steps (Phase 1) eliminate ~300K SNP calls, more than one fifth of the raw novel calls, with quality metrics consistent with more than 90% of these SNPs being false positives (Table 2).
Table 2.
Raw to recalibrated, imputed SNP calls HiSeq, Exome, and 61 sample low-pass data sets. Part one of each section summarizes the impact of local realignment and base quality recalibration by comparing SNP calls on reads with raw quality scores and alignments to those made on the realigned, recalibrated reads.
| Site discovery | Comparison to NA12878 variants | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNPs | Ti/Tv | HM3 concordance | HM3 concordance | |||||||
| Call set | All | Known | Novel | dbSNP% | Known | Novel | NR sensitivity |
NRD rate | NR sensitivity |
NRD rate |
| HiSeq | ||||||||||
| Raw reads, all calls | 4.43M | 3.49M | 941K | 78.77 | 2.05 | 1.29 | 99.74 | 0.10 | 99.57 | 0.20 |
| Unique to raw read calls | 263K | 37K | 226K | 13.95 | 1.37 | 0.70 | 0.02 | 37.97 | 0.09 | 12.64 |
| Unique to +recal/+MSA calls | 9.8K | 1.8K | 8.0K | 18.08 | 1.38 | 1.39 | 0.00 | 18.18 | 0.00 | 9.93 |
| +recal/+MSA, all calls | 4.18M | 3.45M | 722K | 82.71 | 2.06 | 1.57 | 99.72 | 0.09 | 99.48 | 0.19 |
| Filtered by variant recalibration | 595K | 235K | 360K | 39.44 | 1.19 | 1.21 | 0.67 | 3.00 | 2.2 | 4.31 |
| Final call set | 3.58M | 3.22M | 362K | 89.89 | 2.15 | 2.05 | 99.05 | 0.07 | 97.28 | 0.10 |
| Low-pass | ||||||||||
| Raw reads, all calls | 13.4M | 6.5M | 6.9M | 48.77 | 2.05 | 1.13 | 83.97 | 20.34 | 80.45 | 22.53 |
| Unique to raw read calls | 670K | 32K | 638K | 4.74 | 1.19 | 0.67 | 0.01 | 49.21 | 0.02 | 52.57 |
| Unique to +recal/+MSA calls | 45K | 2.5K | 42K | 5.62 | 0.94 | 0.68 | 0.00 | N/A | 0.00 | 38.89 |
| +recal/+MSA, all calls | 12.8M | 6.5M | 6.3M | 50.92 | 2.06 | 1.18 | 83.97 | 20.33 | 80.43 | 22.52 |
| Filtered by variant recalibration | 5.5M | 706K | 4.8M | 12.84 | 1.31 | 1.01 | 0.95 | 26.54 | 3.44 | 32.91 |
| Variant recalibrated call set | 7.3M | 5.8M | 1.5M | 79.7 | 2.18 | 2.05 | Itemized below | |||
| Sample variant calls for NA12878 only | ||||||||||
| Variant recalibrated NGS reads only | 2.44M | 2.30M | 140K | 94.28 | 2.15 | 2.06 | 83.02 | 20.26 | 76.99 | 22.01 |
| Recalibrated with Beagle imputation | 3.20M | 3.01M | 191K | 94.03 | 2.18 | 2.09 | 96.72 | 3.32 | 91.21 | 3.35 |
| Exome capture | ||||||||||
| Raw reads, all calls | 18.9K | 16.8K | 2.1K | 88.83 | 3.20 | 1.16 | 99.10 | 0.09 | 99.12 | 0.12 |
| Unique to raw read calls | 483 | 39 | 444 | 8.07 | 2.55 | 0.31 | 0.04 | 25.00 | 0.03 | 33.33 |
| Unique to +recal/+MSA calls | 81 | 40 | 41 | 49.38 | 3.44 | 1.73 | 0.01 | 0.00 | 0.04 | 16.67 |
| +recal/+MSA, all calls | 18.5K | 16.8K | 1.7K | 90.77 | 3.20 | 1.61 | 99.07 | 0.08 | 99.13 | 0.11 |
| Filtered by variant recalibration | 1274 | 609 | 665 | 47.8 | 1.85 | 0.84 | 0.59 | N/A | 0.76 | N/A |
| Final call set | 17.2K | 16.2K | 1039 | 93.96 | 3.27 | 2.57 | 98.49 | 0.08 | 98.38 | 0.11 |
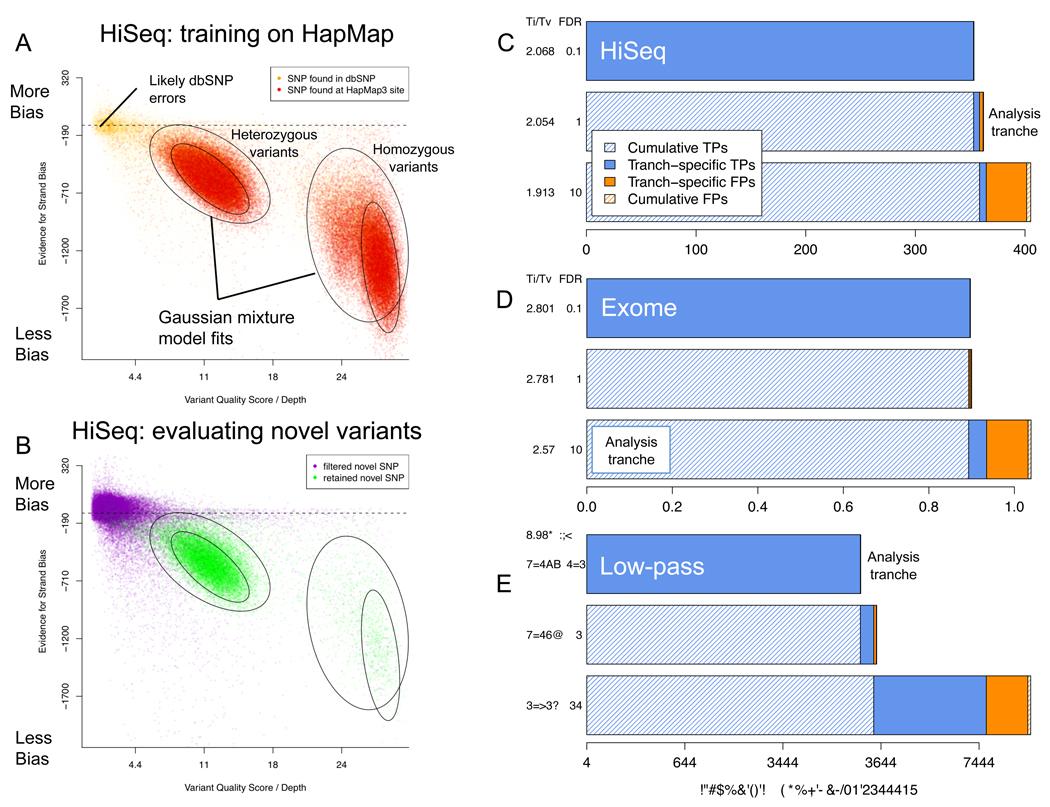
The initial 4.2M confidently called non-reference sites include 99.7% and 99.5% of the HapMap3 and 1KG Trio sites genotyped as non-reference in NA12878; at these variant sites the sequencing and genotyping calls are concordant 99.9% of the time (Table 2). Variant quality score recalibration of these initial calls identifies a tranche of SNPs with estimated FDR of <1% containing 3.2M known variants and 362K novel variants, a 90% dbSNP rate, and Ti/Tv ratios of 2.15 and 2.05, respectively, consistent with our genome-wide expectations (Box 1). While the variant recalibrator removed ~595K total variants with a Ti/Tv ratio of ~1.2, it retained 99% and 97.3% of the HapMap3 and 1KG Trio non-reference sites. The discordant sites have 100× higher genotype discrepancy rates, suggesting that the sites themselves may be problematic. Almost all of the variants in the 1% tranche are already present in the even higher stringency 0.1% FDR tranche, while analysis of the 10% FDR tranche suggest that some more variants could be obtained, at the cost of many more false positives (Figure 4).
Figure 4.
(a) Relationship in the HiSeq call set between strand bias and quality by depth, for genomic locations in HapMap3 (red) and dbSNP (yellow) used for training the variant quality score recalibrator (left) and the same annotations applied to differentiate likely true positive (green) from false positive (purple) novel SNPs. (b,c,d) Quality tranches in the recalibrated HiSeq (b), exome (c), and low-pass CEU (d) calls beginning with (top) the highest-quality but smallest call set with an estimated false positive rate among novel SNP calls of <1/1000 to a more comprehensive call set (bottom) that includes effectively all true positives in the raw call set along with more false positive calls for a cumulative false positive rate of 10%. Each successive call set contains within it the previous tranche’s true and false positive calls (shaded bars) as well as tranche-specific calls of both classes (solid bars). The tranche selected for further analyses here is indicated.
Applying the analysis pipeline to 28Mb exome capture at ˜150× of NA12878
The raw data processing tools here eliminated ~450 novel call sites from the pre-MSA/pre-recal call set, representing more than 20% of all the novel calls, with a Ti/Tv of 0.30 - fully consistent with all being false positives - while adding several sites present in HapMap3 and the 1KG Trio. The raw whole exome data call set, at ~150× coverage (Table 1), includes >99% of both the HapMap3 and 1KG Trio non-reference sites within the 28Mb exome target region, with >99.8% genotype concordance at these sites. As with HiSeq, even with recalibration and local realignment, however, the Ti/Tv ratio of the novel sites in the initial SNP calls indicates that more than 50% of these calls are false positives. Variant quality score recalibration, using only ~5400 SNPs for training, identifies a high-quality subset of calls that capture >98% of the HapMap3 and 1KG Trio sites in the target regions. The value of the tranches is more pronounced in the whole exome (Figure 4d), where 900 of the 1039 novel calls come from tranches with FDRs under 1%, despite needing to reach into the 10% FDR tranche to include most true positive SNPs.
The Hiseq WGS and exome capture datasets differ drastically in their sequencing protocols (WGS vs. hybrid capture), the sequencing machines (HiSeq vs. GA), and the initial alignment tools (BWA vs. MAQ). Neverthless, the exome call set is remarkably consistent the subset of calls from HiSeq that overlap the target regions of the hybrid capture protocol. 94% of the HiSeq calls are also called in the final exome set sliced at 10% FDR (data not shown), and at these sites the non-reference discrepancy rate is extremely low (<0.4%). Mapping differences between the aligners used for HiSeq (BWA) and exome (MAQ) data sets account for vast the majority of these discordant calls, with the remainder of the differences due to limited coverage in the exome, and only a small minority of sites due to differential SNP calling or variant quality score recalibration. Overall, despite the technical differences in the capture and sequencing protocols of the HiSeq and Exome data sets, the data processing pipeline presented here uncovers a remarkably consistent set of SNPs in exomes with excellent genotyping accuracy.
Applying the analysis pipeline to low-pass (4×) sequencing of NA12878 with 60 unrelated CEPH individuals
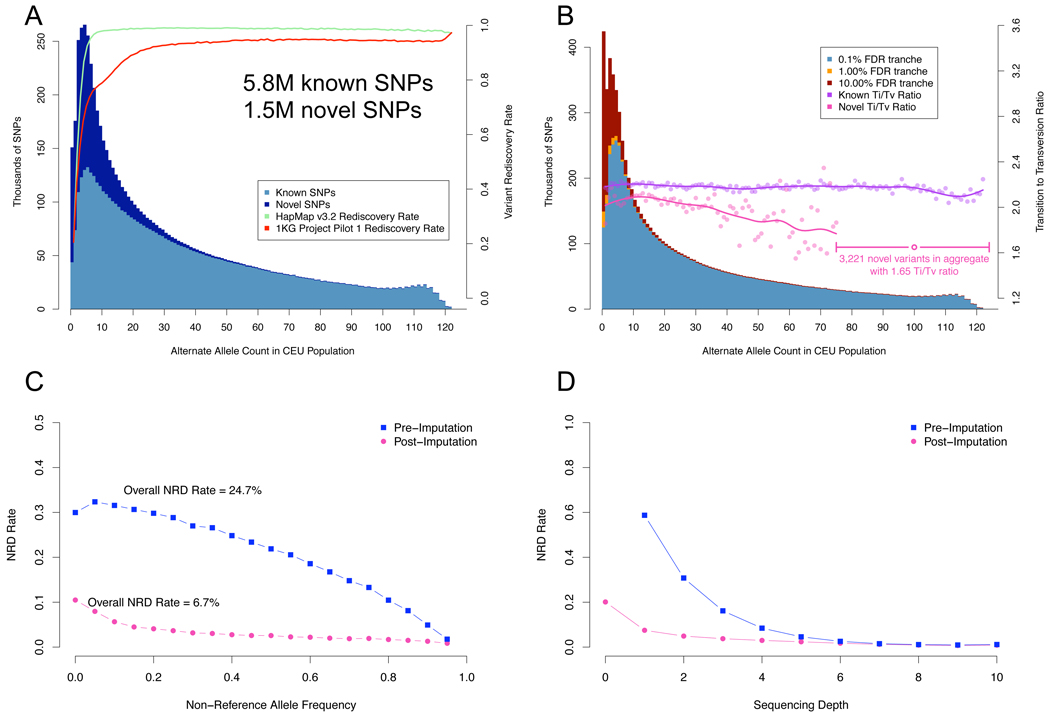
Multi-sample low-pass resequencing poses a major challenge for variant discovery and genotyping because there is so little evidence at any particular locus in the genome for any given sample (Table 1). Consequently, it is in precisely this situation where there is little signal from true SNPs that our data processing tools are most valuable, as can be seen from the progression of call sets in Table 2. Local realignment and base quality recalibration eliminate ~650K false positive SNPs among 13M sites, 4× more sites than in the HiSeq data set, with an aggregate Ti/Tv of 0.7. The initial low-pass CEU set includes over 13M called sites among all individuals, of which nearly 7M are novel. NA12878 herself has 2.9M variants, of which 430K are novel. The 4× average coverage limits the sensitivity and concordance of this call set, with only 84% and 80% of HapMap3 and 1KG Trio sites assigned a non-reference genotype in the NA12878 sample, both with a ~20% NRD rate.
The variant quality recalibrator identifies from the 13M potential variants ~6M known and 1.5M novel sites in tranches from 0.1% to 10% FDR. Figure 5a highlights several key features of the data: the allele frequency distribution of these calls closely matches the population genetics expectation and the vast majority of HapMap3 and 1000 Genomes official CEU call sites are recovered, with the proportion nearing 100% for more common variant sites (Figure 5a). Although we selected a 0.1% FDR tranche for analysis here, which contains the bulk of HapMap3, 1KG Trio, and HiSeq sites, there are another ~700K true sites can be found in the 1 and 10% FDR tranche, albeit among many more false positives. This highest quality tranche includes nearly all variants observed more than 5 times in the samples and 1.4M novels, with the SNPs in the tranches at 1% and 10% generally occupying the lower alternate allele frequency range (Figure 5b). The overall picture is clear: calling multiple samples simultaneously, even with only a handful of reads spanning a SNP for any given sample, enables one to detect the vast majority of common variant sites present in the cohort with a high degree of sensitivity.
Figure 5.
Variation discovered among 60 individuals from the CEPH population from 1000 Genomes pilot phase plus low-pass NA12878. (a) Discovered SNPs by non-reference allele count in the 61 CEPH cohort, colored by known (light blue, striped) and novel (dark blue, filled) variation, along with non-reference sensitivity to CEU HapMap3 and 1000 Genomes low-pass variants. (b) Quality and certainty of discovered SNPs by non-reference allele count. The histogram depicts the certainty of called variation broken out into 0.1, 1, and 10% novel FDR tranches. The Ti/Tv ratio is shown for known and novel variation for each allele count, aggregating the novel calls with allele count > 74 due to their limited numbers. (c,d) Genotyping accuracy for NA12878 from reads alone (blue circles) and following genotype-likelihood based imputation (pink squares) called in the 61 sample call set as assessed by the NRD rate to HiSeq genotypes, as a function of allele count (c) and sequencing depth (d).
While the bulk properties of the 61-sample call set are good, we expect the low-pass 4× design to limit variation discovery and genotyping in each sample relative to deep re-sequencing. In the 61 sample call set we discover ~80% of the non-reference sites in NA12878 according to HapMap3, 1KG Trio, and HiSeq call sets (Table 2). The ~20% of the missed variant sites from these three data sets had little to no coverage in the NA12878 sample in the low-pass data and, therefore, could not be assigned a genotype using only the NGS data, a general limitation of the low-pass sequencing strategy (Table 2, Figure 5c/d). The multi-sample discovery design, however, affords us the opportunity to apply imputation to refine and recover genotypes at sites with little or no sequencing data. Applying genotype-likelihood based imputation with Beagle 28 to the 61 sample call set recovers an additional 15–20% of the non-reference sites in NA12878 that had insufficient coverage in the sequencing data (Table 2) as well as vastly improving genotyping accuracy (Figure 5c/d).
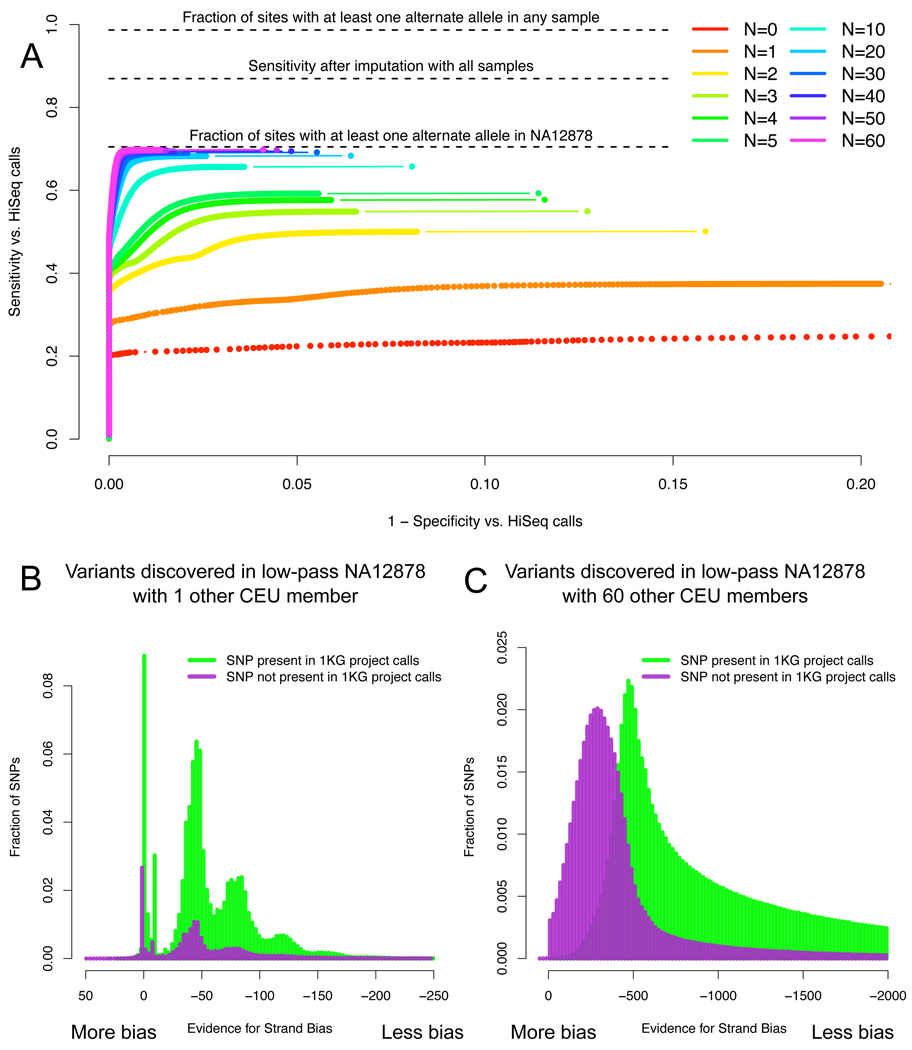
We further characterize the quality of our low-pass call set as a function of the number of samples included during the discovery process in addition to NA12878 herself. Increasing the number of samples in the cohort rapidly improves both sensitivity and specificity of the call set. As evidence mounts with more samples that a particular site is polymorphic, our confidence in the call increases and the site is more likely to be called (Figure 6a). Distinguishing true positive variants from sequencing and data processing artifacts is more difficult with few samples and, consequently, low aggregated coverage; adding more reads empowers the error covariates to identify sites as errors by the variant recalibrator (Figure 6b and 6c).
Figure 6.
Sensitivity and specificity of multi-sample discovery of variation in NA12878 with increasing cohort size for low-pass NA12878 read sets processed with N additional CEPH samples. (a) Receiver operating characteristic (ROC) curves for SNP calls relating specificity and sensitivity to discover non-reference sites from the NA12878 HiSeq call set. The maximum callable sensitivity, 66%, is the percent of sites from the HiSeq call set where at least one read carries the alternate allele in the low-pass data for NA12878; it reflects both differences in the sequencing technologies (36–76bp GAII for the low-pass NA12878 sample vs. 101bp HiSeq) as well as the vagaries of sampling at 4× coverage. Because most of these missed sites are common and are consequently called in the other samples, imputation recovers ~50% of these sites. (b,c) Increasing power to identify strand-biased, likely false positive SNP calls with additional samples. Histograms of the Strand Bias annotation at raw variant calls discovered in the low-pass CEU data using NA12878 at 4× combined with one other CEU individual (b) and with 60 other individuals (c) stratified into sites present (green) and not (purple) in the 1000 Genomes CEU trio.
The combination of multi-sample SNP calling, variant quality recalibration using error covariates, and imputation allows one to achieve a high-quality call set, both in aggregate and per-sample, with astoundingly little data. The aggregated 61-sample set at 4× coverage includes only four times as much sequencing data as the HiSeq data, yet we discover 3.2M polymorphic sites in NA12878, which includes 97%, 91%, and 87% of the variants in HapMap3, 1000 Genomes Trio, and HiSeq call sets, respectively, while also finding ~5M additional variants among the 60 other samples.
Comparison of hard filtering to variant quality score recalibration
Supplemental Table 3 lists the quality of call sets derived using our previous filtering approaches on all three data sets relative to the adaptive recalibrator described here. In all cases the adaptive approach outperforms the manually optimized hard filtering previously developed for this calling system for the 1000 Genomes pilot data. This highlights two important points – first, that a principled integration of all covariates (which may have a complex correlation structure) should and does outperform single manually defined thresholds on covariates independently, with the added benefit of not requiring human intervention; second, that an accurate ranking of discovered putative variants by the probability that each represents a true site permits the definition of tranches for specificity or sensitivity (Figure 4c–e) as appropriate to the needs of the specific project. Although the most permissive tranche includes almost all sites that have any chance of being true polymorphisms – critical for projects looking for single large effect mutations – the vast majority of true polymorphisms are present in the highest quality tranche of data (not shown).
Comparison of this calling pipeline to Crossbow
To calibrate the additional value of the tools described here we contrast our results with SNPs called on our raw NA12878 exome data using Crossbow 29, a package combining bowtie, a gapless read mapping tool based on the Burrows-Wheeler transformation 30 and SoapSNP for SNP detection 16. We chose to perform this analysis on the exome data because its wide range of read depths and complex error modes make SNP calling a challenge, especially given the small number of novel variants (~1000 per sample) expected in this 28Mb target. In Supplemental Table 4 the high-level results of the GATK and Crossbow calling pipelines are compared and contrasted. Key metrics such as the number of novel SNP calls, their Ti/Tv ratio, the number of calls not seen in either the 1000G trio or the HiSeq data, and the high nonsense/read-through rates indicate that the Crossbow call set has lower specificity than the GATK pipeline. This is the case despite applying a aggressive P-value threshold (P < 0.01) for the base quality rank sum test 16 to filter false positive variants, which reduces the sensitivity to HM3, 1000G, and the HiSeq call sets by >3%. As usual, the intersection set between GATK and Crossbow is more specific but less sensitive than the calls unique to each pipeline (Table 1), a clear sign that despite the advances presented here significant work remains in perfecting calling in data sets like single sample exome capture. Although the value of the data processing and error modeling presented here is also clear, applying local realignment and base quality score recalibration -- publicly available, easy-to-use modules in the GATK -- are likely to improve the results of the Crossbow pipeline.
Discussion
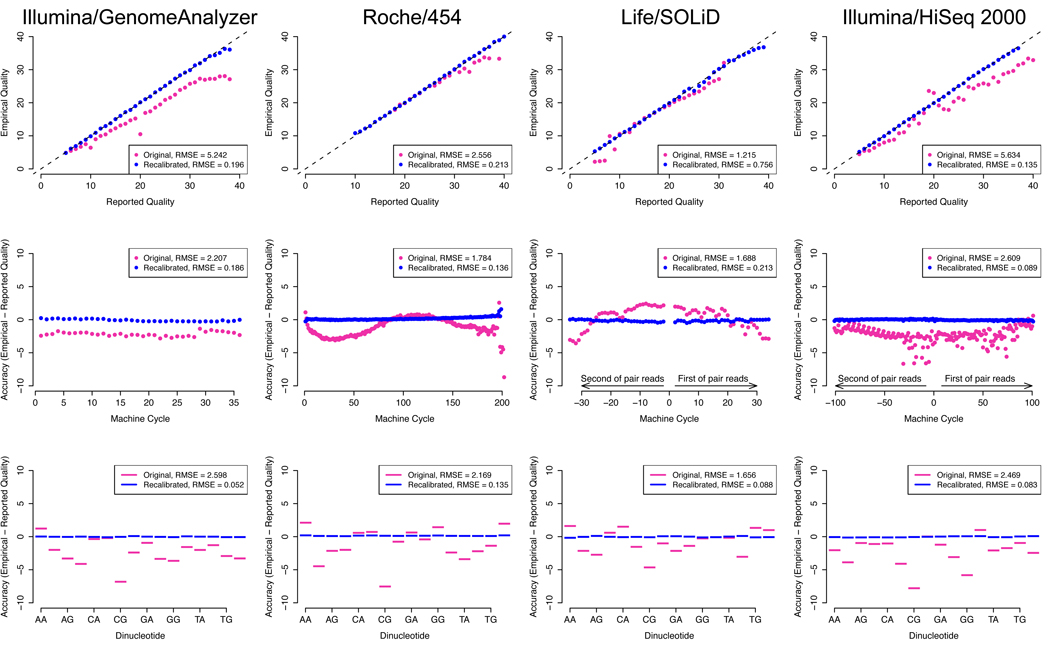
The inaccuracy and covariation patterns differ strikingly between sequencing technologies (Figure 3), which if uncorrected can propagate into downstream analyses. Accurately recalibrated base quality scores eliminates these sequencer-specific biases (Figure 3) and enables integration of data generated from multiple systems. Although developed for early NGS data sets like those from the 1000 Genomes Project pilot, the impact of recalibration is still significant even for data emerging today on newer sequencers like the HiSeq 2000. Together with local realignment, these two data processing methods eliminate millions of mostly false positive variants while preserving nearly all truly variable sites, such as those in HapMap3 and 1KG Trio sites (Table 2). In single sample data sets, such as HiSeq and exome, without realignment and recalibration these false variants account for more than a fifth of all of the novel calls.
Figure 3.
Raw (violet) and recalibrated (blue) base quality scores for NGS paired end read sets of NA12878 of (a) Illumina/GA (b) Life/SOLiD and (c) Roche/454 lanes from 1000 Genomes, and (d) Illumina/HiSeq. For each technology: top panel: shows reported base quality scores compared to the empirical estimates (Methods); middle panel: the difference between the average reported and empirical quality score for each machine cycle, with positive and negative cycle values given for the first and second read in the pair, respectively; bottom panel: the difference between reported and empirical quality scores for each of the 16 genomic dinucleotide contexts. For example, the AG context occurs at all sites in a read where G is the current nucleotide and A is the preceding one in the read. Root-mean-square errors (RMSE) are given for the pre- and post-recalibration curves.
Even with very deep coverage, the na‘ve Bayesian model for SNP calling results in an initial call set with a surprisingly large number of false-positive calls. While we expect 3.3M known and 330K novel non-reference sites in a single European sample sequenced genome-wide, the initial HiSeq call set contains 3.5M known and 800K novel calls. The excessive number of variable sites, and the low Ti/Tv ratio in particular among the novel calls, implies that ~600K of these variants are likely errors resulting from stochastic and systemic sequencing and alignment errors. The same calculations suggest that a similar fraction of the initial exome calls are likely false positives, while more than 80% of the initial novel low pass SNP calls are likely errors. The adaptive error modeling developed here enables us to identify these false positive variants based on their dissimilarity to known variants, despite error rates of 50–80% among the novel variants.
In each step of the pipeline, the improvements derive from the correction of systematic errors made in base calling or read mapping/alignment. By characterizing the specific NGS machine error processes and capturing our certainty, or lack thereof, that a putative variant is truly present in the sample or population, we deliver not a single concrete call set but a continuum from confident to less reliable variant calls for use as appropriate to the specific needs of downstream analysis. Mendelian disease projects can select a more sensitive set of calls with a higher error rate to avoid missing that single, high-impact variant, while community-resource projects like the 1000 Genomes Project can place a high premium on specificity.
The division between SNP discovery and preliminary genotyping and genotype refinement (columns 2 and 3, Figure 1) avoids embedding in the discovery phase assumptions about population structure, sample relationships, and the linkage disequilibrium relationships between variants. Consequently, our calling approach applies equally well to population samples in Hardy-Weinberg equilibrium like mother-father-child trios or interbreeding families suffering from Mendelian disorders. Critically, our framework produces highly sensitive and specific variation calls without the use of linkage disequilibrium and so can be applied in situations where LD information is unavailable or weak (many organisms) or would confound analytic goals such as studying LD patterns themselves or comparing Neanderthals and modern humans 31. Where appropriate, however, imputation can be applied to great value, as we demonstrate in the 61 sample CEU low-pass call set.
The analysis results presented here clearly indicate that even with our best current approaches we are still far from obtaining a complete and accurate picture of genetic variation of all types in even a single sample. Even with the HiSeq 101bp paired-end reads nearly 4% (~100 Mb) of the potentially callable genome is considered poorly mapped (Suppl. Mats) and analysis of variants within these regions requires care. Nearly two-thirds of the differences between the HiSeq and exome call sets can be attributed to different read mappings between BWA and MAQ.
The challenge of obtaining accurate variant calls from NGS data is substantial. We have developed an analysis framework for NGS data that achieves consistent and accurate results from a wide array of experimental design options including diverse sequencing machinery and distinct sequencing approaches. We have introduced here an integrated approach to data processing and variation discovery from NGS data that is designed to meet these specifications. Using data generated both at the Broad Institute and throughout the 1000 Genomes project, we have demonstrated that the introduction of improved calibration of base quality scores, local realignment to accommodate indels, the simultaneous evaluation of multiple samples from a population, and finally an assessment of the likelihood that an identified variable site is a true biological DNA variant significantly improves the sensitivity and specificity of variant discovery from NGS data. The impending arrival of yet more NGS technologies makes even more important modular, extensible frameworks like ours that produce high-quality variant and genotype calls despite distinct error modes of multiple technologies for many experimental designs.
Supplementary Material
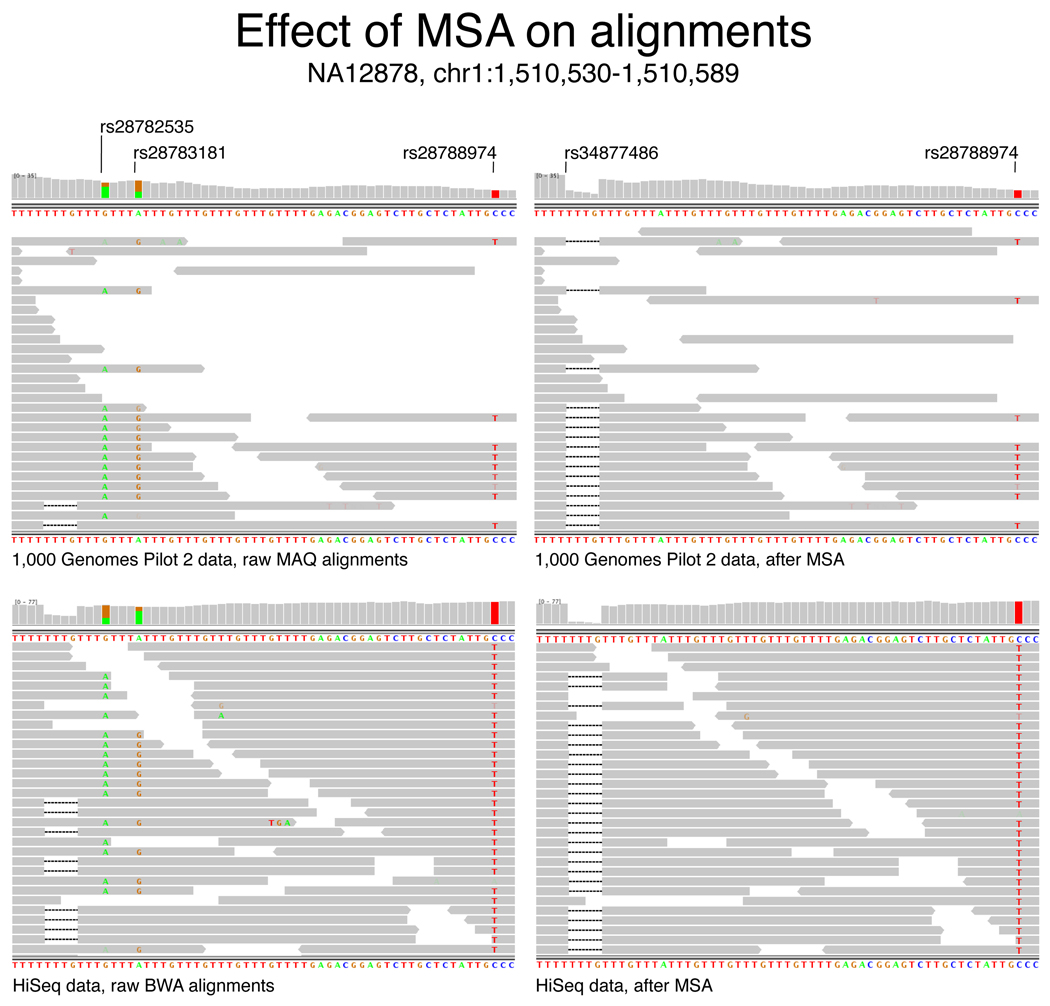
Figure 2.
IGV visualization of alignments in region chr1:1,510,446–1,510,622 from the (a) Trio NA12878 Illumina reads from 1000 Genomes and (b) NA12878 HiSeq reads before (left) and after (right) multiple sequence realignment. Reads are depicted as arrows oriented by increasing machine cycle; highlighted bases indicate mismatches to the reference: A is green, G is orange, T is red, and deleted bases are dashes; a coverage histogram per base is shown above the reads. Both the 4bp indel (rs34877486) and the C/T polymorphism (rs28788874) are present in dbSNP, as are the artifactual A/G polymorphisms (rs28782535 and rs28783181) resulting from the mis-modeled indel, indicating that these sites are common misalignment errors.
Acknowledgments
Many thanks to our colleagues in Medical and Population Genetics and Cancer Informatics and the 1000 Genomes Project who encouraged and supported us during the development of the Genome Analysis ToolKit and associated tools. This work was supported by grants from the National Human Genome Research Institute, including the Large Scale Sequencing and Analysis of Genomes grant (54 HG003067) and the Joint SNP and CNV calling in 1000 Genomes sequence data grant (U01 HG005208). We would also like to thank our excellent anonymous reviewers for their thoughtful comments.
Footnotes
Author contributions
M.A.D., E.B., R.E.P., K.V.G., J.R.M, C.H., A.A.P., G.d.A., M.A.R., T.J.F., A.Y.S., K.C. conceived of, implemented, and performed analytic approaches. M.A.D., E.B., R.E.P., K.V.G., G.d.A., A.M.K., M.J.D. wrote the manuscript. M.A.D., M.H., A.M. developed Picard and GATK infrastructure underlying the tools implemented here. M.A.D, S.B.G, D.A., M. J. D. lead the team.
References
- 1.The 1000 Genomes Project Consortium. A map of human genome variation from population scale sequencing. Nature. 2010 doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi X, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2009 doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 5.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009 doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Submitted. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Research. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Research. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- 14.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Research. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Nordborg M, Li LM. Adjust quality scores from alignment and improve sequencing accuracy. Nucleic Acids Res. 2004;32:5183–5191. doi: 10.1093/nar/gkh850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Research. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drmanac R, et al. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 18.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koboldt D, Chen K, Wylie T, Larson D. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. 2009 doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 21.Mokry M, et al. Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Res. 2010:1–9. doi: 10.1093/nar/gkq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Research. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoberman R, et al. A probabilistic approach for SNP discovery in high-throughput human resequencing data. Genome Research. 2009;19:1542–1552. doi: 10.1101/gr.092072.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhis N, Jones S. High quality SNP calling using Illumina data at shallow coverage. Bioinformatics. 2010;26:1029. doi: 10.1093/bioinformatics/btq092. [DOI] [PubMed] [Google Scholar]
- 25.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nature Genetics. 2011 doi: 10.1038/ng.768. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna AH, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010 doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng S, Turner E, Robertson P, Flygare S. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009 doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mckernan KJ, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Research. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebersberger I, Metzler D, Schwarz C, Pââbo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freudenberg-Hua Y, et al. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Research. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: Probabilistic models of proteins and nucleic acids. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 38.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.HUGO Consortium et al. Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 40.Bishop C. Pattern recognition and machine learning. Springer: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.