Jasmonate is essential for diverse biological processes, including male fertility and plant defense in Arabidopsis. This work shows that the R2R3-MYB transcription factors MYB21 and MYB24 function as direct targets of JAZ proteins to mediate jasmonate-regulated stamen development.
Abstract
The Arabidopsis thaliana F-box protein CORONATINE INSENSITIVE1 (COI1) perceives jasmonate (JA) signals and subsequently targets the Jasmonate-ZIM domain proteins (JAZs) for degradation by the SCFCOI1-26S proteasome pathway to mediate various jasmonate-regulated processes, including fertility, root growth, anthocyanin accumulation, senescence, and defense. In this study, we screened JAZ-interacting proteins from an Arabidopsis cDNA library in the yeast two-hybrid system. MYB21 and MYB24, two R2R3-MYB transcription factors, were found to interact with JAZ1, JAZ8, and JAZ11 in yeast and in planta. Genetic and physiological experiments showed that the myb21 myb24 double mutant exhibited defects specifically in pollen maturation, anther dehiscence, and filament elongation leading to male sterility. Transgenic expression of MYB21 in the coi1-1 mutant was able to rescue male fertility partially but unable to recover JA-regulated root growth inhibition, anthocyanin accumulation, and plant defense. These results demonstrate that the R2R3-MYB transcription factors MYB21 and MYB24 function as direct targets of JAZs to regulate male fertility specifically. We speculate that JAZs interact with MYB21 and MYB24 to attenuate their transcriptional function; upon perception of JA signal, COI1 recruits JAZs to the SCFCOI1 complex for ubiquitination and degradation through the 26S proteasome; MYB21 and MYB24 are then released to activate expression of various genes essential for JA-regulated anther development and filament elongation.
INTRODUCTION
The plant hormone jasmonate (JA), including jasmonic acid and its oxylipin derivatives, is ubiquitous in the plant kingdom (Creelman and Mullet, 1997; Sasaki et al., 2001; Feussner and Wasternack, 2002; Cheong and Choi, 2003; Farmer et al., 2003; Wasternack, 2007). It acts as regulatory molecule to influence many plant developmental processes, such as stamen development (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000; Cheng et al., 2009; Chua et al., 2010), root growth (Staswick et al., 1992; Feys et al., 1994; Pauwels et al., 2010), anthocyanin accumulation (Franceschi and Grimes, 1991; Shan et al., 2009), and senescence (Ueda and Kato, 1980; Schommer et al., 2008; Shan et al., 2011). In addition, JA also functions as a defense signal to activate stress (Maslenkova et al., 1992; Rao et al., 2000) and wound responses (Farmer and Ryan, 1992; Reymond et al., 2000; Schilmiller and Howe, 2005; Robson et al., 2010) and to mediate plant defense responses against insect attacks (Howe et al., 1996; McConn et al., 1997; Farmer, 2001) and pathogen infections (Reymond and Farmer, 1998; Vijayan et al., 1998; Farmer et al., 2003; Xiao et al., 2004; Wasternack, 2007; Browse, 2009).
Characterization of JA biosynthesis-deficient mutants revealed that JA is essential for stamen development in Arabidopsis thaliana. JA-deficient mutants fad3 fad7 fad8 (McConn and Browse, 1996), dad1 (Ishiguro et al., 2001), aos (Park et al., 2002), and opr3/dde1 (Sanders et al., 2000; Stintzi and Browse, 2000; Mandaokar et al., 2003) are male sterile due to short stamen filament, failure of anther dehiscence, and unviable pollens. Application of exogenous jasmonic acid can restore the male fertility of the JA-deficient mutants.
The essential role of JA signaling in regulating stamen development was uncovered through characterization of CORONATINE INSENSITIVE1 (COI1). coi1-1 with an early stop codon at W467 exhibits complete male sterility due to retarded stamen development, including short filaments, delayed anther dehiscence, and unviable pollen (Feys et al., 1994; Xie et al., 1998). Exogenous application of JA cannot restore the fertility in coi1-1. The coi1-1 mutant is also defective in many other JA-regulated responses, including JA-mediated root growth inhibition, anthocyanin accumulation, and defense against insect attack and pathogen infection (Feys et al., 1994; Xie et al., 1998; Wang et al., 2005; Shan et al., 2009). Further study revealed that COI1 encodes an F-box protein that forms SCFCOI1 complexes with Cullin1, Rbx1, and ASK1 or ASK2 to mediate diverse JA-regulated responses in Arabidopsis (Xu et al., 2002; Liu et al., 2004; Ren et al., 2005; Shan et al., 2007). Upon perception of a JA signal (Yan et al., 2009), COI1 recruits the Jasmonate-ZIM domain proteins (JAZs), which include 12 members and function as the substrates of the SCFCOI1 complex, for degradation through the 26S proteasome (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). Overexpression of truncated JAZ1 in which the Jas domain is deleted (Thines et al., 2007), or the alternatively spliced form JAZ10.4 lacking the Jas domain, leads to male sterility (Chung and Howe, 2009), indicating that JAZ proteins repressed JA-regulated male fertility (Thines et al., 2007; Chung and Howe, 2009).
It was hypothesized that JAZ proteins may exert repression on JA responses via their interactions with a series of transcription factors and that degradation of JAZs would disrupt these interactions, leading to activation of these transcription factors that mediate various JA-regulated biological processes (Katsir et al., 2008; Browse, 2009; Fonseca et al., 2009; Santner and Estelle, 2009). The transcription factor MYC2, a direct target of JAZs (Chini et al., 2007), affects particular aspects of JA-regulated biological functions, such as JA inhibition of root growth (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007). Although JA-regulated male fertility is modulated by degradation of JAZs through the SCFCOI1-26S proteasome pathway, little is known about the molecular basis of JAZ function in JA-regulated male fertility.
In this study, we identified MYB21 and MYB24, two R2R3-transcription factors, as interacting with JAZ1, JAZ8, and JAZ11 in yeast and in planta. Genetic and physiological experiments demonstrated that the myb21 myb24 double mutant exhibited defects specifically in pollen maturation, anther dehiscence, and filament elongation leading to male sterility. Transgenic expression of MYB21 in the coi1-1 mutant was able to rescue male fertility partially but not other JA-regulated processes. These results demonstrate that MYB21 and MYB24 function as direct targets of JAZs to mediate specifically JA-regulated anther development and filament elongation.
RESULTS
Isolation of JAZ-Interacting Proteins in the Yeast Two-Hybrid System
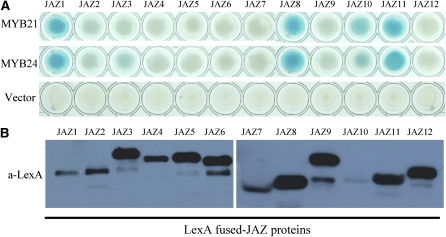
The JAZ8 protein was used as a bait to screen an Arabidopsis cDNA library in the yeast two-hybrid (Y2H) system. Sequence analysis of putative interacting clones revealed that two R2R3-MYB transcription factors, MYB21 and MYB24 (see Supplemental Figure 1 online), interacted with JAZ8 in the Y2H system.
We further investigated interactions of MYB21 and MYB24 with all 12 Arabidopsis JAZs in the Y2H system. As a control, we examined the expression levels of the twelve JAZs by immunoblot analysis with anti-LexA antibody in yeast cells (Chung and Howe, 2009). The result showed that all LexA-JAZ fusion proteins were expressed in yeast (Figure 1B) and that both MYB21 and MYB24 interacted with JAZ1, JAZ8, JAZ11, as well as with JAZ10 to a lower degree, whereas no obvious interaction was observed for other JAZs (JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ7, JAZ9, and JAZ12) in yeast (Figure 1A).
Figure 1.
Interactions of JAZs with MYB21 and MYB24 in the Y2H System.
(A) Y2H assay to detect interactions of JAZs with MYB21 and MYB24. Twelve Arabidopsis JAZs were individually fused with the LexA DNA binding domain (BD) in pLexA. MYB21 and MYB24 were individually fused with the activation domain (AD) in pB42AD. Interactions of JAZs with the AD domain in the pB42AD empty vector were used as negative controls. Interactions (represented by blue color) were assessed on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium.
(B) Immunoblot analysis of JAZ proteins expressed in yeast strains from (A). Total proteins were extracted from the yeast strains of (A) and analyzed by immunoblot using anti-LexA antibody. Expression of the JAZ proteins was detected as expected (Chung and Howe, 2009).
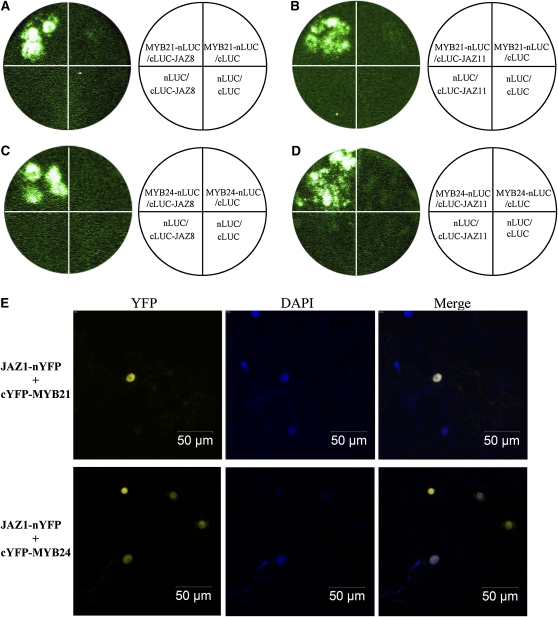
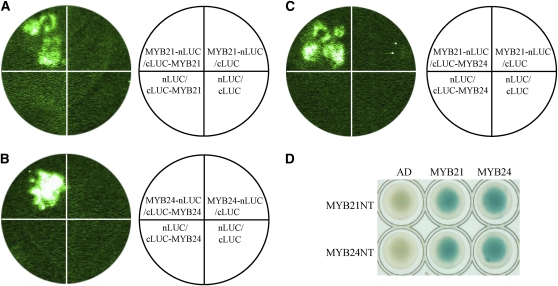
Interactions of JAZs with MYB21 and MYB24 in planta were further investigated with a firefly luciferase (LUC) complementation imaging (LCI) assay in Nicotiana benthamiana (Luker et al., 2004; Chen et al., 2008). JAZ8 and JAZ11 were used as representatives in the LCI assay. The results showed that coinfiltration of MYB21, which was fused to N-terminal part of LUC (MYB21-nLUC), with JAZ8 fused to C-terminal part of LUC (cLUC-JAZ8) lead to strong LUC activity in N. benthamiana leaves, whereas no obvious LUC activity was detected in negative controls (the combinations of MYB21-nLUC/cLUC, cLUC-JAZ8/nLUC, and nLUC/cLUC; Figure 2A; see Supplemental Figure 2A online). Similar results were also observed for interaction of MYB21 with JAZ11 (Figure 2B; see Supplemental Figure 2B online) and for interaction of MYB24 with JAZ8 or JAZ11 (Figures 2C and 2D; see Supplemental Figures 2C and 2D online).
Figure 2.
MYB21 and MYB24 Interact with JAZ1, JAZ8, and JAZ11 in N. benthamiana.
(A) to (D) LCI assays show that Arabidopsis MYB21 and MYB24 interact with Arabidopsis JAZ8 and JAZ11 in N. benthamiana. MYB21 and MYB24 were fused with N-terminal fragment of LUC (nLUC) to generate MYB21-nLUC and MYB24-nLUC, respectively. JAZ8 and JAZ11 were fused with the C-terminal fragment of LUC (cLUC) to produce cLUC-JAZ8 and cLUC-JAZ11, respectively. The leaves of N. benthamiana were infiltrated with Agrobacterium strains containing the indicated construct pairs. The data were collected 50 h after infiltration.
(E) BiFC assay indicates that Arabidopsis MYB21 and MYB24 interact with Arabidopsis JAZ1 in N. benthamiana. MYB21 and MYB24 were fused with the C-terminal fragment of yellow fluorescence protein (cYFP) to form cYFP-MYB21 and cYFP-MYB24. JAZ1 was fused with N-terminal fragment of YFP (nYFP) to form JAZ1-nYFP. YFP fluorescence was detected in N. benthamiana leaves coinfiltrated with combinations of JAZ1-nYFP/ cYFP-MYB21, and JAZ1-nYFP/ cYFP-MYB24. The positions of nuclei were shown by 4′,6-diamidino-2-phenylindole (DAPI) staining.
We also employed a yellow fluorescence protein (YFP) bimolecular fluorescence complementation (BiFC) system (Weinthal and Tzfira, 2009) to test the interactions of JAZ1 with MYB21 and MYB24 in planta. JAZ1-nYFP, in which JAZ1 was fused to N-terminal part of YFP, and cYFP-MYB21 (MYB21 fused with C-terminal part of YFP) were coexpressed in leaf epidermal cells in N. benthamiana through Agrobacterium tumefaciens–mediated transformation. As shown in Figure 2E, coexpression of JAZ1-nYFP and cYFP-MYB21 reconstructed YFP signal in the nucleus, which demonstrated that JAZ1 interacted with MYB21 in planta. Similar results were also observed for interaction of JAZ1 with MYB24 (Figure 2E).
Taken together, the Y2H, LCI, and BiFC assays demonstrate that MYB21 and MYB24 interact with JAZs, implying that MYB21 and MYB24 may function as direct targets of JAZs.
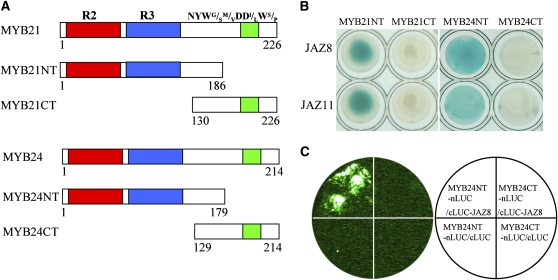
The N-Terminal R2R3 DNA Binding Domain of MYB21 and MYB24 Is Involved in Interaction with JAZs
Phylogenetic analysis showed that MYB21 and MYB24 belong to subgroup 19 of the R2R3-MYB transcription factor family (Kranz et al., 1998). MYB21 shares 67.7% identity with MYB24 at the amino acid level (see Supplemental Figure 1 online). Both MYB21 and MYB24 contain an N-terminal R2R3 repeat domain, which is responsible for DNA binding, and a NYWG/SM/VDDI/LWS/P motif in the C terminus (see Supplemental Figure 1 online).
To study which domain in these two transcription factors is responsible for interaction with JAZs, MYB24 and MYB21 were divided into the N-terminal parts (MYB24NT and MYB21NT, respectively) containing the R2R3 domain and the C-terminal parts (MYB24CT and MYB21CT, respectively) including the NYWG/SM/VDDI/LWS/P motif (Figure 3A). As shown in Figure 3B, JAZ8 and JAZ11 interacted with MYB24NT but not with MYB24CT in yeast. JAZ8 and JAZ11 also interacted with N-terminal part of MYB21 but not the C-terminal part of MYB21 in yeast (Figures 3A and 3B). LCI assays also showed that coinfiltration of MYB24NT-nLUC with cLUC-JAZ8 resulted in strong LUC signal in N. benthamiana leaves (Figure 3C; see Supplemental Figure 2E online). These results demonstrate that the N-terminal R2R3 domain of MYB21 and MYB24 is essential for interaction with JAZs.
Figure 3.
JAZ8 and JAZ11 Interact with the N Terminus of MYB21 and MYB24.
(A) Schematic diagram of MYB21 and MYB24 domain constructs. MYB21 and MYB24 were separated into two parts: the MYB21NT/MYB24NT part containing conserved R2 (red) R3 (blue) domain, and the MYB21CT/MYB24CT part containing the NYWG/SM/VDDI/LWS/P motif (green). MYB21NT/MYB24NT and MYB21CT/MYB24CT were ligated into pB42AD vector for fusion with the AD domain individually.
(B) Y2H assays show that JAZ8 and JAZ11 interact with MYB21NT and MYB24NT in yeast. The interactions were observed on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium.
(C) LCI assays show that JAZ8 interacts with MYB24NT in N. benthamiana. MYB24NT and MYB24CT shown in (A) were fused with nLUC to produce MYB24NT-nLUC and MYB24CT-nLUC, respectively. JAZ8 was fused with cLUC to produce cLUC-JAZ8. Signal was detected in N. benthamiana leaves 50 h after coinfiltration with the construct pairs indicated in the white circle.
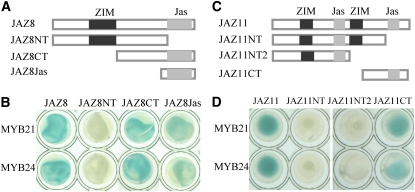
The Jas Domain of JAZs Is Responsible for Interaction with MYB21 and MYB24
To investigate further which domain of JAZs is responsible for interaction with MYB21 and MYB24, JAZ8 was divided into the N-terminal part (JAZ8NT) and C-terminal part (JAZ8CT), and the JAZ8CT was further truncated into the Jas domain (JAZ8Jas) (Figure 4A). As shown in Figure 4B, MYB21 and MYB24 interacted with JAZ8CT as well as JAZ8Jas but not with JAZ8NT in yeast.
Figure 4.
The Jas Domain in the C-Terminal Part of JAZ Proteins Is Required for Interaction with MYB21 and MYB24.
(A) Schematic diagram of JAZ8 domain constructs. The diagram shows the conserved ZIM (black) and Jas (gray) domains. Different domains of JAZ8 were fused with the BD domain.
(B) Y2H assay for interactions of JAZ8 domain constructs with MYB21 and MYB24. MYB21 and MYB24 were fused with the AD domain individually.
(C) Schematic diagram of JAZ11 domain constructs. There are two ZIM domains (black) and two Jas domains (gray) in JAZ11.
(D) Y2H assay for interactions of JAZ11 domain constructs with MYB21 and MYB24.
[See online article for color version of this figure.]
JAZ8 is composed of one ZIM domain in its N-terminal part and one Jas domain in its C-terminal part. However, JAZ11 contains two ZIM domains and two Jas domains, with the first Jas domain between the two ZIM domains and the second Jas domain in the C-terminal part (Figure 4C). We divided JAZ11 into the N-terminal part (JAZ11NT) harboring two ZIM domains and the first Jas domain, and the C-terminal part (JAZ11CT) containing the second Jas domain (Figure 4C). JAZ11NT was further truncated into JAZ11NT2 in which the first Jas domain was exposed (Figure 4C). We found that both MYB21 and MYB24 interacted with JAZ11CT but not with JAZ11NT and JAZ11NT2, indicating that the second Jas domain in the C-terminal part of JAZ11 is required for interaction with MYB21 and MYB24 (Figure 4D). In conclusion, our results demonstrate that the C-terminal Jas domain of JAZs is responsible for interaction with MYB21 and MYB24.
Transgenic Expression of MYB21 Partially Rescues the Male Fertility of coi1-1
MYB21 and MYB24 were previously shown to regulate stamen development (Mandaokar et al., 2006; Cheng et al., 2009). Consistent with this, we found that the myb21-3 mutant displayed a severe reduction in fertility due to shorter filaments (see Supplemental Figure 3 online), whereas no obvious defect was observed in myb24-t1. The myb21-3 myb24-t1 double mutant was male sterile and unable to set seeds (see Supplemental Figure 3D online). We further found that anthers in myb21-3 were able to dehisce to release viable pollen (see Supplemental Figure 3G online). However, the myb21-3 myb24-t1 double mutant exhibited a clear delay in anther dehiscence, and their pollen grains were unable to germinate in vitro (see Supplemental Figure 3G online) and unviable when used for manually pollination of wild-type stigma. Together with previous observations (Mandaokar et al., 2006; Cheng et al., 2009), these results demonstrate that MYB21 plays a dominant role in stamen filament elongation and that MYB21 and MYB24 function redundantly in regulation of anther dehiscence and pollen maturation in Arabidopsis.
JAZ proteins, as negative regulators that accumulate in the coi1-1 mutant, repress JA responses, including JA-regulated male fertility (Katsir et al., 2008; Browse, 2009; Fonseca et al., 2009). Interactions of JAZs with MYB21 and MYB24 (Figures 1A and 2) may attenuate the function of these transcription factors essential for stamen development (see Supplemental Figure 3 online), thereby repressing male fertility in coi1-1. To verify this speculation, we investigated whether transgenic expression of the R2R3-MYB transcription factor MYB21 could restore male fertility in the coi1-1 mutant.
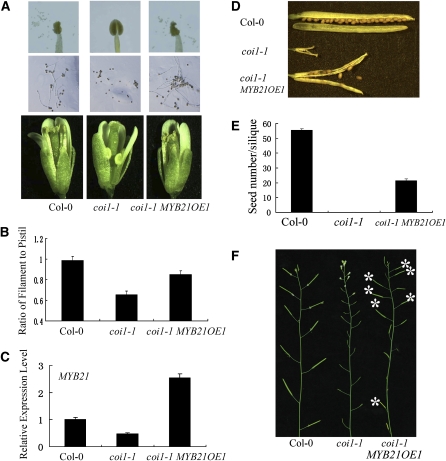
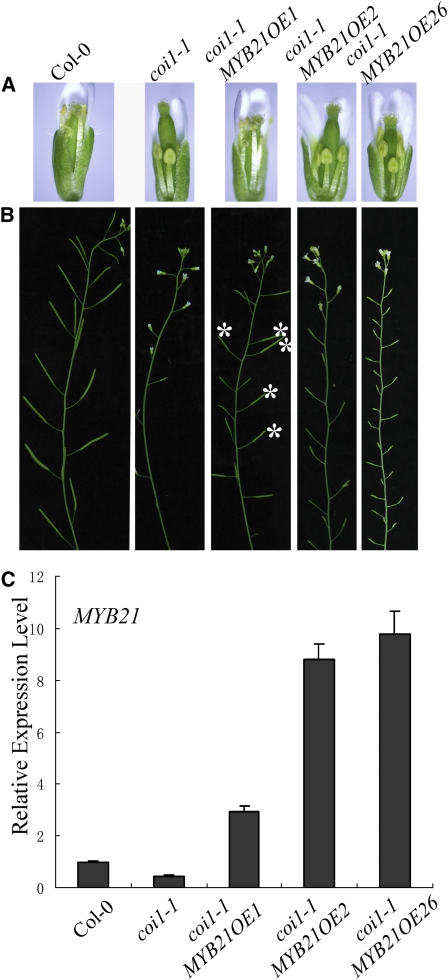
We created transgenic expression of MYB21 in the coi1-1 mutant via genetic transformation of COI1/coi1-1 heterozygous plants and subsequent identification of the coi1-1 homozygous plants transgenic for MYB21. Three individual lines with transgenic expression of MYB21 in the coi1-1 mutant background were found to be partially fertile (see Methods) (Figure 5). As expected, anthers in coi1-1 failed to dehisce, and filament elongation was arrested in the coi1-1 mutant flowers with the ratio of filament length to pistil length at ~0.66 on average (Figures 5A and 5B). The transgenic line coi1-1 MYB21OE1, with ~3-fold of the wild-type level of MYB21 (Figure 5C), contained fertile flowers that harbored dehisced anthers (Figure 5A) and elongated filaments with the ratio of filament length to pistil length at ~0.85 (Figure 5B). Mature plants of the coi1-1 MYB21OE1 line exhibited partial fertility, and they were able to set small amounts of seeds (Figures 5D to 5F). These results demonstrate that transgenic expression of MYB21 could partially restore male fertility in the coi1-1 mutant.
Figure 5.
Transgenic Expression of MYB21 Partially Rescues the Male Fertility of coi1-1.
(A) Comparison of anthers and flowers in different genotypes as indicated. The flower in coi1-1 MYB21OE1 shows dehisced anther (top panel) and elongated filaments (bottom panel) compared with coi1-1. Pollen grains from coi1-1 fail to germinate in vitro (middle), whereas transgenic expression of MYB21 rescues the pollen germination of coi1-1 (right middle).
(B) The ratio of filament length to pistil length in Columbia-0 (Col-0), coi1-1, and coi1-1 MYB21OE1. Error bars represent se (n = 10).
(C) Quantitative real-time PCR analysis of MYB21 transcription level in young flower buds from Col-0, coi1-1, and coi1-1 MYB21OE1 using ACTIN8 as the internal control. Error bars represent se.
(D) Comparison of seed set in different genotypes as indicated.
(E) Seed numbers per silique in Col-0, coi1-1, and coi1-1 MYB21OE1. For coi1-1 MYB21OE1, the seed numbers per silique were calculated from 10 randomly selected fertile siliques. Error bars represent se (n = 10).
(F) Main inflorescences in Col-0, coi1-1, and coi1-1 MYB21OE1. Asterisks indicate fertile siliques in coi1-1 MYB21OE1.
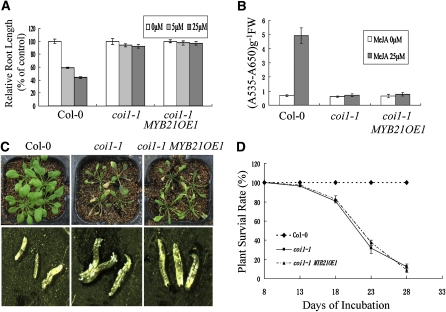
We examined whether MYB21 was involved in other JA responses. Measurement of root length of seedlings treated with various concentrations of methyl jasmonate (MeJA) showed that the coi1-1 MYB21OE1 line, similar to coi1-1, was insensitive to JA inhibition of root growth (Figure 6A), indicating that transgenic expression of MYB21 was unable to rescue the JA sensitivity of root growth in coi1-1 (Figure 6A). JA also can induce anthocyanin accumulation in Arabidopsis seedlings (Shan et al., 2009). We found that, similar to coi1-1, anthocyanin content in the coi1-1 MYB21OE1 line was not induced by treatment with 25 μM MeJA (Figure 6B).
Figure 6.
MYB21 Is Not Required for JA-Regulated Root Growth Inhibition, Anthocyanin Accumulation, and Defense against Insect Attack.
(A) Relative root length of 9-d-old seedlings grown on MS medium containing indicated concentrations of MeJA. Relative root length is shown as a percentage of root length on MS medium. Error bars represent se (n = 20).
(B) Anthocyanin contents of seedlings grown on MS medium containing indicated concentrations of MeJA. Error bars represent se. FW, fresh weight.
(C) Top panel shows the phenotype of 35-d-old seedlings grown in a B. impatiens–infested growth chamber. Bottom panel shows young B. impatiens from indicated soil pots.
(D) Plant survival rates of the indicated plants after incubation with B. impatiens at the indicated time points.
[See online article for color version of this figure.]
JA is required for plant defense against insect attack (McConn et al., 1997). We tested whether transgenic expression of MYB21 could rescue plant defense against Bradysia impatiens in coi1-1. Although a few rosette leaves in the wild type were gnawed by B. impatiens larvae, no wild-type plant died from the insect attack (Figures 6C and 6D). Most of the coi1-1 and coi1-1 MYB21OE1 plants died from B. impatiens larvae attack ~28 d after incubation in the B. impatiens–infested growth room (Figures 6C and 6D), demonstrating that coi1-1 MYB21OE1 was also susceptible to B. impatiens attack.
Taken together, these results demonstrate that MYB21 is essential for JA-regulated male fertility but not for other JA responses such as JA-regulated root growth inhibition, anthocyanin accumulation, and plant defense response.
Excess Expression of MYB21 Results in Retarded Stamen Development
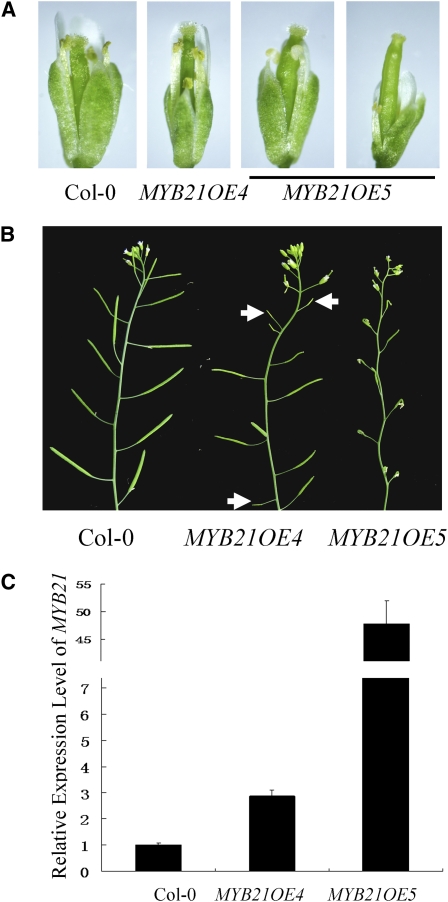
As the COI1/coi1-1 heterozygous plants were used in genetic transformation for transgenic expression of MYB21, we characterized transgenic plants in various backgrounds (wild type, COI1/coi1-1, or coi1-1). We noticed that many primary transgenic plants (29 out of 59) with the wild-type or COI1/coi1 background showed severe or complete reduction in male fertility. Further observation revealed that the reduction in male fertility in these sterile transgenic plants resulted from defects of filament elongation and failure of anther dehiscence (Figure 7A).
Figure 7.
Excess Expression of MYB21 Causes Male Sterility.
(A) Flowers of Col-0 and two MYB21 overexpression lines (MYB21OE4 and MYB21OE5).
(B) Main inflorescences. Arrows indicate sterile siliques in MYB21OE4.
(C) Quantitative real-time PCR analysis of the MYB21 expression level in young flower buds of the indicated plants using ACTIN8 as the internal control. Error bars represent se.
We used quantitative real-time PCR to analyze the expression level of MYB21 among these transgenic plants. We found that the transgenic plants with severe or complete loss in fertility contained a very high level of MYB21 (20- to ~100-fold of the wild-type level). The male sterility in the transgenic lines was positively correlated with the high expression level of MYB21 (Figure 7; see Supplemental Figure 4 online). As a representative example shown in Figure 7, the MYB21OE5 plant (COI1/COI1 background) was completely male sterile and contained ~48-fold the wild-type level of MYB21 (Figures 7B and 7C). However, in transgenic plants with mild or undetectable reduction in fertility, the expression level of MYB21 was ~1- to ~5-fold of the wild-type level. As a representative example shown in Figure 7, the MYB21OE4 line (COI1/COI1 background) exhibited a mild reduction in fertility and contained ~3-fold the wild-type level of MYB21.
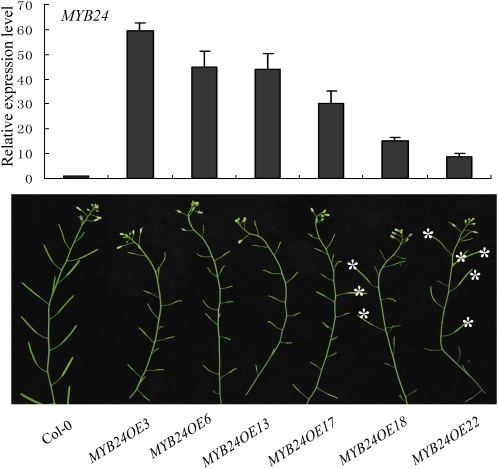
Similar results were observed for transgenic plants overexpressing MYB24 (Figure 8). The severe disruption or complete abolition of stamen development occurred in transgenic plants with excess expression of MYB24 (>15-fold of the wild-type level) (Figure 8). Taken together, these data suggest that a suitable expression level of MYB21 and MYB24 is important for stamen development but that excess expression of MYB21 and MYB24 may function to attenuate stamen development.
Figure 8.
Excess Expression of MYB24 Causes Male Sterility.
Quantitative real-time PCR analysis (top panel) of the MYB24 expression level in young flower buds of Col-0 and six MYB24 transgenic lines as indicated (bottom panel). ACTIN8 was used as the internal control. Error bars represent se. Asterisks in the bottom panel indicate fertile siliques in MYB24 transgenic lines.
[See online article for color version of this figure.]
Consistent with this, we found that excess expression of MYB21 failed to restore the male fertility in coi1-1 (Figure 9). We observed that many primary MYB21 transgenic plants in the coi1-1 background were completely male sterile. Quantitative real-time PCR analysis of seven male sterile transgenic coi1-1 plants showed that they all contained >8-fold the wild-type level of MYB21 (Figure 9), whereas the coi1-1 transgenic plants with partial fertility all expressed MYB21 at ~3-fold of the wild-type level (Figures 5 and 9). These results indicate that MYB21 acts downstream of COI1 specifically to regulate JA-mediated stamen development in a manner requiring a suitable level of MYB21.
Figure 9.
Excess Expression of MYB21 Fails to Rescue Male Fertility in coi1-1.
(A) Flowers of Col-0, coi1-1, and three MYB21 transgenic lines in the coi1-1 background as indicated.
(B) Main inflorescences of (A). Asterisks indicate fertile siliques in coi1-1 MYB21OE1.
(C) Quantitative real-time PCR analysis of the MYB21 expression level in young flower buds. Seven male-sterile coi1-1 plants transgenic for MYB21 contain >8-fold wild-type level of MYB21, coi1-1MYB21OE2, and coi1-1MYB21OE26 are shown as representatives. ACTIN8 was used as the internal control. Error bars represent se.
Dimeric interaction of proteins is a key regulatory mechanism in signal transduction (Klemm et al., 1998). Y2H assays showed strong interactions among MYB21NT, MYB24NT, MYB21, and MYB24 (Figure 10D). LCI assays indicated that MYB21 and MYB24 might display homo- and heteromeric interactions in planta (Figures 10A to 10C; see Supplemental Figures 2F to 2H online). It remains to be elucidated whether excess expression of MYB21 or MYB24 would promote dimeric interactions to repress stamen development.
Figure 10.
Homomeric and Heteromeric Interactions of MYB21 and MYB24.
(A) to (C) LCI assays show the interactions of MYB21-MYB21 (A), MYB24-MYB24 (B), and MYB21-MYB24 (C) in N. benthamiana leaves. Leaf regions were coinfiltrated with Agrobacterium strains containing the indicated constructs. The data were collected 50 h after infiltration.
(D) Y2H assays show interactions among MYB21NT, MYB24NT, MYB21, and MYB24 in yeast. The interactions were observed on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium.
DISCUSSION
JAZ proteins were identified as substrates of the SCFCOI1 complex (Chini et al., 2007; Thines et al., 2007) to repress JA responses including male fertility, root growth, anthocyanin accumulation, senescence, and plant defense. Previous studies showed that the transcription factor MYC2 is a direct target of JAZs (Chini et al., 2007) and functions in some aspects of JA-regulated biological functions, such as JA inhibition of root growth, but not in JA-regulated male fertility (Lorenzo et al., 2004). Recent studies showed that NINJA, DELLA, and MYC2 homologs (MYC3/4) interact with JAZs to regulate JA inhibition of root growth and JA-induced gene expression but not JA-regulated stamen development (Hou et al., 2010; Pauwels et al., 2010; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). Little is known about the JAZ-containing signaling cascade that is required for JA-regulated male fertility. Here, we demonstrate that JAZs interact with MYB21 and MYB24, two R2R3-MYB transcription factors essential for filament elongation, anther dehiscence, and pollen maturation, to mediate JA-regulated male fertility in Arabidopsis.
MYB21 and MYB24 were identified through Y2H screening for interaction with JAZ8. They also showed strong interaction with JAZ1 and JAZ11 as well as weak interaction with JAZ10 in yeast (Figure 1A). LCI and BiFC assays showed that MYB21 and MYB24 interact with JAZ1, JAZ8, and JAZ11 in planta (Figure 2). The R2R3 DNA binding domain of MYB21 and MYB24 is responsible for interaction with JAZs (Figure 3), which is different from the MYC2-JAZ interactions, where the bHLH DNA binding domain of MYC2 is unable to interact with JAZs (Chini et al., 2007). The Jas domain in the C-terminal part of JAZs is required for JAZ interaction with the R2R3-MYB transcription factors MYB21 and MYB24 (Figure 4), the bHLH transcription factor MYC2 (Chini et al., 2009), and the F-box protein COI1 (Chini et al., 2009). It would be very interesting to investigate the mechanism and affinity of the interactions between JAZs and the various proteins.
R2R3 transcription factors (MYB21, MYB24, MYB57, and MYB108) were previously shown to be involved in regulating stamen development (Mandaokar et al., 2006; Cheng et al., 2009; Mandaokar and Browse, 2009). The auxin response factors were found to act through JA biosynthesis to regulate anther dehiscence (Nagpal et al., 2005). We previously showed that gibberellin acts through JA to regulate MYB21, MYB24, and MYB57, which function redundantly in influencing filament elongation (Cheng et al., 2009). Here, we demonstrate that the R2R3 transcription factors, including MYB21 and MYB24, function as direct targets of JAZs to affect specifically JA-regulated filament elongation, anther dehiscence, and pollen maturation: MYB21 and MYB24 interacted strongly with JAZ1, JAZ8, and JAZ11 and weakly with JAZ10 (Figures 1 and 2); the myb21 myb24 double mutant plants were male sterile due to defects in filament elongation, anther dehiscence, and pollen viability (see Supplemental Figure 3 online); transgenic expression of MYB21 partially rescued the defects of coi1-1 in filament elongation, anther dehiscence, and pollen maturation but not in JA inhibition of root growth, pigmentation, and defense response (Figures 5 and 6). In addition, we observed the interaction of JAZ with MYB57, but not with MYB108 (see Supplemental Figure 5 online; data not shown), implying that MYB57 might also be a direct target of JAZ1 whereas MYB108 may function downstream. We further detected the expression of JAZ1, JAZ8, JAZ10, and JAZ11 in flowers (see Supplemental Figure 6 online), which is consistent with their role in the flower to interact with R2R3 transcription factors, including MYB21 and MYB24, to repress stamen development (Shin et al., 2002; Yang et al., 2007; Cheng et al., 2009).
Overexpression of JAZ1Δ3A (truncated JAZ1 without the Jas domain) or JAZ10.4 (an alternatively spliced form of JAZ10 lacking the Jas domain) caused male sterility in Arabidopsis (Thines et al., 2007; Chung and Howe, 2009; Chung et al., 2010). In these transgenic plants, JAZ1Δ3A and JAZ10.4 may interact with the endogenous JAZ proteins through the TIFY domain to prevent degradation of the endogenous JAZ proteins (Chini et al., 2007, 2009; Thines et al., 2007; Chung and Howe, 2009). These endogenous JAZ proteins are speculated to interact with the R2R3 transcription factors to inhibit functions of MYB21 and MYB24, resulting in male sterility.
Transcription factors MYC2 and TT8 were found to bind to their own promoters to regulate their own expression (Baudry et al., 2006; Dombrecht et al., 2007; Chung et al., 2008). As shown in Supplemental Table 1 online, the promoter regions of the R2R3 transcription factors MYB21 and MYB24 harbor the binding sites (gttaggaa and gttaggtt) for Antirrhinum majus R2R3 transcription factor MYB305 (Sablowski et al., 1994), which shares 55.5 and 57.9% identity with MYB21 and MYB24, respectively, at the amino acid level, indicating that MYB21 and MYB24 might be able to bind their own promoters to promote their own transcription. MYB24 expression was downregulated in myb21 flowers (Mandaokar and Browse, 2009), indicating a regulatory role for MYB21 in expression of MYB24. Accumulation of JAZs (Thines et al., 2007) in coi1 or opr3 attenuates function of MYB21 and MYB24 and probably also inhibit their expression, leading to low levels of MYB21 and MYB24 in flowers of coi1 or opr3 (Cheng et al., 2009) (Figure 5). JA treatment of the JA-deficient mutant opr3 induced degradation of JAZs, which probably released MYB21 and MYB24, promoting expression of MYB21 and MYB24 themselves and leading to an obvious increase of 24.64- and 30.48-fold, respectively, in opr3 stamens within 2 h (Mandaokar et al., 2006).
We propose a model to elucidate the molecular mechanism of JA-regulated male fertility in Arabidopsis (Figure 11). In wild-type flowers, COI1 recruits JAZs for degradation through the 26S proteasome pathway, and the R2R3 transcription factors MYB21 and MYB24 are then released to activate expression of various genes essential for JA-regulated stamen development. In coi1 and the JA-deficient mutants, JAZ proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to attenuate transcriptional function of MYB21 and MYB24, which are required for stamen development. In the transgenic plants overexpressing MYB21 or MYB24 in the coi1 background, the elevated MYB21 or MYB24 may escape from the JAZ interaction to execute their transcriptional function in modulation of male fertility.
Figure 11.
A Simplified Model for the Molecular Mechanism of JA-Regulated Male Fertility.
The Jas domain of JAZ proteins (JAZ) interact with the R2R3 domain of MYB21 and MYB24 (MYB) to attenuate transcriptional function of MYB21 and MYB24 and inhibit expression of downstream genes (Early genes) leading to male sterility; upon perception of the JA signal, COI1 recruits JAZs to the SCFCOI1 complex for ubiquitination and degradation through the 26S proteasome. MYB21 and MYB24 (MYB) are then released to activate expression of downstream genes essential for JA-regulated stamen development.
[See online article for color version of this figure.]
Gene expression at the proper level is essential for function in some cases. A previous study showed that excess expression of the MYB305 gene caused shorter anther length similar to MYB305-RNAi mutant plants in ornamental tobacco (Nicotiana langsdorffii × Nicotiana sanderae) (Liu et al., 2009). Here, we found that suitable levels of MYB21 and MYB24 are critical for stamen development. Transgenic expression of MYB21 with ~3-fold of the wild-type level could partially restore male fertility in coi1-1, whereas excess expression of MYB21 (>8-fold) failed to rescue the stamen development in coi1-1 (Figure 9). Consistent with this, excess expression of MYB21 or MYB24 in wild-type plants caused retarded stamen development leading to male sterility (Figures 7 and 8; see Supplemental Figure 4 online) (Yang et al., 2007). The observation of MYB21/MYB21, MYB24/MYB24, and MYB21/ MYB24 interactions in Y2H and LCI assays indicates that MYB21 and MYB24 might form homo- and heterodimers in planta (Figure 10). It remains to be elucidated whether the excess expression of MYB21 or MYB24 would promote formation of the homo- and heterodimers to repress (or activate) downstream genes that initiate (or attenuate) stamen development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were sterilized with 20% bleach, plated on Murashige and Skoog medium (MS; Sigma-Aldrich), chilled at 4°C for 3 d, and transferred to a growth room with a 16-h (22 to 24°C)/8-h (19°C) light/dark photoperiod.
Nicotiana benthamiana was grown under a 16-h (28°C)/8-h (22°C) light/dark photoperiod.
The coi1-1 mutant was described as previously (Xie et al., 1998). myb21-3 (CS311165) and myb24-t1 (Salk_017221) (Cheng et al., 2009) were obtained from the ABRC.
Bioinformatics Prediction of Promoter Binding Sites
Promoter binding site analysis for MYB21 and MYB24 was performed on http://Arabidopsis.med.ohio-state.edu/AtcisDB (Sablowski et al., 1994).
Measurement of Root Length
Seeds were grown on MS medium with 0, 5, or 25 μM MeJA, chilled at 4°C for 3 d, and transferred to the growth room. Root lengths of twenty 9-d-old seedlings for each genotype and treatment were measured. The experiment was repeated three times.
Anthocyanin Measurement
For anthocyanin measurement, 11-d-old Arabidopsis seedlings grown on MS medium with 0 or 25 μM MeJA were measured as described previously (Shan et al., 2009). The anthocyanin content is presented as (A535-A650) /g fresh weight. The experiment was repeated three times.
Measurement of Pistil and Filament Length
To measure the pistil and filament length, 10 flowers at stage 13 (Smyth et al., 1990) for each genotype were harvested. Micrographs of the stamens and carpels were taken. One of the four longer filaments and the pistil were measured under the microscope for each flower. The average ratio of filament to pistil is presented. The experiment was repeated three times.
Pollen Germination Assay
Pollen germination assays were done according to the method of Boavida and McCormick (2007). The germination medium contained 10% (w/v) Suc, 0.01% boric acid, 1 mM MgSO4, 5 mM CaCl2, and 5 mM KCl at pH 7.5, then 1.5% agar. The germination medium was dropped on a glass plate. Pollen grains were then released from the mature anthers onto the medium, subsequently incubated in the dark at 22°C for 10 h, and finally observed under a microscope. Pollen from coi1-1 or myb21-3 myb24-t1 was isolated manually.
Insect Defense Trials
Seven-day-old seedlings were transferred into the growth room that was infested with Bradysia impatiens. The plant survival rate for each genotype was recorded at the indicated time points. Fifty plants for each genotype (the wild type, coi1-1, or coi1-1 MYB21OE1) were investigated. The experiment was repeated three times.
Generation of Transgenic Plants
The Arabidopsis MYB21 CDS was amplified and cloned through XbaI and SacI sites into the pCAMBIA1301 vector under control of the cauliflower mosaic virus 35S promoter (Cheng et al., 2009) and subsequently introduced into the COI1/coi1-1 heterozygous plants using Agrobacterium tumefaciens–mediated flower dip method (Clough and Bent, 1998). The cleaved-amplified polymorphic sequence marker specific for coi1-1 (Xie et al., 1998) was used to analyze the genetic background for T1 primary transgenic plants (COI1/COI1, COI1/coi1-1, or coi1-1/coi1-1). Three independent MYB21 overexpression lines with coi1-1 background were able to set seeds, and one of them (coi1-1MYB21OE1) was representatively shown in Figure 5. MYB24 overexpression transgenic plants were generated similarly. Primers used for vector construction are listed in Supplemental Table 2 online.
LCI and BiFC Assays
An LCI assay with pCAMBIA-nLUC and pCAMBIA-cLUC vectors (Chen et al., 2008) was used to detect protein–protein interaction in N. benthamiana leaves. The firefly LUC enzyme was divided into the N-terminal part (nLUC) and C-terminal part (cLUC). MYB21, MYB24, MYB24NT, and MYB24CT were fused with nLUC in pCAMBIA-nLUC vector, respectively. JAZ8, JAZ11, MYB21, and MYB24 were fused with cLUC in pCAMBIA-cLUC vector, respectively. Primers used for the vector construction were shown in Supplemental Table 2 online.
For BiFC assays, full-length coding sequences of Arabidopsis JAZ1, MYB21, MYB24, MYB57, and MYB108 were cloned into the binary nYFP or cYFP vector through Gateway reaction with pDONR207 vector system (Invitrogen). Primer pairs for the construction of vectors are listed in Supplemental Table 2 online.
Agrobacterium strains transformed with indicated nLUC, cLUC, nYFP, or cYFP vector were incubated, harvested, and resuspended in infiltration buffer (0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES) to identical concentrations (OD600 = 0.5). Equal concentrations and volumes of Agrobacterium strains were mixed and coinfiltrated into N. benthamiana leaves by a needleless syringe. After infiltration, plants were placed at 24°C for 50 h before observation.
For LCI assays, LCI images were obtained with a low-light cooled CCD imaging apparatus (Andor iXon). Luciferin (100 μM) was sprayed onto leaves. Before fluorescence observation, the leaves were kept in dark for 15 min. The experiment was repeated three to eight independent biological replicates.
For BiFC assays, fluorescence was detected with a Zeiss microscope (LSM710). Leaves were infiltrated with 5 μg/mL 4′,6-diamidino-2-phenylindole for nuclei staining. The experiment was repeated three to eight independent biological replicates.
Y2H Assay
JAZ8 was fused to DNA binding domain (BD) in pLexA vector (Xu et al., 2002) for the Y2H screening of the pB42AD-based Arabidopsis cDNA library, which was constructed with mRNAs isolated from mixed Arabidopsis materials, including seedlings, flowers, stems, and roots treated with or without JA (Xu et al., 2002). Primers used in vector construction are listed in Supplemental Table 2 online. Yeast transformants were exhaustively selected on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal according to the manufacturer’s instructions (Clontech). Putative JAZ-interacting clones were characterized and sequenced. MYB21 and MYB24 were identified through the screening.
To verify the interactions of JAZs and their domains with MYB21 and MYB24, 11 other JAZs (JAZ1, JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ7, JAZ9, JAZ10, JAZ11, and JAZ12) and the related domains were fused with the BD domain in pLexA vector individually. Primers used for the constructs are in Supplemental Table 2 online. Yeast (Saccharomyces cerevisiae; EGY48) transformants with the indicated construct pairs were cultured at 30°C in 1.5 mL SD medium with -His/-Trp/-Ura DO supplement (Clontech) to an OD600 of 1.2 to 1.5 and then harvested and resuspended in distilled water. Five microliters of the resuspended yeast were spread in a well on the 96-well plates containing 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal medium. The interactions were observed after 3 d of incubation at 30°C. Expression of the JAZ-BD fusion proteins was detected by immunoblot analysis using anti-LexA antibody (Upstate). The experiment was repeated three times.
Real-Time PCR and RT-PCR Analysis
Total RNA was extracted from young flower buds or rosette leaves of 40-d-old plants. Quantitative real-time PCR analysis was performed using the ABi7500 real-time PCR system with the RealMasterMix (SYBR Green I) (Takara); ACTIN8 was used as the internal control. The real-time PCR experiment was repeated for three independent biological replicates. For RT-PCR analysis of Arabidopsis gene expression (see Supplemental Figure 3B online), ACTIN1 was used as the normalization control.
For RT-PCR analysis in the LCI assay (see Supplemental Figure 2 online), total RNA was extracted from N. benthamiana leaves coinfiltrated with Agrobacterium containing the indicated vectors after 50 h. Actin (EU938079) from Nicotiana was used as the normalization control for RT-PCR analysis.
All the real-time PCR and RT-PCR analyses were repeated with more than three independent biological replicates. Primer pairs used for real-time PCR and RT-PCR analyses are listed in Supplemental Table 3 online.
Accession Numbers
The Arabidopsis Genome Initiative numbers for genes mentioned in this article are as follows: JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ4 (At1g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10 (At5g13220), JAZ11 (At3g43440), JAZ12 (At5g20900), MYB21 (At3g27810), MYB24 (At5g40350), MYB57 (At3g01530), MYB108 (At3g06490), MYC2 (At1g32640), TT8 (At4g09820), ACTIN1 (At2g37620), and ACTIN8 (At1g49240). Other genes referred in this study are AmMYB305 (P81391) and Nicotiana Actin (EU938079).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Alignment of MYB21 and MYB24.
Supplemental Figure 2. RT-PCR Analysis of Expression of Constructs Coinfiltrated for LCI Assays.
Supplemental Figure 3. MYB21 and MYB24 Function Redundantly in Regulating Male Fertility.
Supplemental Figure 4. Excess Expression of MYB21 Causes Male Sterility.
Supplemental Figure 5. MYB57 Interacts with JAZ1 in Nicotiana benthamiana.
Supplemental Figure 6. Expression of JAZ1, JAZ8, JAZ10, and JAZ11 in Arabidopsis Rosette Leaves and Flowers.
Supplemental Table 1. Bioinformatics Analysis of Am-MYB305 Binding Sites in Promoter Region of MYB21 and MYB24.
Supplemental Table 2. Primers Used for Vector Construction.
Supplemental Table 3. Primers Used for Real-Time PCR and RT-PCR.
Supplementary Material
Acknowledgments
We thank Jianmin Zhou for providing vectors for LCI assays and the ABRC for the myb21 and myb24 mutant seeds. This work was financially supported by the grants from the Ministry of Science and Technology (2011CB915404), the National Science Foundation of China (91017012), and the Ministry of Agriculture (2008ZX08011-006 and 2011ZX08011-006) to D.X.
References
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Cheong J.J., Choi Y.D. (2003). Methyl jasmonate as a vital substance in plants. Trends Genet. 19: 409–413 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chua L., Shan X., Wang J., Peng W., Zhang G., Xie D. (2010). Proteomics study of COI1-regulated proteins in Arabidopsis flower. J. Integr. Plant Biol. 52: 410–419 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Creelman R.A., Mullet J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E. (2001). Surface-to-air signals. Nature 411: 854–856 [DOI] [PubMed] [Google Scholar]
- Farmer E.E., Alméras E., Krishnamurthy V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Farmer E.E., Ryan C.A. (1992). Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4: 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Franceschi V.R., Grimes H.D. (1991). Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc. Natl. Acad. Sci. USA 88: 6745–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Howe G.A., Lightner J., Browse J., Ryan C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J.K., Howe G.A. (2008). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J.D., Schreiber S.L., Crabtree G.R. (1998). Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 16: 569–592 [DOI] [PubMed] [Google Scholar]
- Kranz H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Liu F., Ni W., Griffith M.E., Huang Z., Chang C., Peng W., Ma H., Xie D. (2004). The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Ren G., Guirgis A., Thornburg R.W. (2009). The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21: 2672–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K.E., Smith M.C., Luker G.D., Gammon S.T., Piwnica-Worms H., Piwnica-Worms D. (2004). Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA 101: 12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Browse J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Kumar V.D., Amway M., Browse J. (2003). Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol. Biol. 52: 775–786 [DOI] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Maslenkova L.T., Miteva T.S., Popova L.P. (1992). Changes in the polypeptide patterns of barley seedlings exposed to jasmonic acid and salinity. Plant Physiol. 98: 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Creelman R.A., Bell E., Mullet J.E., Browse J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.V., Lee H., Creelman R.A., Mullet J.E., Davis K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C.M., Pan J.W., Peng W., Genschik P., Hobbie L., Hellmann H., Estelle M., Gao B., Peng J.R., Sun C.Q., Xie D.X. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42: 514–524 [DOI] [PubMed] [Google Scholar]
- Reymond P., Farmer E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Reymond P., Weber H., Damond M., Farmer E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R.W., Moyano E., Culianez-Macia F.A., Schuch W., Martin C., Bevan M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., et al. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Howe G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Wang J., Chua L., Jiang D., Peng W., Xie D. (2011). The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Shan X.Y., Wang Z.L., Xie D.X. (2007). Jasmonate signal pathway in Arabidopsis. J. Integr. Plant Biol. 49: 81–86 [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B., Choi G., Yi H., Yang S., Cho I., Kim J., Lee S., Paek N.C., Kim J.H., Song P.S., Choi G. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30: 23–32 [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W.P., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Ueda J., Kato J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C.A., Cook R.J., Browse J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dai L., Jiang Z., Peng W., Zhang L., Wang G., Xie D. (2005). GmCOI1, a soybean F-box protein gene, shows ability to mediate jasmonate-regulated plant defense and fertility in Arabidopsis. Mol. Plant Microbe Interact. 18: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal D., Tzfira T. (2009). Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 14: 59–63 [DOI] [PubMed] [Google Scholar]
- Xiao S., Dai L., Liu F., Wang Z., Peng W., Xie D. (2004). COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L.H., Liu F.Q., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D.F., Xie D.X. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.Y., Li J.G., Pei M., Gu H., Chen Z.L., Qu L.J. (2007). Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep. 26: 219–228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.