Abstract
High density lipoproteins (HDL) mediate cholesterol transport and protection from cardiovascular disease. Although synthetic HDLs have been studied for 30 years, the structure of human plasma-derived HDL, and its major protein apolipoprotein (apo)A-I, is unknown. We separated normal human HDL into 5 density subfractions and then further isolated those containing predominantly apoA-I (LpA-I). Using cross-linking chemistry and mass spectrometry, we found that apoA-I adopts a structural framework in these particles that closely mirrors that in synthetic HDL. We adapted established structural models for synthetic HDL to generate the first detailed models of authentic human plasma HDL in which apoA-I adopts a symmetrical cage-like structure. The models suggest that HDL particle size is modulated via a twisting motion of the resident apoA-I molecules. This understanding offers insights into how apoA-I structure modulates HDL function and its interactions with other apolipoproteins.
The inverse correlation between plasma high density lipoprotein (HDL) cholesterol levels and the risk for cardiovascular disease (CVD) is well established 1. As its most abundant protein, apolipoprotein, (apo)A-I mediates many HDL functions. It is a key player in the HDL-mediated process of reverse cholesterol transport, and exhibits cardioprotective anti-inflammatory and anti-oxidative properties 2. Unfortunately, the conformational plasticity of apoA-I combined with HDL's staggering compositional heterogeneity has precluded an understanding of how apoA-I structure mediates these critical functions.
Most structural studies of apoA-I in HDL have utilized reconstituted particles (rHDL) that mimic in vivo intermediates between lipid-free apoA-I and mature spherical HDL (reviewed in 3, 4). Recent work on these phospholipid-containing ‘discoidal’ particles suggests that apoA-I molecules adopt a ‘double-belt’ orientation in which two apoA-I molecules wrap around a patch of phospholipid bilayer in an antiparallel fashion. They maintain a registry, stabilized by intermolecular salt bridges, in which the 5th amphipathic helix of each molecule overlaps 5. The basic tenets of this model have been supported by numerous experimental studies 6-10, though several variations on the model have been proposed. Given such colorful names as the ‘belt-buckle’ 6, 11, the ‘looped belt’ 12 and ‘solar-flares’ 13, these propose localized conformational features that may be of key importance to apoA-I interaction with plasma enzymes or exposure to oxidants. Another model 14 postulates an elongated phospholipid micelle instead of a bilayer, though two recent molecular dynamics simulations have questioned the stability of such an arrangement 15, 16. Nevertheless, all these models maintain the basic intermolecular interactions of the 5/5 double belt.
Unlike reconstituted ‘discs’, most circulating HDL contain a cholesteryl ester and triglyceride-rich core resulting in a spherical shape. Instead of a bilayer, spherical HDLs exhibit a highly curved phospholipid monolayer stabilized by surface apolipoproteins. Despite the differences in lipid structure, some have suggested that apoA-I conformations are related on both types of particles 17, 18, while others point out differences in fluorescence emission, charge or proteolytic sensitivity 19, 20. Recently, our laboratory addressed apoA-I structure in spherical particles by applying chemical cross-linking and mass spectrometry to both discoidal and spherical reconstituted HDL particles. We found apoA-I cross-linking patterns that strongly supported the 5/5 double-belt model in the discs, though a minor subset were consistent with a shifted 5/2 registry 10. Interestingly, the cross-linking patterns in reconstituted spheres were highly similar to the discs, regardless of whether they contained three molecules of apoA-I on each particle or only two 21.
How can three apoA-I molecules on spherical HDL adopt the same intermolecular contacts as only two in the discs? After considering numerous possibilities, our solution was a modification of the double belt called the trefoil model 21. This invokes a bending of the planar belts and the intercalation of a similarly bent third apoA-I molecule so that all three adopt identical intermolecular contacts as in the disc. The resulting cage-like structure can stabilize surface phospholipids, contain the neutral lipid core and offers a parsimonious solution to the similarity of the cross-links between rHDL discs and spheres. Thus, apoA-I adopts a common structural framework regardless of particle shape, at least in reconstituted HDL.
Although the use of reconstituted particles for structural studies has been repeatedly validated by functional studies, the ultimate goal is to understand the authentic particles in circulation. However, inherent sample heterogeneity has precluded systematic studies of apoA-I structure in human plasma derived HDL. Here, we took advantage of the fact that the cross-linking/mass spectrometry approach is not limited by the requirement of a homogeneous sample. We found that apoA-I adopts intermolecular interactions in plasma HDL that are strikingly similar to those of the double belt and trefoil models derived in reconstituted systems. Furthermore, our analysis supports assertions from molecular dynamics studies 22, 23 that apoA-I can twist across the particle surface to accommodate a range of HDL particle diameters. The resulting models are the first to describe apoA-I on authentic plasma HDL particles in detail.
Results
HDL isolation
To study apoA-I in isolated human HDL, we used a one-step gradient ultracentrifugation method to obtain 5 density subfractions from normal human plasma that have been extensively characterized previously 24 (defined here as HDL2b-HDL3c). ApoA-I comprises between 60-70% of total protein in HDL, with apoA-II the second most abundant at about 20%. Although the cross-linking technique can provide structural information in complex systems, we felt it was important to simplify the HDL particles as much as possible. Therefore, for each HDL density subfraction, we further isolated particles containing apoA-I but not apoA-II (LpA-I) using sulfhydryl covalent chromatography 25. Briefly, reduced HDL was passed down a sulfhydryl resin. The free disulfides at Cys 6 in human apoA-II interact with the column to retain any particle containing apoA-II (or other disulfide-containing proteins, i.e. apoJ) while non Cys-containing particles pass through. The products, defined as LpA-I2b – LpA-I3c, were analyzed by SDS PAGE in Fig. 1a. The apoA-II content of the LpA-I fractions was reduced by 70-80% compared to the original HDL sample. The separation was most effective in the HDL3c, HDL3b and HDL2b fractions but moderately less so in the HDL3a and HDL2a fractions where we typically find the most apoA-II 26. Nevertheless, apoA-I represented some 80-90% of the coomassie stainable protein in the LpA-I particles. We recognize that these particles also contained trace levels of other proteins, the majority of which likely include the apoC's 27, 28.
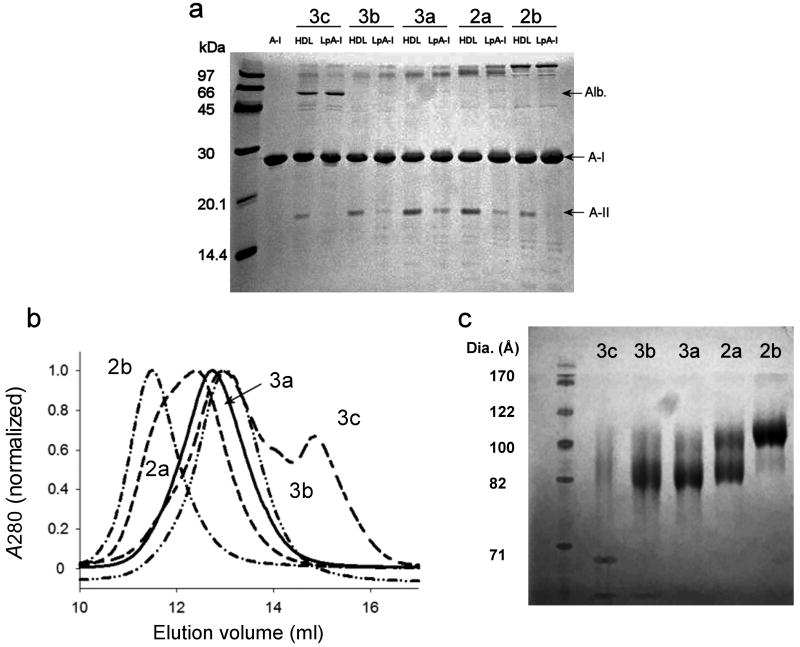
Figure 1. Isolation and characterization of human plasma LpA-I HDL particles.
(a) An 18% SDS PAGE analysis of the density-isolated HDL particles before (denoted as HDL2b-3c) and after (denoted as LpA-I2b-3c) sulfhydryl covalent chromatography. Lipid-free apoA-I is shown in lane 2 as a reference. (b) The indicated LpA-I subfractions were analyzed on a calibrated Superdex 200 gel filtration column. The figure shows a representative result from 3 analyses of 3 independently prepared samples. (c) An 8-25% native PAGE analysis of BS3 cross-linked LpA-I subfractions. See Table 1 for calculated diameters from panels b and c. All gels were stained with coomassie blue.
Once separated, we measured the LpA-I particle sizes on a calibrated gel filtration column (Fig. 1b) and by native gel electrophoresis (Fig. 1c). The diameter estimates from both techniques were consistent and ranged from 11.2 to 8.8 nm (Table 1). Native PAGE indicated that the LpA-I2a fraction contained two major populations, consistent with the non-Gaussian peak shape by gel filtration. The LpA-I particle diameters were similar to the corresponding HDL particles, although these had evidence of at least two populations in all subfractions, consistent with the removal of apoA-II containing particles (Supplementary Fig. 1a). Negative stain electron microscopy (EM) showed that all particles were spheres and confirmed the size trends in Fig. 1 (Supplementary Fig. 1b). However, the measured EM diameters were notably smaller than those from gel filtration and native PAGE. This could be due difficulties in resolving particle edges at this size 29 or deformation of particles during drying 23. Given that gel filtration and native PAGE offer hydrated diameters that are likely more physiologically relevant than EM, we averaged the diameters from these two techniques for the model building described below. Some albumin contamination was apparent in the LpA-I3c fraction in Fig. 1a, as a shoulder eluting at 15 ml in the gel filtration trace (Fig. 1b) and as a separate band in the native gel (Fig. 1c). The latter result suggests that albumin is probably not associated with the HDL particles.
Table 1. Experimental characterization of LpA-I density subfractions.
| Particle: | Densitya (g ml-1) | GF Dia. (Å) b | PAGE Dia. (Å) c | Avg. Dia. (Å) | A.A. (%)d | PL (%) | CE (%) | TG (%) | FC (%) |
|---|---|---|---|---|---|---|---|---|---|
| LpA-I2b | 1.09±0.01 | 114±3 | 110±1 | 112 | 34.7 | 32.5 | 23.0 | 4.7 | 5.1 |
| LpA-I2a | 1.11±0.01 | 101±8 | 95±1 | 98 | 41.3 | 29.5 | 20.9 | 4.7 | 3.6 |
| LpA-I3a | 1.14±0.01 | 90±3 | 90±3 | 90 | 49.3 | 25.6 | 17.5 | 4.7 | 2.8 |
| LpA-I3b | 1.16±0.01 | 88±2 | 89±3 | 88 | 56.0 | 21.9 | 15.4 | 4.4 | 2.3 |
| LpA-I3c | 1.19±0.01 | 88±2 | 88±4 | 88 | 65.5 | 16.8 | 11.7 | 4.1 | 1.9 |
Measured after the density gradient (mass / volume) from 3 independent preparations ± 1 S.D.
Determined by relative elution volume on a Superdex 200 gel filtration column calibrated with HMW standards (GE Healthcare) from 3 independent preparations ± 1 S.D.
Determined by native polyacrylamide gel electrophoresis against HMW standards (GE Healthcare) from three runs on one particle preparation ± 1 S.D.
The LpA-I3c fractions contained a detectable level of contaminating albumin (apparent in Fig. 1). To minimize artifacts in our composition calculations due to albumin contamination, we estimated the approximate ratio of albumin to all other proteins by densitometry and corrected our protein concentration values by this factor. Compositional data was obtained from two independent experiments and averaged. All percentages are w/w.
LpA-I compositional analysis
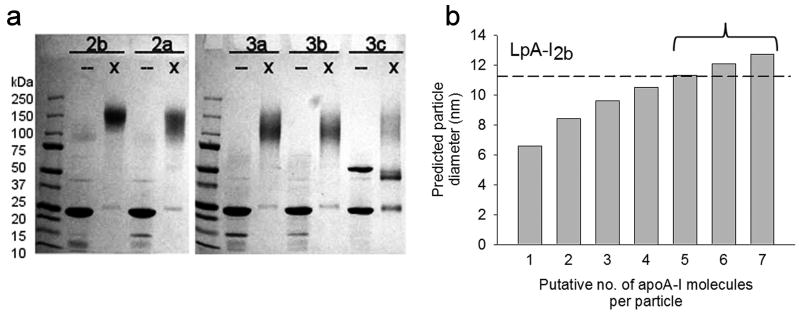
We next characterized the particle chemical compositions (Table 1). Supplementary Fig. 2 shows that the predicted particle densities, calculated from the weight percentages of the various components, correlated tightly with experimentally determined particle densities. To estimate the average number of apoA-I molecules per particle we cross-linked the samples with the soluble homobifunctional cross-linker BS3. Supplementary Fig. 3 compares the susceptibility of LpA-I3a, LpA-I2b and a control reconstituted discoidal particle to cross-linking as a function of increasing cross-linker to protein ratio. All particles responded identically to the cross-linker, indicating that the extent of intermolecular contacts were similar between all three particles. The cross-linked LpA-I particles were analyzed by SDS-PAGE (Fig. 2a). All subfractions generated a diffuse band that decreased in size only slightly between LpA-I2b and LpA-I3c. The diffuse bands are typical of cross-linked proteins as the reagent adds variable masses and the point(s) at which cross-linking occurs can affect migration on SDS gels 11. By measuring the smallest and largest band boundaries, we estimated that LpA-I2b contains 5-7 molecules of apoA-I, whereas LpA-I2a through LpA-I3c contain 4-6, 4-6, 3-5, and 3-5 apoA-I molecules, respectively.
Figure 2. Estimation of the number of apoA-I molecules per particle in LpA-I subfractions.
(a) A 4-15% SDS PAGE analysis of unmodified (–) and cross-linked (x) LpA-I subfractions. The gel was stained with coomassie blue. (b) Predicted diameters for the LpA-I2b particle given the experimentally derived particle compositions calculated at various numbers of apoA-I molecules per particle (see text). The dashed line shows the experimental particle diameter (averaged from gel filtration and native PAGE, Table 1) for this particle. The bracket indicates the range of possible apoA-I molecules/per particle determined by SDS PAGE analysis of cross-linked LpA-I2b particles (e.g. lane 2 in panel (a)).
Before building structural models, it is critical to determine the average molar stoichiometry of protein and lipid components within each species. Knowing the molar ratio of each lipid component with respect to apoA-I (calculated from Table 1), we summed the partial specific volumes for each component to derive a predicted diameter for each particle (see Methods). This was calculated for various numbers of apoA-I molecules per particle and then compared to the experimentally measured hydrodynamic diameter averaged from native PAGE and gel filtration in Table 1. An example calculation is shown in Fig. 2b for the LpA-I2b particle. As the putative number of apoA-I molecules was increased, the predicted volume (and hence diameter) of the particle increased. At 5 apoA-I molecules per particle, the theoretical diameter matched closely with the experimentally determined diameter for LpA-I2b (dotted line). This also fell within the range of expected apoA-I molecules determined by cross-linking (Fig. 2b, bracket). Performing this calculation for all LpA-I particles, we were surprised that LpA-I2a-3c all averaged 4 molecules per particle. The concordance between calculated and experimental particle diameters are shown in Supplementary Fig. 4. This suggests that a transition from a diameter of 11.2 nm to 9.8 nm involves the loss of one molecule of apoA-I, on average. However changes in diameter from 9.8 to 8.8 nm can all be accommodated with four molecules remaining on the particle. We point out that these determinations represent averages for each subfraction and do not preclude the possibility that the LpA-I3c fraction, for example, contains some particles with only three or even two molecules of apoA-I per particle. Based on these calculations, Supplementary Table 1 shows the number of molecules of each major component of the particles studied. It should be noted that our previous estimates of apoA-I molar content in native HDL particles were slightly lower and range from 2.9 in HDL3c to 4.3 in HDL2b 30, likely due to the removal of apoA-I-poorer LpA-I:A-II particles by covalent chromatography.
We also analyzed the contributions of lipid and protein to the total surface area of the particles. Using data from surface balance experiments on HDL lipids indicating molecular surface areas of 65 Å2 per phospholipid molecule 31 and 40 Å2 per cholesterol 31, total surface lipids accounted for only 33, 26, 23, 18, and 12% of the total surface area of LpA-I2b through LpA-I3c, respectively. They accounted for even less area when values from molecular dynamics simulations were assumed (27, 21, 19, 15 and 10%, based on 55 Å2 for phospholipid and 27 Å2 for cholesterol 32). Thus, by any measure, the outer shell of these particles is dominated by apoA-I.
Cross-linking and mass spectrometry
Each subfraction was cross-linked, delipidated, subjected to exhaustive trypsin digestion, and then analyzed by ESI-MS. Table 2 summarizes all MS/MS verifiable cross-links found in three separate experiments (see Methods). We identified 39 cross-links among the 5 subfractions. Of these, 25 cross-links had been identified in our previous studies of both discoidal and spherical reconstituted HDL particles. In addition, we identified a set of 14 cross-links that were not found in reconstituted HDL particles. Surprisingly, most cross-links were distributed equally across the 5 subfractions, indicating that apoA-I contacts do not vary substantially among them, despite differences in particle diameter.
Table 2. BS3 cross-links detected in human plasma LpA-I density subclasses.
| X-linka | Th. mass (Da) |
Exp. mass (Da) |
LpA-I subfraction b | Consistent 5/5 or 5/2 DB or TF model? | ||||
|---|---|---|---|---|---|---|---|---|
| 2b | 2a | 3a | 3b | 3c | ||||
| Lys239-Lys239* | 1342.777 | 1342.793 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys118-Lys239* | 1608.972 | 1608.987 | 3 | 3 | 3 | 3 | 3 | No |
| Lys94-Lys239* | 1670.962 | 1670.975 | 3 | 3 | 3 | 2 | 1 | No |
| Lys88-Lys94 | 1671.838 | 1671.888 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys96-Lys106 | 1716.908 | 1716.976 | 2 | 0 | 0 | 0 | 0 | Yes |
| Lys208-Lys239* | 1751.985 | 1752.059 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys182-Lys239 | 1897.026 | 1897.110 | 2 | 3 | 3 | 3 | 3 | Yes |
| Nt-Lys239* | 1965.950 | 1965.980 | 2 | 2 | 2 | 3 | 2 | Yes |
| Lys12-Lys23 | 2015.093 | 2015.135 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys238-Lys239 | 2108.103 | 2108.150 | 1 | 3 | 3 | 3 | 3 | Yes |
| Lys118-Lys133 | 2158.210 | 2158.297 | 2 | 3 | 3 | 3 | 3 | Yes |
| Lys208-Lys208 | 2161.210 | 2161.305 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys96-Lys239* | 2191.160 | 2191.220 | 1 | 3 | 1 | 1 | 0 | No |
| Nt-Lys118 | 2232.117 | 2232.198 | 3 | 3 | 3 | 3 | 3 | Yesc |
| Nt-Lys94 | 2294.106 | 2294.184 | 3 | 3 | 3 | 3 | 3 | Yesc |
| Lys94-Lys96 | 2302.173 | 2302.223 | 2 | 2 | 2 | 3 | 3 | Yes |
| Lys133-Lys140 | 2302.198 | 2302.236 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys182-Lys208* | 2306.340 | 2306.342 | 3 | 3 | 3 | 3 | 3 | No |
| Lys206-Lys208 | 2346.242 | 2346.313 | 3 | 3 | 3 | 3 | 3 | No |
| Lys107-Lys118 | 2417.241 | 2417.346 | 2 | 2 | 3 | 3 | 3 | Yes |
| Lys133-Lys182* | 2446.325 | 2446.425 | 1 | 1 | 0 | 0 | 0 | No |
| Lys182-Lys182* | 2451.315 | 2451.410 | 3 | 3 | 3 | 1 | 0 | No |
| Lys96-Lys118 | 2457.326 | 2457.356 | 1 | 1 | 1 | 1 | 1 | Yes |
| Lys12-Lys94 | 2530.414 | 2530.461 | 2 | 3 | 3 | 2 | 3 | Yesc |
| Nt-Nt* | 2589.193 | 2589.221 | 3 | 2 | 0 | 0 | 0 | Yes |
| Lys40-Lys239 | 2736.433 | 2736.509 | 3 | 3 | 3 | 3 | 3 | Yes |
| Nt-Lys106 | 2743.312 | 2743.389 | 0 | 3 | 3 | 3 | 0 | Yesc |
| Lys182-Lys238 | 2808.490 | 2808.587 | 3 | 3 | 2 | 2 | 2 | Yes |
| Nt-Lys12 | 2825.448 | 2825.509 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys226-Lys238 | 2863.562 | 2863.640 | 0 | 1 | 0 | 1 | 1 | Yes |
| Lys118-Lys140 | 2914.558 | 2914.656 | 3 | 3 | 2 | 2 | 2 | Yes |
| Lys88-Lys96 | 2923.452 | 2923.545 | 3 | 3 | 3 | 3 | 3 | Yes |
| Lys59-Lys208 | 3030.602 | 3030.688 | 2 | 0 | 0 | 0 | 0 | Yes |
| Nt-Lys40* | 3359.630 | 3359.730 | 2 | 1 | 0 | 0 | 0 | Yes |
| Lys23-Lys59* | 3668.957 | 3669.006 | 2 | 3 | 2 | 2 | 2 | Yes |
| Lys40-Lys45 | 3727.861 | 3727.949 | 0 | 0 | 1 | 1 | 0 | Yes |
| Lys182-Lys226* | 3892.148 | 3892.191 | 3 | 3 | 3 | 3 | 3 | Yes |
| Nt-Lys77* | 3980.948 | 3980.970 | 2 | 2 | 2 | 2 | 2 | Yesc |
| Lys77-Lys195 | 4782.366 | 4782.496 | 2 | 3 | 2 | 2 | 2 | Yes |
Cross-links marked with an asterisk are unique to native human plasma LpA-I particles (compared to both discoidal and spherical reconstituted particles).
The number of times that an MS/MS verifiable spectrum was observed in a total of three experiments (n=3).
Since the gel filtration analysis in Fig. 1b shows considerable size overlap among the LpA-I species, even between LpA-I2b and LpA-I3c, we were concerned that the similarity in cross-linking patterns may have occurred because of particle cross-contamination. To test this, we isolated LpA-I2b and LpA-I3b fractions by ultracentrifugation and disulfide chromatography as described above and then subjected the fractions to high-resolution gel filtration chromatography using three Superdex 200 columns connected in series 33. LpA-I3b was selected rather than LpA-I3c to avoid albumin contamination (Fig. 1). We then isolated the largest fractions of LpA-I2b and the smallest fractions of LpA-I3b. Supplementary Fig. 5a shows a native PAGE analysis of these fractions indicating no size overlap between the samples. Cross-linking of both samples again showed a nearly identical pattern consistent with Table 2 (Supplementary Fig. 5b). Therefore, our data strongly indicate that apoA-I molecular contacts do not vary substantially between the smallest/densest and the largest/lightest HDL particles in human plasma.
ApoA-I conformation in LpA-I particles
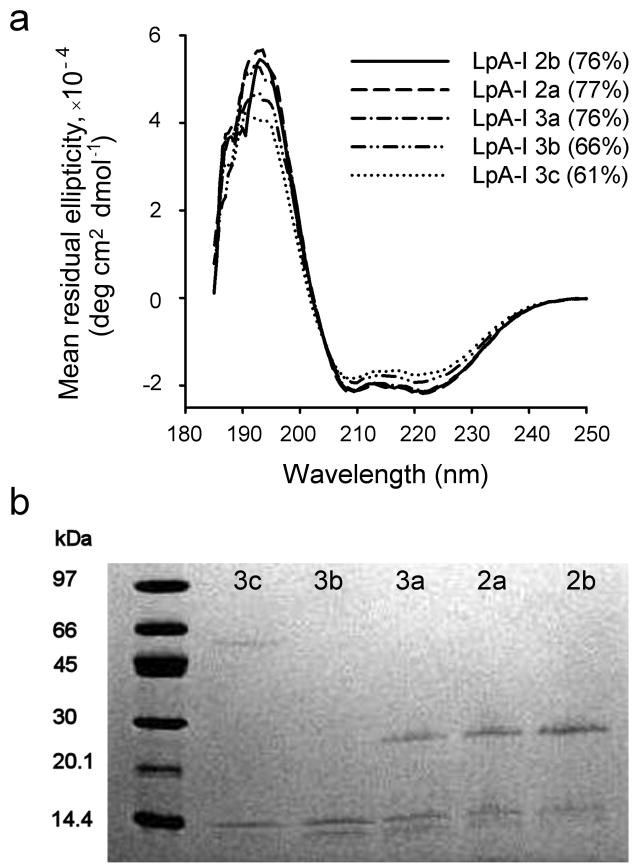
We next evaluated apoA-I structure in the subfractions by three independent methods. First, we used circular dichroism (CD) spectroscopy. Ordinarily, one would not attempt this on physiologically isolated HDL particles because the presence of diverse apolipoproteins would provide only an averaged readout. However, given the dominance of apoA-I in these separated particles (Fig. 1), we reasoned that CD should provide a suitable estimate of its secondary structure. The CD spectral shapes (Fig. 3a) were indicative of highly helical proteins with characteristic minima at 208 and 222 nm. LpA-I2b exhibited a helical content of 76%, consistent with measurements of reconstituted particles 21. LpA-I2b through LpA-I3a showed no difference in spectral shape indicating similar secondary structures. However, LpA-I3b and LpA-I3c exhibited reduced helical content, suggesting that certain apoA-I helical domains are unfolded in the smaller particles. We also evaluated the particles by susceptibility to limited proteolysis (Fig. 3b). ApoA-I molecules on LpA-I2b, 2a, and 3a were relatively resistant to trypsin under these conditions but were substantially more susceptible in the two densest fractions. Finally, we measured the binding of a monoclonal antibody (A-115) that is specific for the central domain of apoA-I (residues 115-126) 34. Supplementary Fig. 6 shows that the association of this antibody was minimal in the HDL2b through HDL3a particles, but increased dramatically in HDL3c and trended upwards in HDL3b. Taken together, the CD, proteolysis and antibody binding studies indicate that apoA-I conformation is similar in the LpA-I2b, LpA-I2a and LpA-I3a particles, but localized conformational differences likely exist in LpA-I3b and LpA-I3c.
Figure 3. Probing apoA-I conformation in LpA-I particles.
(a) Far UV circular dichroism spectra for the indicated LpA-I subfractions. The inset shows the calculated percent helicity of apoA-I as determined by the SELCON algorithm with a typical standard deviation of ± 5%. (b) An 8-25% SDS PAGE analysis of LpA-I subfractions subjected to limited trypsin digestion, stained with coomassie blue. Both panels show representative results from two independent experiments.
Evaluation of the trefoil model in LpA-I
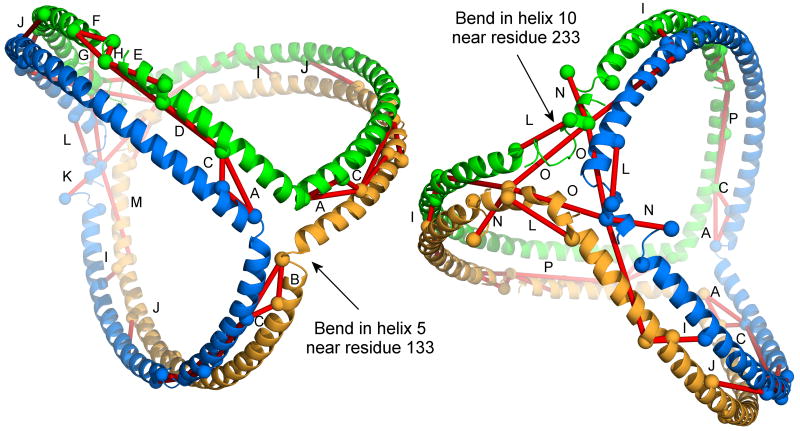
We next determined the plausibility of all cross-links in Table 2 with respect to the trefoil model of apoA-I in spherical HDL 21 (see Introduction). Interestingly, of the 39 identified cross-links, 32 were judged to be consistent with the 5/5 (or 5/2) forms of the double belt/trefoil models, i.e. the side chain - NH2 groups of the involved lysines could be within the 11.4 Å spacer arm of the cross-linker 10. These include the critical long range cross-links such as Lys118-Lys140, Lys59-Lys208 and Lys77-Lys195 that distinguish the antiparallel belt model 7. The 5/5 trefoil model is shown in Fig. 4 with applicable cross-links from Table 2 shown in red, illustrating the concordance of the model with the data.
Figure 4. Trefoil model of apoA-I on spherical particles with experimental cross-links derived from human plasma HDL particles.
The model contains three apoA-I molecules (modeled with residues 40-243) shown as ribbons, each in a different color. No lipids are shown for clarity. The same model is shown from two views: (left) looking from the intersection of helix 5 (residue 133) in each molecule, and (right) looking down from helix 10 (residue 233). Cross-links derived from physiological HDL in Table 2 judged to fit the 5/5 form of the double belt/trefoil model (see text) are shown as red lines connecting the α-carbons of the involved Lys residues (shown as spheres). Lysines were considered plausible if the α-carbons were within 25 Å (11.4 Å for BS3, and 6.8 Å for each lysine side chain). Clearly visible cross-links are labeled with a letter and refer to: A) 133-118, B) 133-140, C) 140-118, D) 107-118, E) 96-106, F) 88-94, G) 88-96, H) 94-96, I) 208-59, J) 195-77, K) 40-239, L) 226-239, M) 239-208, N) 40-239, O) 239-239, and P) 96-118. Additional molecules of apoA-I can be added to the trefoil as illustrated in Fig. 5 without affecting the crosslinking patterns, but were not shown for clarity.
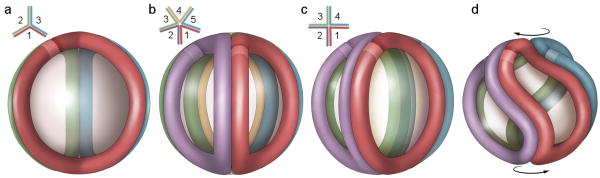
To further refine how the base trefoil model might apply to the different sized LpA-I species, we began by considering the largest particle LpA-I2b. In its fully extended form, the trefoil model defines a spherical particle with a diameter of about 10.8 nm, reasonably close to the experimentally determined diameter of LpA-I2b of 11.2 nm. However, LpA-I2b contains, on average, 5 apoA-I molecules vs. three in the trefoil. This discrepancy can be resolved by intercalating two additional apoA-I molecules with a corresponding decrease in the apoA-I bend angles so that each creates a 72° biangle vs. 120° for the trefoil (Fig. 5a,b). This “pentafoil” markedly increases the percentage of the particle surface area occupied by protein, but the particle diameter remains consistent with LpA-I2b. Importantly, all apoA-I molecules make similar intermolecular contacts, consistent with the cross-linking data. It should be noted that the original trefoil model was built without the N-terminal 43 amino acids of apoA-I. Thus, the diameter predicted by the trefoil is likely smaller than that generated by full-length apoA-I and may account for the small difference in size between the trefoil prediction and the measured LpA-I2b diameter. We also point out that models related to the double belt such as the ‘belt and buckle’ model 6 (in which the N-terminus lays back across the belts) is consistent with this arrangement. Indeed several of the cross-links in Table 2 (see footnotes) support this idea.
Figure 5. Incorporation of additional apoA-I molecules to the trefoil model and apoA-I adaptation to smaller particle diameters.
(a) Schematic representation of the three molecule trefoil model as originally proposed with each molecule of apoA-I shown in a different color (see Fig. 4 for more detail). The lighter color band on each molecule represents the N-terminus (residue 44 since the model was built in the absence of residues 1-43). The inset shows a schematic top view showing the bend angles of each apoA-I. (b) Pentameric complex proposed for the structure of LpA-I2b. (c) An idealized, fully extended tetrameric complex. (d) Twisted tetrameric complex with a reduced particle diameter as proposed for LpA-I2a. The figures were generated using Adobe Photoshop.
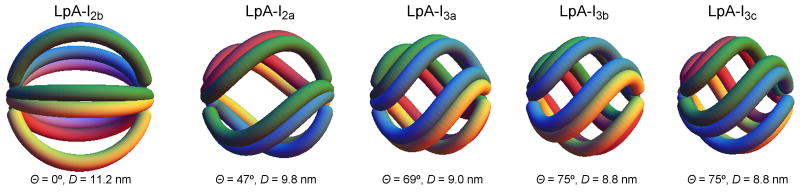
In the case of LpA-I2a, only one molecule needs to be added to the trefoil. However, the predicted diameter is larger (10.8 nm) than the experimentally measured 9.8 nm (Fig. 5c). We can envision two general ways in which the smaller diameter can be accommodated. First, regions of the resident apoA-I molecules could fold off the particle surface to form a hinge domain that reduces the distance between the particle poles 35,36. However, one expects that these exposed sites should be hypersensitive to proteolysis. Since the experiments in Fig. 3 showed no remarkable differences in proteolytic sensitivity or secondary structure between LpA-I2b and LpA-I2a (or LpA-I3a), we favor a second explanation for varying particle size that involves the twisting of apoA-I molecules on the particle surface (Fig. 5d). One can imagine turning the poles of the particle in opposite directions by an angle Θ, shrinking the particle diameter. Using a mathematical modeling program, we calculated the degree of twist required to account for the diameters of LpA-I2a through LpA-I3c (Fig. 6). By inducing moderate degrees of twist we could easily account for the observed experimental diameters of all LpA-I particles while maintaining the required number of apoA-I molecules. This predicts that the LpA-I3c particle surface is composed almost entirely of protein with only room for a small amount of surface lipids in each biangle, consistent with the relative contributions of protein and PL surface areas measured experimentally. Importantly, this twisting should not result in major changes in the intermolecular registry of apoA-I molecules, consistent with the similarities in cross-linking patterns among the LpA-I species.
Figure 6. Molecular twist required to attain the experimentally LpA-I particle diameters.
The helical domains of apoA-I molecules are represented as tubes extending across the particle surface with each molecule in a different color. In LpA-I2b, apoA-I helical domains from 5 molecules are all maximally extended. The smaller particles contain 4 apoA-I molecules per particle and each pole has been twisted by an angle Θ such that the complex diameter (D) matches the experimentally determined values (average of the diameter values from gel filtration and native PAGE in Table 1). The simulation was performed in Mathematica 7 (Wolfram Research).
Discussion
Our laboratory has taken advantage of chemical cross-linking and mass spectrometry to derive distance constraints for apoA-I molecules in HDL with both discoidal and spherical shapes of various sizes prepared in vitro. In each case, our data supported the molecular contacts proposed for the double belt model 10 in particles that have two molecules of apoA-I and the related trefoil model 21 for those with three. In addition, we interpreted a smaller subset of cross-links to be indicative of an alternative 5/2 registry 10. Since little is known about apoA-I structure in plasma-derived HDL particles and high resolution structural techniques such as NMR and X-ray crystallography will likely never be applicable to these particles, we extended this methodology to authentic LpA-I HDL. We found that the majority of the cross-links identified in reconstituted particles were also present in plasma-derived particles. Thus, one important finding of this study is that apoA-I adopts a common general structural organization, characterized by distinct intermolecular contacts, in virtually all lipid-containing particles, regardless of size and shape or natural vs. synthetic method of production. However, it is also clear from Fig. 3 and Supplementary Fig. 6 that apoA-I can undergo conformational changes within this framework, particularly in the smaller particles.
Given the dominance of apoA-I in human HDL, particularly after its enrichment in LpA-I, these data provide a new opportunity to evaluate models for apoA-I organization in human plasma HDL. In our view, a successful model must account for the following: a) the presence of multiple apoA-I molecules, both even and odd numbers, on a given particle, b) the ability to adapt to different HDL particle diameters, and c) pursuant to our cross-linking data, must maintain similar intermolecular contacts in all cases. We proposed the modified forms of the trefoil model described above that fit these criteria (Figs. 5 and 6). Below, we discuss the strengths and weaknesses of this model.
A key feature of the trefoil model that allows for intercalation of additional apoA-I molecules is the clamshell-like bending motion at two points, one near residue 133 and the other near 233 (Fig. 4). There is ample experimental 12 and theoretical evidence 37 for a hinge-like action of the sequence surrounding residue 133 in helix 5. Although less information exists on the flexibility of the region near residue 233 in helix 10, this site is known to be susceptible to V8 protease digestion in reconstituted HDL particles 38, potentially consistent with a hinge. However, more work will be required to conclusively demonstrate that this motion occurs at these sites.
The concept of apoA-I molecular twisting to modulate particle size has been proposed previously by Segrest et al. in discoidal particles 22 23. In molecular dynamics simulations, as phospholipids were incrementally removed from discoidal particles, they adopted a twisted, saddle-shape. Both associated apoA-I molecules also twisted, but remained attached to the lipids. More recent studies applied atomistic and course-grained simulation methods to simple spherical particles containing a cholesteryl oleate core and two molecules of apoA-I 39. These also demonstrated the potential for apoA-I molecules to twist around the surface of a sphere while still maintaining intermolecular contacts consistent with the double belt. Furthermore, the Borhani crystal structure depicted four apoA-I molecules (PDB: 2A01) that twisted around each other in the absence of lipid 40. When amino acids 40-243 of apoA-I were plotted as a single idealized helix, the hydrophobic face made a 360° turn around the helical axis 5. Given that the hydrophobic face interacts with a fixed lipid surface, it is quite reasonable that the entire molecule twists around a sphere.
Wu et al. recently proposed a double super helix model (DSH) of apoA-I (PDB: 3K2S) in reconstituted particles where two antiparallel apoA-I molecules, making similar intermolecular contacts as the double belt, form an elongated micellar arrangement instead of a bilayer disc 14. Although this model can be adapted to a spherical surface (see Supplementary Fig. 7 for a possibility), we favor the trefoil-based model for human plasma LpA-I particles because it allows for both even and odd numbers of apoA-I molecules that all adopt the same conformation. It is not clear how the DSH model can accommodate odd numbers of apoA-I molecules without splitting the super helix or adding a molecule in a different conformation. Second, Table 2 clearly shows N-terminus to N-terminus as well as C-terminus to C-terminus cross-links in native LpA-I particles that are predicted in the closed trefoil, but not the DSH.
One limitation of the analysis performed here is that we were unable to distinguish between intramolecular and intramolecular apoA-I cross-links in the native HDL samples. In our previous work in simple reconstituted particles, we made this distinction by isolating monomeric and dimeric apoA-I from cross-linked particles and analyzing them separately 10. This allowed us to rule out certain hairpin arrangements in favor of the antiparallel double belt in the rHDL particles. Unfortunately, this approach became prohibitively complex in particles containing 3 or more molecules of apoA-I. Thus, our extrapolations between the double belt and the trefoil-based models proposed here rely on the assumption that cross-links identified as intermolecular in the reconstituted particles are also intermolecular in the authentic LpA-I particles. However, we recognize the possibility that hairpins or certain combinations of belts and hairpins could exist in the LpA-I particles in arrangements that are consistent with our cross-linking data. Nevertheless, considering that the extent of multimer formation as a function of cross-linking reagent concentration is identical between reconstituted discs and LpA-I particles of various size (Supplementary Fig. 3), we believe it is unlikely that there are substantial differences between apoA-I contacts on rHDL discs vs. LpA-I particles. One would expect this relationship to be quite different if the discs adopted the double belt and the LpA-I particles adopted alternate arrangements such as hairpins because the degree of intermolecular contact varies dramatically between the two models.
As shown in Table 2, we found 7 cross-links that are not predicted by the trefoil-based models. These exclusively involved residues 182 and 239. The nature of these are unclear at this point, but they may reflect alternate apoA-I conformations within native HDL particles. Indeed, the recent studies of Lund-Katz et. al. 41 demonstrated that apoA-I can adopt a distinct conformation on HDL particles that may represent a partially-associated transition state. It is also possible that novel conformations of apoA-I can be induced by the presence of non-apoA-II proteins on some of the particles. We also acknowledge that we may not have yet conceived of a comprehensive model that explains all the cross-linking data, if one exists.
Implications of the new models for HDL functionality
The models shown in Fig. 6 clearly lack enough resolution to allow speculation of how apoA-I mediates interactions with other proteins. However, our demonstration that apoA-I overwhelmingly dominates the surface of these HDL particles may provide clues to how other HDL-associated proteins interact with the particle. HDL has been shown to contain some 35-50 minor proteins in addition to apoA-I and apoA-II 42 and we recently showed that a majority of these are associated with the smallest 28 or most dense HDL3c fractions 27. The presumption has been that these proteins associate with the phospholipid surface to coexist with apoA-I. However, it is clear from Fig. 6 and our composition calculations that about 85% of the LpA-I3c particle surface is covered by apoA-I. Thus, it is difficult to imagine how other proteins can find room to bind. One implication of this is that additional proteins may associate directly with apoA-I and not the lipid surface, or they may form their own separate particles which happen to co-purify in this density range.
Finally, our limited proteolysis, CD and antibody binding data indicate that, although all subfractions exhibit a similar global structure, the smallest/densest particles have localized structural differences vs. the larger/lighter ones. Under high degrees of apoA-I twist, localized areas of apoA-I may be envisioned to unfold without necessarily leaving the particle surface. This concept can be likened to twisting a rubber band until local areas begin to distend. A similar idea was previously proposed by Gu et. al in discoidal particles studied by molecular dynamics simulated annealing 23. They hypothesized that two localized regions, one near the N-terminus and the other in helix 8 of apoA-I, lose helicity when discs transition between the planar and saddle-shape morphologies. Our data may suggest that a similar transition occurs in physiological spherical particles at the LpA-I3a and LpA-I3b boundary. Whether these conformational changes occur at the same locations in physiological spherical HDL and how they might affect HDL metabolism awaits further study.
In summary, we have provided the most comprehensive set of distance constraints for native human plasma HDL particles achieved to date. The results unambiguously show that apoA-I contacts found in model reconstituted particles occur in authentic human plasma HDL particles. We extended models originally derived in reconstituted particles to these native species. Our favored scheme, based on our proposed trefoil arrangement, is a parsimonious solution that is consistent with the majority of experimental, geometric and molecular simulation data from this study and others in which changes in the particle neutral lipid core are mediated by twisting motions of the surface apoA-I molecules. It remains to be seen how these changes affect important physiological interactions with plasma lipid remodeling enzymes and cell surface receptors responsible for HDL metabolism and function.
Methods
HDL subfractionation and LpA-I isolation
Normolipidemic human EDTA plasma was subjected to a one-step gradient density ultracentrifugation procedure detailed by Chapman et al. 43. Lipoprotein fractions (VLDL, LDL and HDL) of 0.8 ml each were collected from top to bottom of the tube and the fractions corresponding to the five HDL subclasses 26: HDL2b (d 1.063 to 1.090 g ml-1), HDL2a (d 1.090 to 1.120 g ml-1), HDL3a (d 1.120 to 1.150 g ml-1), HDL3b (d 1.150 to 1.180 g ml-1) and HDL3c (d 1.180 to 1.210 g ml-1) from multiple centrifuge tubes were pooled, concentrated by ultrafiltration and dialyzed into standard Tris buffer (STB) for further LpA-I HDL isolation. These density ranges were slightly different from previous reports 44 and likely represent slight intralab variations between the French and U.S. laboratories. Three individual human plasma preparations were used for the study (n=3).
To isolate LpA-I from LpA-I/A-II HDL density subfractions, we used the sulfhydryl covalent chromatography technique developed by Rosales et al 25 with modifications (details are in Supplementary Methods).
Particle composition and characterization
The protein was determined by Markwell modified Lowry assay 45, the phospholipid (PL), total cholesterol (TC), free cholesterol (FC) and triglyceride (TG) contents were determined using enzymatic assay kits from Wako Diagnostics (Richmond, VA). Cholesterol ester (CE) concentrations were calculated by subtraction of FC concentrations from TC concentration and then multiplied by 1.67 (MW of CE divided by MW of FC). Particle size distribution was assessed by gel filtration using a calibrated Superdex 200 column (Amersham). Particle Stokes diameters were measured on cross-linked particles by native PAGE (Phast system, Amersham Pharmacia). Negative stain electron microscopy (EM) (Supplementary Methods) was also performed on LpA-I subfractions as independent approach to assess particle sizes.
Circular dichroism, limited proteolysis and surface plasmon resonance with monoclonal antibody experiments are described in Supplementary Methods.
Cross-linking and MS measurements and data analysis
LpA-I subfractions (1 mg ml-1) dialyzed into phosphate buffered saline (PBS) were cross-linked with a homo-functional cross-linker, bis(sulfosuccinimidyl) suberate (BS3) at a protein to BS3 molar ratio of 1:50 that was optimized for the maximum formation of the high-order oligomers (Supplementary Fig. 3). We carried out the cross-linking reaction, chloroform/methanol delipidation and trypsin digestion for MS analysis using our optimized protocols (Supplementary Methods).
MS measurements of cross-linked particles were performed on a Sciex/Applied Biosystems QSTAR XL coupled with an Agilent on-line capillary HPLC (Supplementary Methods). The MS data analysis was carried out using the Mascot Script in the instrument software and home built software using our previously described criteria for cross-links (Supplementary Methods).
Building the twisted particle models
The models in Fig. 6 were produced using the Mathematica 7 software package. In each case, the apoA-I proteins are represented by a pair of like-colored tubes lying on the surface of a sphere. The leftmost panel shows 5 molecules of apoA-I lying in a symmetric pentafoil arrangement on the LpA-I2b particle. The molecules have bend angles slightly less than 72° (the reduction being necessary to enable separation of the molecules from each other). The remaining panels represent twisted quatrefoil arrangements of four apoA-I molecules on the LpA-I2a, LpA-I3a, LpA-I3b and LpA-I3c particles. The strands have been twisted in such a way that the twist at the poles is ± Θ and the intermediary twisting is a linear function of distance along the polar axis. Thus, for instance, the axis of one strand is given by the vector function
| (1) |
The path of the axes of the remaining strands is obtained by rotating this curve around the polar axis. The twist Θ is determined by requiring that the strands retain a constant length. The length of such a strand as a function of the twist angle is given by
| (2) |
In the models, the helical diameter of the apoA-I proteins is assumed to be 11 Å. Supplementary Table 2 shows the total particle diameter, A; the radius R of the sphere on which the axes of the apoA-I helices lie; and the twist angle Θ required for the proteins to fit on the particle in this fashion.
Particle volume, diameter and surface area calculations
We calculated the particle volume from the experimentally determined molar ratio of various lipid components and derived particle diameter from the calculated particle volume. Particle surface area calculations were taken from compositional data for phospholipids and free cholesterol. Details are described in Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by a RO1 (HL67093) grant to WSD, an American Heart Association Great Rivers Post-doctoral fellowship to RH (3880030), a RO1 (HL48148) to WGJ and a R00 (HL087561) award to RAGDS from the NHLBI. Negative stain EM was carried out in the Vanderbilt University Research Electron Microscopy Resource of the Cell Imaging Core. This resource is partially supported by NIH grants CA68485, DK20593, and DK58404. AK was supported by INSERM, FRM and CODDIM (France). The images in Figure 6 were rendered by Marcia Hartsock (marcia@hartsockillustration.com). Any use of these images is subject to copyright law and should be negotiated with the artist. We also thank Cali Smith for excellent administrative assistance.
Footnotes
Author Contributions: RH, MJC, and WSD designed the research plan. RH, RAGDS, LKC, WGJ, and AK performed experiments. RH, WSD and TJH analyzed data and RH and WSD wrote the manuscript.
Structure Accession Numbers: Borhani crystal structure of apoA-I(Δ1-43)40: PDB: 2A01
Wu double super helix model of full length apoA-I in rHDL (hypothesized structure) 14: PDB: 3K2S
Silva trefoil model of apoA-I(Δ1-43) in a spherical rHDL particle (hypothesized structure) 21: Protein Model database, http://mi.caspur.it/PMDB/main.php (accession no. PM0075240).
References
- 1.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 2.Rye KA, Bursill CA, Lambert G, Tabet F, Barter PJ. The metabolism and anti-atherogenic properties of HDL. J Lipid Res. 2009;50(Suppl):S195–S200. doi: 10.1194/jlr.R800034-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson WS, Thompson TB. The structure of apolipoprotein A-I in high density lipoproteins. J Biol Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MJ, Bhat S, Sorci-Thomas MG. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J Lipid Res. 2008;49:1875–1883. doi: 10.1194/jlr.R800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segrest JP, Harvey SC, Zannis V. Detailed molecular model of apolipoprotein A-I on the surface of high-density lipoproteins and its functional implications. Trends Cardiovasc Med. 2000;10:246–252. doi: 10.1016/s1050-1738(00)00078-5. [DOI] [PubMed] [Google Scholar]
- 6.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 7.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: A mass spectrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 8.Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. The structure of human lipoprotein A-I. Evidence for the “belt” model. J Biol Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 9.Panagotopulos SE, Horace EM, Maiorano JN, Davidson WS. Apoliprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J Biol Chem. 2001;276:42965–42970. doi: 10.1074/jbc.M106462200. [DOI] [PubMed] [Google Scholar]
- 10.Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 11.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, et al. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, et al. Double superhelix model of high density lipoprotein. J Biol Chem. 2009;284:36605–36619. doi: 10.1074/jbc.M109.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gogonea V, et al. Congruency between biophysical data from multiple platforms and molecular dynamics simulation of the double-super helix model of nascent high-density lipoprotein. Biochemistry. 2010;49:7323–7343. doi: 10.1021/bi100588a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones MK, et al. Assessment of the validity of the double super helix model for reconstituted high density aipoproteins: A combined computational-experimental approach. J Biol Chem. 2010 doi: 10.1074/jbc.M110.187799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J Biol Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- 18.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. Review. [DOI] [PubMed] [Google Scholar]
- 19.Li HH, et al. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 20.Sparks DL, Phillips MC, Lund-Katz S. The conformation of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. 13C NMR studies of lysine ionization behavior. J Biol Chem. 1992;267:25830–25838. [PubMed] [Google Scholar]
- 21.Silva RA, et al. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc Natl Acad Sci U S A. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catte A, et al. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu F, et al. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J Biol Chem. 2010;285:4652–4665. doi: 10.1074/jbc.M109.069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontush A, Chantepie S, Chapman MJ. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler Thromb Vasc Biol. 2003;23:1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 25.Rosales C, Gillard BK, Courtney HS, Blanco-Vaca F, Pownall HJ. Apolipoprotein modulation of streptococcal serum opacity factor activity against human plasma high-density lipoproteins. Biochemistry. 2009;48:8070–8076. doi: 10.1021/bi901087z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontush A, et al. Preferential sphingosine-1-phosphate enrichment and sphingomyelin depletion are key features of small dense HDL3 particles: relevance to antiapoptotic and antioxidative activities. Arterioscler Thromb Vasc Biol. 2007;27:1843–1849. doi: 10.1161/ATVBAHA.107.145672. [DOI] [PubMed] [Google Scholar]
- 27.Davidson WS, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brouillette CG, Anantharamaiah GM. Structural models of human apolipoprotein A-I. Biochim Biophys Acta. 1995;1256:103–129. doi: 10.1016/0005-2760(95)00018-8. Review. [DOI] [PubMed] [Google Scholar]
- 29.Jerome WG. Department of Pathology, Vanderbilt University Medical Center, Nashville, Tennessee, USA, Personal communication. 2010 [Google Scholar]
- 30.de Souza JA, et al. Metabolic syndrome features small, apolipoprotein A-I-poor, triglyceride-rich HDL3 particles with defective anti-apoptotic activity. Atherosclerosis. 2008;197:84–94. doi: 10.1016/j.atherosclerosis.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Ibdah JA, Lund-Katz S, Phillips MC. Molecular packing of high-density and low-density lipoprotein surface lipids and apolipoprotein A-I binding. Biochemistry. 1989;28:1126–1133. doi: 10.1021/bi00429a029. [DOI] [PubMed] [Google Scholar]
- 32.Hofsass C, Lindahl E, Edholm O. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys J. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon SM, Deng J, Lu LJ, Davidson WS. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res. 2010;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtiss LK, Bonnet DJ, Rye KA. The conformation of apolipoprotein A-I in high-density lipoproteins is influenced by core lipid composition and particle size: a surface plasmon resonance study. Biochemistry. 2000;39:5712–5721. doi: 10.1021/bi992902m. [DOI] [PubMed] [Google Scholar]
- 35.Corsico B, Toledo JD, Garda HA. Evidence for a central apolipoprotein A-I domain loosely bound to lipids in discoidal lipoproteins that is capable of penetrating the bilayer of phospholipid vesicles. J Biol Chem. 2001;276:16978–16985. doi: 10.1074/jbc.M011533200. [DOI] [PubMed] [Google Scholar]
- 36.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 37.Jones MK, Catte A, Li L, Segrest JP. Dynamics of activation of lecithin:cholesterol acyltransferase by apolipoprotein A-I. Biochemistry. 2009;48:11196–11210. doi: 10.1021/bi901242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts LM, et al. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 1997;36:7615–7624. doi: 10.1021/bi962952g. [DOI] [PubMed] [Google Scholar]
- 39.Catte A, et al. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys J. 2008;94:2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lund-Katz S, et al. Surface plasmon resonance analysis of the mechanism of binding of apoA-I to high density lipoprotein particles. J Lipid Res. 2010;51:606–617. doi: 10.1194/jlr.M002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisar T, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman MJ, Goldstein S, Lagrange D, Laplaud PM. A density gradient ultracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J Lipid Res. 1981;22:339–358. [PubMed] [Google Scholar]
- 44.Zerrad-Saadi A, et al. HDL3-mediated inactivation of LDL-associated phospholipid hydroperoxides is determined by the redox status of apolipoprotein A-I and HDL particle surface lipid rigidity: relevance to inflammation and atherogenesis. Arterioscler Thromb Vasc Biol. 2009;29:2169–2175. doi: 10.1161/ATVBAHA.109.194555. [DOI] [PubMed] [Google Scholar]
- 45.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.