Abstract
Gene regulation by external signals requires access of transcription factors to DNA sequences of target genes, which is limited by the compaction of DNA in chromatin. Although we have gained insight into how core histones and their modifications influence this process, the role of linker histones remains unclear. Here we show that, within the first minute of progesterone action, a complex cooperation between different enzymes acting on chromatin mediates histone H1 displacement as a requisite for gene induction and cell proliferation. First, activated progesterone receptor (PR) recruits the chromatin remodeling complexes NURF and ASCOM (ASC-2 [activating signal cointegrator-2] complex) to hormone target genes. The trimethylation of histone H3 at Lys 4 by the MLL2/MLL3 subunits of ASCOM, enhanced by the hormone-induced displacement of the H3K4 demethylase KDM5B, stabilizes NURF binding. NURF facilitates the PR-mediated recruitment of Cdk2/CyclinA, which is required for histone H1 displacement. Cooperation of ATP-dependent remodeling, histone methylation, and kinase activation, followed by H1 displacement, is a prerequisite for the subsequent displacement of histone H2A/H2B catalyzed by PCAF and BAF. Chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) and expression arrays show that H1 displacement is required for hormone induction of most hormone target genes, some of which are involved in cell proliferation.
Keywords: chromatin remodeling, histone H1, hormonal gene activation, hormone receptors
In eukaryotic cells, chromatin presents a barrier to proteins that must access DNA, but also offers an opportunity for modulating gene expression. The basic unit of chromatin, the nucleosome core particle, consists of an octameric disc composed of two copies of each of the four core histone (H3, H4, H2A, and H2B), around which 147 base pairs (bp) of DNA are wrapped in 1.65 left-handed superhelical turns. However, the repeating unit of metazoan chromatin is the nucleosome, which includes a linker histone of the H1 family and some additional 20–30 bp of linker DNA. Histone H1 is localized at the dyad of the nucleosome and contacts asymmetrically the linker DNA at the nucleosomal entry and exit sites, limiting the mobility of the nucleosome core particle and contributing to the folding and stabilization of the 30-nm chromatin fiber (Brown 2003; Bustin et al. 2005).
Regulation of eukaryotic gene expression implies mechanisms that facilitate access of transcription factors to DNA sequences in chromatin. This is the function of chromatin remodeling complexes, which either chemically modify the core histones, mainly in their N-terminal tails, or use the energy of ATP hydrolysis to weaken the interaction of histones with DNA. Four different families of ATP-dependent chromatin remodeling complexes have been described, all sharing similar ATPases: the SWI/SNF, the ISWI (imitation switch), the CHD, and the INO80 (Clapier and Cairns 2009). The chromatin remodelers of the ISWI family contain two to four subunits and include NURF, CHRAC, and ACF complexes, initially purified from Drosophila melanogaster, and the human WICH or NoRC. Many ISWI family complexes (ACF and CHRAC) optimize nucleosome spacing to promote proper chromatin folding and the repression of transcription. NURF, the founding member of this family, has been isolated from human cells and has been shown to catalyze energy-dependent nucleosome sliding (Xiao et al. 2001; Barak et al. 2003), but its function in gene regulation is not clear.
Histone-modifying enzymes are also key actors in chromatin remodeling, and there are indications that they cooperate with ATP-dependent remodelers. Initial studies focused on the role of histone acetylation and, to a lesser extent, on histone phosphorylation and histone arginine methylation (Bartsch et al. 1996; Spencer et al. 1997; Li et al. 2003; Metivier et al. 2003; Vicent et al. 2006b), while lysine methylation has not been studied extensively (Metzger et al. 2005; Wissmann et al. 2007). Although H3 trimethylated at Lys 4 (H3K4me3) is associated with transcription start sites (TSSs) of active genes (Santos-Rosa et al. 2002, 2003; Schneider et al. 2004), the role of this modification during mouse mammary tumor virus (MMTV) activation has been questioned (Kinyamu and Archer 2007). The largest subunit of NURF, BPTF/NURF301, has been shown to recognize H3K4me3 by means of a plant homeodomain (PHD) located close to a bromodomain (Li et al. 2006), thus stabilizing the NURF complex on chromatin (Wysocka et al. 2005).
Unlike histone acetylation and phosphorylation, the reversibility of histone methylation was debated for many years due to the low turnover rate of methylated histones (Byvoet et al. 1972). However, recent studies have demonstrated that histone methylation can also be removed enzymatically by histone demethylases that include PADI4 (peptidyl arginine deiminase, type 4), LSD1 (lysine-specific demethylase 1), and the JmjC (Jumonji C) domain-containing proteins (Klose and Zhang 2007). Within the seven subfamilies of the JmjC domain-containing proteins, the JARID1 family members are highly similar in domain architecture and contain JmjN, BRIGHT, JmjC, zf-C5HC2, and PHD domains (Yamane et al. 2007). RBP2/JARID1A and JARID1B/PLU-1/KDM5B are the most thoroughly characterized JARID1 family members, and both have proven roles in cancer cell proliferation (Lu et al. 1999; Benevolenskaya et al. 2005). KDM5B is an H3K4-specific histone demethylase abnormally expressed in breast cancer cell lines and tissues (Lu et al. 1999; Barrett et al. 2002; Yamane et al. 2007).
Steroid hormones regulate gene activity by binding of their receptors to promoter and/or enhancer regions of their target genes and recruitment of protein kinases, histone-modifying enzymes, and ATP-dependent remodeling complexes. In addition to these transcriptional effects, steroid hormones also signal via cytoplasmic kinases that ultimately impinge on and are needed for transcriptional regulation. In breast cancer cells, a small subpopulation of estrogen receptor (ER) and progesterone receptor (PR) are palmitoylated and attached to the cell membrane (Pedram et al. 2007). Upon binding of its ligand in breast cancer cells, the membrane anchored PR cross-talks and activated the Src/Ras/Erk signaling pathway, eventually leading to Erk-mediated phosphorylation of PR at S294 and of Msk1 at T581, and to the formation of a ternary complex, pPR–pErk–pMsk1 (Vicent et al. 2006a). The activated ternary complex is targeted to the regulated promoters where Msk1 phosphorylates histone H3 at S10, leading to displacement of a repressive complex containing HP1γ (Vicent et al. 2006a). This enables the recruitment of the ATP-dependent chromatin remodeling complex BAF and the histone acetyl transferase PCAF, which acetylates histone H3 at K14, contributing to the anchoring of BAF (Vicent et al. 2009).
Many studies on gene regulation by hormones have been performed using the MMTV long terminal repeat region, which encompasses a promoter including, within 140 bp, five degenerated hormone-responsive elements (HREs) and a binding site for nuclear factor 1 (NF1). In chromatin, the MMTV promoter is organized into positioned nucleosomes (Richard-Foy and Hager 1987), with a nucleosome located over the five HREs and the NF1-binding site (Truss et al. 1995). On this promoter nucleosome, the binding site for NF1 is not accessible, and only two of the five HREs—the strong palindromic HRE1 and the weak half-palindrome HRE4—can be bound by hormone receptors, while the central HREs are not accessible for receptor binding (Pina et al. 1990). Following hormone induction in vivo, all HREs and the binding site for NF1 are occupied simultaneously on the surface of a nucleosome-like structure, and a functional synergism is observed between PR and NF1 (Truss et al. 1995). In 1984, it was shown that steroid receptors induce chromatin changes at target promoters a few minutes after hormone induction (Zaret and Yamamoto 1984). Since then, the role of SWI/SNF complexes during activation of the MMTV promoter has been widely studied (Trotter and Archer 2004; Vicent et al. 2004, 2009). We showed that BAF catalyzes ATP-dependent displacement of histones H2A and H2B (Vicent et al. 2004), facilitating access of NF1 to its initially hidden binding site (Vicent et al. 2009). On the surface of the resulting histone H3/H4 tetramer, NF1 synergizes with additional PR complexes by facilitating their binding to the central HREs: HRE2 and HR3 (Vicent et al. 2010).
It is unclear whether, in addition to BAF, other ATP-dependent chromatin remodeling complexes are recruited to the promoter after activation and are needed for preparing the chromatin for transcription. In particular, we do not know the mechanisms by which constraints imposed on chromatin dynamics by linker histone binding are managed during gene activation. Incorporation of histone H1 into MMTV minichromosomes inhibits basal transcription by reducing access of general transcription factors to the promoter, but H1 facilitates synergism between PR and NF1, leading to optimal transactivation (Koop et al. 2003). Following PR binding in Drosophila extracts, histone H1 is phosphorylated and subsequently removed from the promoter upon transcription initiation (Koop et al. 2003). Moreover, depletion of histone H1 from target promoters has been reported during hormonal activation of transcription in cultured cells (Bresnick et al. 1992), and a role for histone H1 phosphorylation in modulating MMTV activation has been postulated (Lee and Archer 1998). More generally, histone H1 phosphorylation by Cdk2 has been associated with hormone-dependent transcriptional activation (Bhattacharjee et al. 2001). In MMTV minichromosomes lacking histone H1, dNURF catalyzes the ATP-dependent remodeling needed for simultaneous and synergistic binding of PR and NF1 to the MMTV promoter (Di Croce et al. 1999). However, the role of NURF in physiological gene regulation by steroid hormones has not been addressed so far.
Here, we identify NURF as an ATP-dependent chromatin remodeling complex participating in and required for activation of hormone-dependent genes. The NURF complex is recruited as early as 1 min after induction and is anchored at the promoter chromatin by the H3K4me3 signal produced by the MLL2 and MLL3 components of the ASCOM (ASC-2 [activating signal cointegrator-2] complex) complex. The increase of the H3K4me3 signal is also due to localized displacement of the demethylase KDM5B from target chromatin. In vivo, NURF remodeling allows Cdk2 loading at the promoter and, subsequently, histone H1 phosphorylation and displacement. These steps precede and are required for recruitment of BAF to the promoter. Thus, we identify four additional enzymatic activities (NURF, ASCOM, KDM5B, and Cdk2) involved in the very initial chromatin remodeling events required for progesterone gene activation.
Results
NURF is required for hormonal induction and is recruited by activated PR to the MMTV promoter
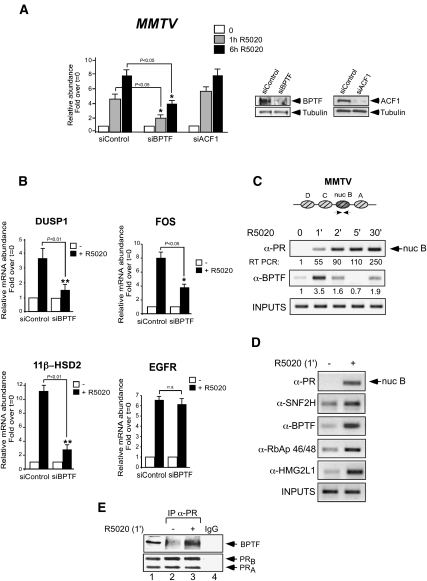
To assess whether SNF2h-containing complexes are involved in hormonal gene induction, T47D-MTVL cells carrying a single copy of the MMTV-luc transgene integrated in their genome (Truss et al. 1995) were transfected with specific siRNAs. In cells transfected with control siRNA, fivefold and eightfold increases in MMTV-luc transcription were observed after 1 h and 6 h of hormone treatment, respectively (Fig. 1A). Transfection of siRNAs against BPTF, a specific subunit of NURF, compromised the induction by 56% and 50%, respectively. In contrast, transfection with siRNAs against ACF1, a subunit present in hACF and in hCHRAC complexes (Corona and Tamkun 2004), failed to reduce the induction of the MMTV promoter, demonstrating an important role of NURF on MMTV promoter activation (Fig. 1A). Progestin induction of three endogenous progesterone target genes (DUSP1, 11β-HSD2, and FOS) was also impaired by the BPTF siRNAs to a level similar or superior to that observed for MMTV-luc (Fig. 1B). However, BPTF siRNAs did not inhibit progestin induction of the EGFR gene (Fig. 1B). We conclude that optimal induction of several, but not all, progesterone target genes depends on the physiological levels of NURF.
Figure 1.
NURF is needed for full activation of progesterone target genes and is recruited to their PR-binding sites. (A) Cells were transfected with control siRNA or siRNA against either BPTF or ACF1 as indicated, and the levels of BPTF, ACF1, and tubulin were determined by Western blotting. (Right panel) BPTF and ACF1 levels showed a marked reduction of 70% and 65%, respectively. (Left panel) Cells maintained 1 d in serum-free conditions were incubated with 10 nM R5020 for 1 and 6 h and total RNA was prepared; cDNA was generated and used as a template for real-time PCR with luciferase oligonucleotides. The values represent the mean and standard deviation from three experiments performed in duplicate. (*) P-value < 0.05. (B) cDNA was generated from cells transfected with control and BPTF siRNAs and used as a template for real-time PCR with gene-specific primers. The values were normalized with GAPDH. The histograms show the mean ± SD of three experiments performed in duplicate. (*) P-value < 0.05; (**) P-value < 0.01. (C) Cells were untreated (0) or treated for 1, 2, 5, and 30 min with 10 nM R5020 and subjected to ChIP assays with α-PR- and α-BPTF-specific antibodies. The precipitated DNA fragments were subjected to PCR analysis to test for the presence of sequences corresponding to the MMTV nucleosome B. Input material (1%) is shown for comparison. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (D) Cells treated with hormone for 1 min were subjected to ChIP assays with α-PR, α-BPTF, α-SNF2H, α-RbAp46/48, and α-HMG2L1. The precipitated DNA fragments were subjected to PCR analysis to test for the presence of sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (E) Cells were untreated (0) or treated for 1 min with R5020, lysed, and immunoprecipitated with either α-PR antibody or normal rabbit IgG as a negative control (IgG). The immunoprecipitates (IP) were analyzed by Western blotting with BPTF- and PR-specific antibodies.
To find out whether NURF is recruited to the MMTV promoter, we performed chromatin immunoprecipitation (ChIP) experiments at different time points after hormone addition (0, 1, 2, 5, and 30 min) using antibodies specific for the BPTF subunit (Barak et al. 2003). We found that BPTF is recruited as early as 1 min after hormone addition and remains bound to the promoter, although to a lesser extent, after 2 min (Fig. 1C). Five minutes after hormone addition, no BPTF is detected in the promoter, but it appears again at 30 min, suggesting a rapid and transient recruitment of NURF, followed by a second slower wave of NURF recruitment (Fig. 1C). PR is also recruited to the MMTV promoter 1 min after hormone addition, but its binding increased with time until 30 min (Fig. 1C). Simultaneously with the recruitment of PR to the MMTV promoter 1 min after hormone treatment, we observed recruitment of the other components of human NURF, such as SNF2H, RbAp 46/48, and the recently identified HMG2L1 (Fig. 1D; Vermeulen et al. 2010). In contrast, we did not observe recruitment of RSF-1, a subunit present in the RSF complex (LeRoy et al. 1998; data not shown). We also performed ChIP with a HMG2L1 antibody in cells transfected with control siRNA and siRNAs against either BPTF or SNF2H in order to test whether HMG2L1 recruitment depends on NURF activity. We found that depletion of any of the two NURF subunits compromised HMG2L1 recruitment (data not shown). Our results imply that, in the absence of BPTF or SNF2H, the complex either is not formed or, if formed, has lost the HMG2L1 chromatin anchoring activity.
Next, we used coimmunoprecipitation experiments to test whether NURF forms a complex with PR in cultured cells. One minute after hormone addition, we detected the presence of PR–BPTF complexes (Fig. 1E, lanes 2,3). Therefore, we conclude that the NURF complex is targeted initially to the MMTV promoter, likely through an interaction with PR, and, after 2 min, leaves the promoter.
Trimethylation of histone H3 at Lys 4 by methyltransferases of the ASCOM complex is necessary for hormonal induction and NURF anchoring
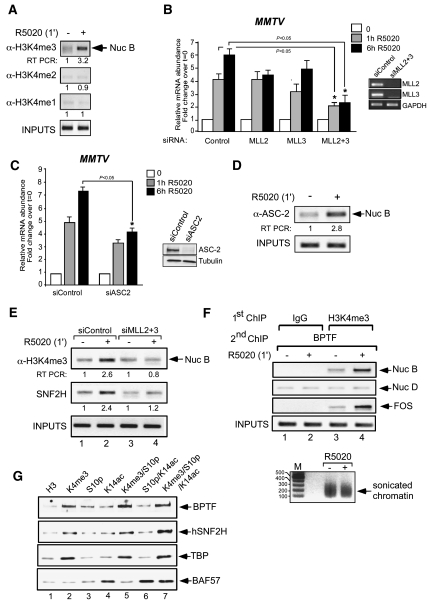
As the major subunit of NURF, BPTF, contains PHD domains that recognize methyl lysines (Wysocka et al. 2006), we wondered about possible roles of histone methyltransferases (HMTs) in hormone induction. Hormone induction increased the H3K4me3 mark at the MMTV promoter without changes in the levels of either H3K4me2 (H3 dimethylated at K4) or H3K4me1 (H3 methylated at K4) (Fig. 2A). Previous reports have described a 2-MDa complex consisting of eight proteins: the methyltransferases MLL2 or MLL3 (MLL3/4 in rodents), ASH2L, RBP5, WDR5, hDPY-30, UTX, PTIP, and the protein ASC2 (Lee et al. 2008). This complex, named ASCOM, belongs to the Set1-like complexes, a conserved family of related H3K4 methyltransferases (Goo et al. 2003; Lee et al. 2006). ASC-2 is a coactivator of many nuclear receptors and transcription factors, including retinoic acid receptor and ERs (Mahajan and Samuels 2005). Its binding to nuclear receptors is dependent on two LXXLL boxes localized in the N-terminal and C-terminal domains of ASC-2 (Li et al. 2007). We tested the function of MLL2 and/or MLL3 in hormone regulation using siRNAs (Goo et al. 2003; Lee et al. 2006). When individual siRNAs against MLL2 or MLL3 were used, there was no striking reduction of the MMTV induction, but the combination of the two siRNAs (MLL2 + 3) decreased induction by 50% and 66% after 1 h and 6 h of hormone addition, respectively (Fig. 2B). We also checked MLL1 and MLL4 siRNAs alone and in combination; they failed to elicit an inhibitory effect on MMTV hormone induction (data not shown). Moreover, in cells transfected with siRNA against ASC-2 and treated with hormone for 1 h or 6 h, we observed a decrease in hormone activation of 40% and 47%, respectively (Fig. 2C). Our results indicate that, in T47D-MTVL cells, the MLL2 or MLL3 of the ASCOM complex are necessary for hormonal activation of the MMTV.
Figure 2.
Trimethylation on histone H3K4 by MLL2/3 of the ASCOM complex is required for full hormonal transactivation and for anchoring of NURF. (A) Control cells or cells treated with R5020 for 1 min were subjected to ChIP assays with α-H3K4me3, α-H3K4me2, and α-H3K4me1. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (B, left) Cells were transfected with the indicated siRNAs and treated with 10 nM R5020 for 1 h and 6 h. RNA was extracted, and cDNA was generated and used as a template for real-time PCR with luciferase oligonucleotides. The values represent the mean and standard deviation from three experiments performed in duplicate. (Right) Cells were transfected with the indicated siRNAs, and the mRNA levels for MLL2, MLL3, and GAPDH were determined by RT–PCR. (*) P-value < 0.05. (C, left) Cells were transfected with siRNAs and treated with 10 nM R5020 as indicated, and the levels of luciferase were analyzed by RT–PCR. The values represent the mean and standard deviation from two experiments performed in duplicate. (Right) Cells were transfected with the siRNAs against ASC-2, and the levels of ASC-2 and tubulin were determined by Western blotting. (*) P-value < 0.05. (D) Cells were treated with R5020 for 1 min as indicated and subjected to ChIP assays with α-ASC-2 antibody. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown. (E) Cells were transfected with either control or a mixture of MLL2 and MLL3 siRNA, treated with hormone as indicated, and subjected to ChIP assays with α-H3K4me3 and α-SNF2H. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. A representative of three independent experiments is shown. (F) Cells were treated with R5020 as indicated and subjected to re-ChIP assays with α-H3K4me3 or IgG (first immunoprecipitation), followed by α-BPTF for the second immunoprecipitation, as described in the Materials and Methods. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to the nucleosome B (Nuc B), nucleosome D (Nuc D), and c-Fos. (Bottom panel) The size of the sonicated chromatin is shown. (G) Nuclear extracts from T47D-MTVL cells were used for pull-down experiments, with beads loaded with H3 N-terminal peptides carrying the indicated modifications. Immunoblotting was performed to detect the presence of components of the NURF complex: TBP and BAF57.
To assess whether the ASCOM complex is recruited to the MMTV promoter, we performed ChIP experiments using a specific antibody against the ASC-2 subunit. We found that ASC-2 is recruited as early as 1 min after hormone addition (Fig. 2D), coinciding with the H3K4me3 signal. Next, we asked whether the increase in H3K4me3 observed after hormone addition is generated by the ASCOM complex. Control and MLL2 + 3 siRNA-transfected T47DMTVL cells were subjected to ChIP assays with antibodies against H3K4me3. In control cells, hormone treatment induced increased H3K4me3, whereas knockdown of MLL2 + 3 abrogated K4 methylation and markedly reduced SNF2H loading on the promoter (Fig. 2E). To test whether H3K4 methylation and NURF binding take place on the same promoter, we performed sequential ChIP (re-ChIP) experiments in the MMTV and FOS promoters. Hormone treatment induced methylation of H3K4 accompanied by recruitment of NURF to the corresponding promoters (Fig. 2F, lanes 3,4), indicating that the H3K4 methylation and NURF recruitment occur in the same genomic region. In contrast, control re-ChIPs performed with irrelevant IgG as the first antibody showed no amplification product (Fig. 2F, lanes 1,2). No change was observed when the adjacent nucleosome D in the MMTV promoter was amplified by PCR (Fig. 2F, lanes 3,4, second row from the top), underlining the localized nature of the H3K4me3 mark and BPTF recruitment. The average sonicated fragment size of the chromatin used for ChIPs and re-ChIPs ranged between 200 and 250 bp (Fig. 2F, bottom panel).
Pull-down experiments with T47D-MTVL nuclear extracts and biotinylated peptides of the H3 tail containing various modifications were used to test whether H3K4 trimethylation influences the binding affinity of the NURF complex for histone tails. We detected binding of the subunits of the NURF complex—namely, BPTF and SNF2H—to histone tails containing H3K4me3 (Fig. 2G, cf. lanes 1 and 2). Binding of NURF subunits to H3K4me3 peptides containing acetylation at K14 and/or phosphorylation at S10 showed a similar affinity as peptides with H3K4me3 alone (Fig. 2G, cf. lanes 2 and 5,7). Phosphorylation or acetylation, alone or in combination, had a weak effect on binding of BPTF to the unmethylated peptides (Fig. 2G, top row, cf. lanes 1 and 3,4,6), likely reflecting the presence of both PHD domains and bromodomains in BPTF (Vermeulen et al. 2007). As a control, we used TBP and BAF57, two well-known interactors with H3K4me3 and H3K14ac, respectively (Fig. 2G, third and fourth rows from the top). These results highlight the role of H3K4me3 as the main modification enhancing NURF binding.
Displacement of the histone demethylase KDM5B contributes to the acquisition of the H3K4me3 mark after hormone induction
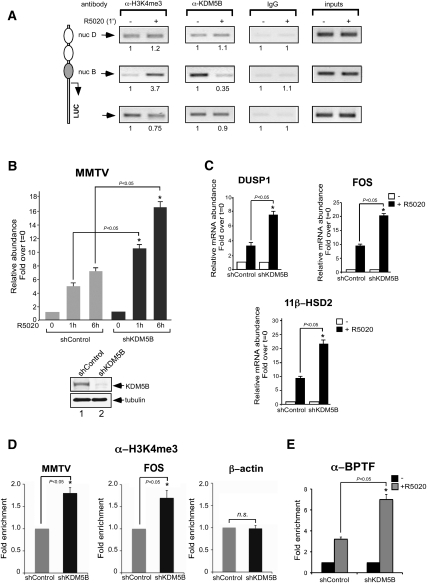
The discovery of histone demethylase enzymes capable of directly removing methyl groups from modified lysine residues has demonstrated that histone methylation is a dynamic modification. JARID1B/PLU1/KDM5B is an H3K4-specific histone demethylase overexpressed in breast cancer cell lines and tissues (Lu et al. 1999; Barrett et al. 2002; Yamane et al. 2007). We asked whether KDM5B could play a role in the increased H3K4me3 signal observed 1 min after hormone treatment in the MMTV promoter nucleosome. Using ChIP, we observed that KDM5B is present at the promoter prior to hormone treatment, and is displaced from the promoter nucleosome already 1 min after hormone addition (Fig. 3A, middle panels). In agreement with the localized nature of the increased H3K4me3 signal, KDM5B displacement is observed only on nucleosome B but not in the adjacent nucleosome D or over the transcribed luciferase gene (Fig. 3A, top and bottom panels, respectively). Knockdown of KDM5B by either siRNA transfection or long-term silencing using shRNA infection increased twofold the hormone-induced activity of the MMTV (Fig. 3B; data not shown) and other progesterone target genes, such as DUSP-1, 11β-HSD2, and FOS (Fig. 3C). Moreover, the level of H3K4me3 over the MMTV and FOS promoters was increased following depletion of KDM5B, while no change was observed in the control β-actin gene (Fig. 3D). Thus, the increase in H3K4me3 mark observed 1 min after hormone induction is generated by a concerted action of a H3K4 methyltransferase recruited to the target promoters and by a concomitant displacement of a H3K4 demethylase.
Figure 3.
Localized displacement of the histone demethylase KDM5B contributes to the acquisition of the H3K4me3 mark after hormone addition. (A) Cells treated with R5020 for 1 min as indicated were subjected to ChIP assays with α-H3K4me3, α-KDM5B, or rabbit IgG as a control. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome D or nucleosome B and Luciferase. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (B) Cells treated with control or KDM5B shRNA were incubated with R5020 for 1 h and 6 h. RNA was extracted, and cDNA was generated and used as a template for real-time PCR with luciferase oligonucleotides. The values represent the mean and standard deviation from three experiments performed in duplicate. (Bottom panel) The levels of KDM5B and tubulin were determined by Western blotting. (*) P-value < 0.05. (C) shControl and shKDM5B cells were incubated with hormone as in B. cDNA was generated and used as a template for real-time PCR using specific primers for DUSP1, c-Fos, 11β-HSD2, and GAPDH. The values represent the mean ± SD of three experiments performed in duplicate. (*) P-value < 0.05. (D) Cells treated with shControl or shKDM5B were subjected to ChIP assays with α-H3K4me3 antibody. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B, c-Fos, and β-actin. The values represent the mean ± SD of three experiments performed in duplicate. (*) P-value < 0.05. (E) Cells treated with shControl or shKDM5B were subjected to ChIP assays with α-BPTF antibody. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. The values represent the mean ± SD of two experiments performed in duplicate. (*) P-value < 0.05.
To explore a possible connection between the levels of KDM5B and NURF loading at target promoters, we checked the recruitment of BPTF in control cells and in cells depleted of KDM5B (Fig. 3E). In the absence of KDM5B, we observed a twofold increase in BPTF recruitment compared with the control cells (Fig. 3E), confirming that chromatin enriched in the H3K4me3 mark constitutes a better platform for NURF recruitment.
NURF is necessary for histone H1 displacement and the subsequent recruitment of PR–BAF complexes
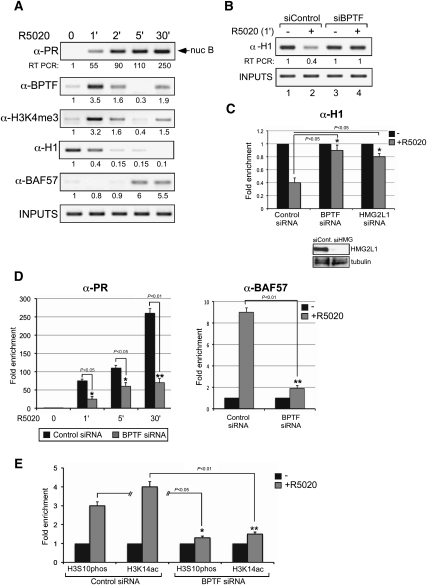
To define the sequence of events during progestin activation of the MMTV promoter, we performed ChIP assays at different times of hormone induction (0, 1, 2, 5, and 30 min) (Fig. 4A). As early as 1 min after hormone addition, PR is recruited to the MMTV promoter; it peaks at 5 min, and remains bound to the promoter until 30 min (Fig. 4A). The BPTF subunit of NURF already reached a maximum after 1 min of induction, decreased at 2 min, and was undetectable 5 min after hormone treatment. However, it reappears weakly at 30 min (Fig. 4A). The initial wave of NURF recruitment correlates very well with the intensity of the H3K4me3 signal, the main epigenetic mark that anchors the complex to chromatin (Fig. 4A). The other remodeling complex involved in hormone action, the BAF complex, is recruited to the MMTV promoter only after 5 min of hormone addition, as indicated by the ChIP behavior of its BAF57 subunit, when H3K4 trimethylation is lost (Fig. 4A).
Figure 4.
Early recruitment of NURF is needed for H1 displacement and facilitates binding of PR–BAF complexes. (A) Untreated cells treated with 10 nM R-5020 for the indicated times were subjected to ChIP assays with specific antibodies to PR, BPTF, H3K4me3, KDM5B, H1, or BAF57. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. Input material (1%) is shown for comparison. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (B) Cells transfected with the indicated siRNAs were treated with hormone and subjected to ChIP assays with α-H1. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome. A representative of three independent experiments is shown. (C, bottom panel) The levels of HMG2L1 and tubulin were determined by Western blotting. (Top panel) Cells transfected with the indicated siRNAs were treated with hormone and subjected to ChIP assays with α-H1. Quantification of the results by real-time PCR from three experiments performed in duplicate is shown. The values represent the mean ± SD. (*) P-value < 0.05. (D) Cells transfected with control siRNAs or siRNA against BPTF were treated with hormone and subjected to ChIP assays with α-PR (left panel) and α-BAF57 (right panel). Quantification of the results by real-time PCR from two experiments performed in duplicate is shown. (*) P-value < 0.05; (**) P-value < 0.01. (E) Cells transfected with control siRNAs or siRNA against BPTF were treated with hormone and subjected to ChIP assays with α-H3S10phos and α-H3K14ac. Quantification of the results by real-time PCR from two experiments performed in duplicate is shown. (*) P-value < 0.05; (**) P-value < 0.01.
What could be the remodeling step catalyzed by NURF during the initial minute of hormone action? In MMTV minichromosomes containing histone H1, binding of PR leads to its phosphorylation followed by H1 displacement (Koop et al. 2003). We first checked whether histone H1 is also displaced after hormone treatment of cultured cells. ChIP experiments with hormone-treated cells showed that H1 starts to be displaced from the promoter already 1 min after hormone addition, concomitant with the recruitment of PR and NURF and the appearance of the H3K4me3 mark and preceding BAF recruitment to the promoter (Fig. 4A). H1 was almost undetectable at the promoter 2 min and 5 min following hormone treatment, and was absent at 30 min (Fig. 4A). Quantification by real-time PCR indicated that 60% of histone H1 is removed after 1 min of hormone induction (Fig. 4C).
To test whether NURF is involved in histone H1 displacement, we used knockdown experiments. In cells transfected with control siRNA, H1 is displaced after 1 min of hormone induction, while displacement was hindered by transfection with specific siRNAs against either BPTF or HMG2L1 (Fig. 4B,C), suggesting that the NURF complex is necessary for the displacement of H1 in response to hormones. We wondered whether a similar displacement of histone H1 is observed in other PR target genes. ChIP assays showed that, in chromatin containing the PR-binding sites of genes that have been shown to be dependent on NURF (see Fig. 1), histone H1 is displaced after hormone activation and displacement is compromised upon BPTF depletion (Supplemental Fig. 1). We conclude that histone H1 displacement could be one of the remodeling steps catalyzed by NURF during the first minute of hormone induction.
Our kinetic results showed that, upon hormone induction, PR and NURF are recruited to the MMTV promoter at 1 min, while BAF and more PR are recruited at 5 min. We therefore tested whether initial remodeling by NURF is needed for the subsequent recruitment of PR and BAF complexes (Vicent et al. 2009). Binding of PR to the MMTV promoter in response to hormone treatment was diminished upon BPTF knockdown, particularly at 30 min (Fig. 4D, left panel). BPTF depletion also abrogated BAF recruitment (Fig. 4D, right panel), indicating that loading of the PR–BAF complex is dependent on the early NURF remodeling. Next, we analyzed the effect of NURF depletion on core histone modifications. We found that NURF knockdown reduced the hormone-dependent increase in H3S10 phosphorylation (H3S10phos) and H3K14 aceylation (H3K14ac) (Fig. 4E). This may be due to the need of NURF for efficient binding of PR, which targets Msk1 and the acetyltransferases SRC1 and PCAF (Vicent et al. 2004, 2006a). On the other hand, no change in H3K9ac and H3K9me3 levels were detected after 1, 2, and 5 min of hormone induction (data not shown). Thus, the NURF complex is required for histone H1 displacement, and for the subsequent binding of PR, PR-associated enzymes, and BAF, which catalyzes H2A/H2B histone displacement and facilitates NF1 binding.
Cdk2 is recruited to target promoters and its loading is facilitated by NURF
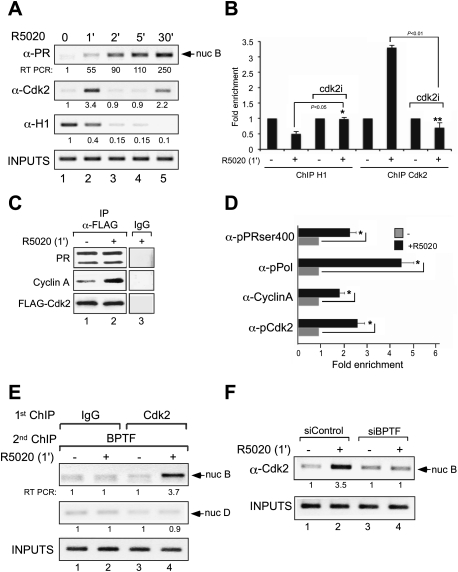
Histone H1 phosphorylation by Cdk2 has been associated with hormone-dependent transcriptional activation. Moreover, SWI/SNF remodeling is inhibited by the presence of histone H1, and this inhibition is alleviated by Cdc2/cyclinB phosphorylation (Horn et al. 2002). Since NURF is required for early histone H1 displacement (Fig. 4B), we hypothesized that NURF remodeling may facilitate Cdk2-dependent displacement of histone H1, as observed in minichromosomes (Koop et al. 2003). Therefore, we checked whether Cdk2 is recruited to the MMTV promoter during hormone activation. ChIP assays showed that Cdk2 is transiently loaded on the promoter as early as 1 min after hormone addition, coinciding with the recruitment of PR and NURF and with histone H1 displacement (Fig. 5A). A second less pronounced recruitment of Cdk2 is observed after 30 min of induction, coinciding also with the presence of NURF and full displacement of histone H1 (Fig. 5A). Inhibition of Cdk2 activity by the specific inhibitor III (Calbiochem) compromised H1 displacement and Cdk2 recruitment, along with the hormone-dependent transcriptional activity of target genes (Fig. 5B; data not shown).
Figure 5.
Cdk2 is rapidly recruited to target promoters in a NURF-dependent process, and its activity is needed for H1 displacement. (A) Cells treated with 10 nM R5020 for the indicated times were subjected to ChIP assays with specific antibodies to PR, Cdk2, or H1. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B. Input material (1%) is shown for comparison. A representative of three independent experiments is shown. The numbers below each panel correspond to the quantification by real-time PCR. (B) Control cells and cells treated with the Cdk2 inhibitor were incubated with hormone and submitted to ChIP assays with antibodies to histone H1 or to Cdk2. Precipitated DNA was analyzed by quantitative PCR for the presence of sequences corresponding to the MMTV nucleosome B region. (*) P-value < 0.05; (**) P-value < 0.01. (C) Cells transfected with Flag-Cdk2 were untreated (−) or treated for 1 min with R5020, lysed, and immunoprecipitated with either α-Flag antibody or normal mouse IgG as a negative control (IgG). The immunoprecipitates (IP) were analyzed by Western blotting with PR-, Cyclin A-, and Flag-specific antibodies. (D) Cells were treated with R5020 as indicated and subjected to ChIP assays with α-ser400pPR, α-phosRNApolymerase, α-cyclinA, and α-phosCdk2. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B region. (*) P-value < 0.05. (E) Cells were treated with R5020 as indicated and subjected to re-ChIP assays with α-Cdk2 and IgG (first immunoprecipitation), followed by α-BPTF for the second immunoprecipitation, as described in the Materials and Methods. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B and nucleosome D. (F) Cells transfected with either control siRNAs or siRNA against BPTF were treated with hormone and subjected to ChIP assays with α-Cdk2. The precipitated DNA fragments were analyzed by PCR for the presence of sequences corresponding to the MMTV nucleosome B.
To study the mechanism of Cdk2 recruitment, we tested whether PR and Cdk2/CycinA form a complex in cultured cells. Coimmunoprecipitation experiments of cells transfected with Flag-Cdk2 showed an interaction of PR with Cdk2 independent of hormone treatment (Fig. 5C). In contrast, a hormone-dependent interaction of Cyclin A with the Cdk2–PR complex was detected 1 min after hormone induction (Fig. 5C). The inverse experiment—using immunoprecipitation with α-PR and blotting with α-Cdk2—was performed, and similar results were obtained (data not shown). These results highlight the role of both cyclin A and Cdk2 as coactivators of PR.
Cdk2 can phosphorylate PR at multiple sites, including Ser 400 (Pierson-Mullany and Lange 2004). Next, we wondered whether the receptor recruited to the target promoter is phosphorylated at this site. ChIP experiments showed that, after 1 min of hormone, the PR phosphorylated at S400 (PR-S400ph) is recruited (Fig. 5D). Along with PR, Cdk2 phosphorylated at T14/T15 and cyclinA are also loaded at the promoter 1 min after adding hormone (Fig. 5D), indicating that the recruited Cdk2 is enzymatically active. We tried to detect phosphorylated histone H1 on the MMTV promoter, but did not obtain consistent results, likely due to the poor quality of the available antibodies or the rapid displacement of the phosphorylated H1 from the promoter. However, transcriptional initiation is indicated by the recruitment of RNA polymerase II phosphorylated at Ser 5 (RNA Pol-II-S5ph) after 1 min of hormone (Fig. 5D).
To test whether NURF and Cdk2 binding take place on the same promoter, we performed re-ChIP experiments in the MMTV promoter. Treatment with hormone for 1 min increased loading of BPTF accompanied by recruitment of Cdk2 (Fig. 5E, lanes 3,4), indicating that the recruitment of both Cdk2 and NURF occur in the same genomic region. In contrast, control re-ChIPs performed with irrelevant IgG as the first antibody showed no amplification product (Fig. 5E, lanes 1,2). Amplification of the adjacent nucleosome D region showed a weak signal that does not change after hormone treatment (Fig. 5E, lanes 1,2, second row from the top), indicating the localized nature of the recruitment of both BPTF and Cdk2.
Finally, we asked whether NURF remodeling activity is required for Cdk2 recruitment, and therefore histone H1 phosphorylation and subsequent displacement. In cells transfected with control siRNA, hormone treatment induced recruitment of Cdk2 accompanied by displacement of histone H1 (Figs. 4B, 5F [lanes 1,2]), whereas knockdown of BPTF abrogated Cdk2 loading (Fig. 5F, lanes 3,4) and markedly reduced H1 removal (Fig. 4B). Hence, NURF activity is necessary for the critical very early events taking place during activation of hormone-dependent promoters; namely, the loading of the kinase Cdk2 and histone H1 displacement.
Genome-wide analysis of NURF recruitment to chromatin and requirement for gene regulation
To test the generality of NURF involvement in gene regulation by progestins, we performed ChIP and sequencing (ChIP-seq) studies in control and hormone-treated cells using specific antibodies against BPTF and the Illumina Genome Analyzer. We aligned the sequence reads to the human genome hg19 assembly by using the Bowtie program (see the Materials and Methods), and used the MACS algorithm to call peaks in the aligned sequence data. We identified 26,069 BPTF peaks or regions occupied by BPTF, located mainly in introns (40.8%) and intergenic regions (46.7%), and only a small fraction in promoter regions (5.1%) (Supplemental Fig. 2A). However, the peak density was higher in proximal promoter regions (−1 kb) (Supplemental Fig. 2B). We found 7394 genomic regions at which BPTF occupancy changed upon hormone treatment. Of those, in 1848 (25%), BPTF was recruited after hormone induction, and these regions were significantly enriched at the proximal promoters (4.3% for BPTF sites vs. 1% for genomic DNA) (Supplemental Fig. 2C, left). High enrichment on BPTF is also observed in the 5′ untranslated region (UTR) and 3′UTR of all genes (Supplemental Fig. 2C, right). Hormone-dependent recruitment of BPTF showed higher density than hormone-dependent displacement of BPTF at the promoters of up-regulated genes (5% vs. 2.6%, respectively). Moreover, the recruited BPTF showed higher peaks upstream of the TSSs of progestin up-regulated genes compared with both down-regulated and unregulated genes (Supplemental Fig. 2D).
NURF accumulates near PR-binding sites and is required for induction of a majority of hormone target genes
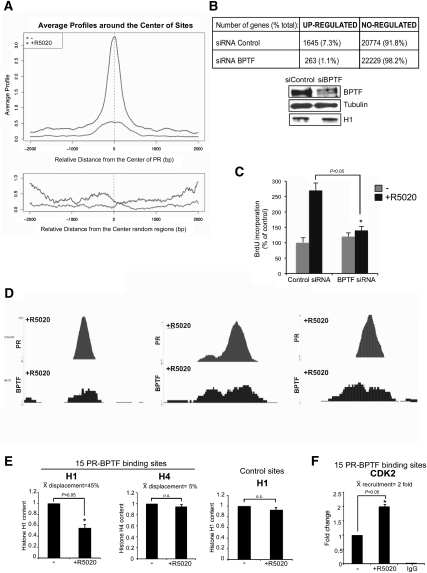
Next, we asked whether the regions where BPTF is bound upon hormone addition are enriched close to PR-binding sites. In order to address this question, we combined the data of the ChIP-seq of BPTF with the ChIP-seq of PR (C Ballare´, L Gaveglia, S Althammer, E Eyras, G Castellano, GP Vicent, and M Beato, unpubl.). Analysis of the average BPTF intensity profiles in the absence and presence of hormone showed a distribution that peaks at the center of PR-binding regions, indicating a preferential location of BPTF overlapping PR sites that is more pronounced after hormone activation (Fig. 6A, top). No peaks in the profiles were observed when random sequences instead of PR-binding sites were used for aligning the BPTF peaks (Fig. 6A, bottom). Globally, a significant overlapping (20.2%) was found between PR and BPTF genomic locations, representing a 40-fold enrichment versus genomic DNA (Supplemental Fig. 3A). These results confirmed and generalize our previous finding that PR and BPTF (as a part of NURF) form a complex in the presence of hormone and are recruited to genomic target regions enriched upstream of TSSs of progestin-regulated genes (Fig. 1).
Figure 6.
Genome-wide analysis of NURF recruitment to chromatin. (A) Genome-wide distribution of BPTF binding around PR-binding sites. (Top panel) Based on the analysis of 40 million unique reads derived from ChIP-seq experiments with antibodies to PR that identified 25,000 PR-binding sites (C Ballaré and M Beato, unpubl.), and on the BPTF-binding sites reported in this study, we generated a graphic of the distribution of BPTF-binding sites centered around the PR-binding sites. Prior to hormone administration (bottom curve), a small preference for PR-binding sequences is observed. After 30 min of hormone treatment (top curve), a much more pronounced accumulation of BPTF binding around the PR sites is observed. (Bottom panel) This is not seen if another random sequence is chosen for aligning the BPTF sites. (B) Effect of BPTF knockdown on hormone response. T47D-MTVL cells transfected with control siRNA or siRNA against BPTF were induced with hormone, and the extracted RNA was hybridized to a Whole Human Genome v2 4x44K oligonucleotide array (Agilent). Three independent knockdown experiments were performed. (Bottom) Western blot showing the effect of siRNA-mediated knockdown on BPTF protein levels. (Bottom panel) The levels of BPTF, H1, and tubulin were determined by Western blotting. (C) siRNA-mediated BPTF knockdown reduces R5020-induced cell proliferation of T47D-MTVL cells. Bars represent mean + SD (n = 5). (*) P-value < 0.05. (D) Three representative examples obtained from the ChIP-seq data where PR and NURF are found together after hormone activation. (E) Cells were treated with R5020 as indicated and subjected to ChIP assays with α-H1 and α-H4. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to 15 regions where PR and NURF are found together after hormone induction. Quantification of the results by real-time PCR from two experiments performed in duplicate is shown. The average of H1 and H4 displacement was calculated. (Right panel) Two control regions (12A1 and Pc) where no change in nucleosome profile is observed were also analyzed. (*) P-value < 0.05. (F) Cells were treated with R5020 as indicated and subjected to ChIP assays with α-Cdk2. Precipitated DNA was analyzed by PCR for the presence of sequences corresponding to 15 regions where PR and NURF are found together after hormone induction. Quantification of the results by real-time PCR from two experiments performed in duplicate is shown. (*) P-value < 0.05.
To further understand the role of NURF in hormone-dependent transcriptional activation, T47D-MTVL cells transfected with control siRNA or siRNA against BPTF were induced with hormone, and the extracted RNA was hybridized to a Whole Human Genome v2 4x44K oligonucleotide array (Agilent). In cells transfected with a control siRNA, 7.3% of all genes in the array were up-regulated in the presence of hormone (Fig. 6B). No significant changes were observed in the basal level of hormone-dependent genes of both control and BPTF knockdown samples (data not shown). Knockdown of BPTF decreased the percentage of up-regulated genes to 1.1% (Fig. 6B). No changes were observed in total H1 protein levels (Fig. 6B, bottom panel). The BPTF-affected genes that we identified are implicated primarily in three broad classes: regulation of transcription, regulation of programmed cell death, and regulation of cell proliferation (Supplemental Fig. 4). Analysis of Gene Ontology (GO) categories revealed that the identified genes are involved in transcription (15.8%, P = 0.02), cell proliferation (6.05%, P = 0.008), and regulation of cell death (either negative or positive, 3.4%, P = 0.006 and 3.6% P = 0.02, respectively). Notably, the genes affected by BPTF knockdown falling within the anti-apoptosis and cell proliferation categories present higher statistical significance (data not shown).
To examine whether NURF regulates proliferation of tumor cells, we monitored progestin-dependent cell growth by quantifying proliferation of cells transfected with BPTF-siRNA. Compared with cells transfected with an unrelated control siRNA, progestin-induced proliferation is reduced by BPTF knockdown (Fig. 6C), thus underlining the importance of the NURF complex in the control of PR-dependent growth of tumor cells.
Next, we asked whether the genes affected by BPTF knockdown are enriched in BPTF-binding regions. Combined analysis of the microarrays with ChIP-seq data showed that 11.2% of the genes whose activation is compromised after BPTF depletion present a BPTF-binding site within 10 kb upstream of the TSS and 5 kb downstream from the TTS (Supplemental Fig. 3B). This percentage is significantly different from the one obtained with either the genes up-regulated upon BPTF depletion or the nonaffected genes: 2.5% and 4.9%, respectively (Supplemental Fig. 3B). Globally, 94% of the genes that either are dependent on BPTF or contain a BPTF-binding region also present a PR-binding site. Taken together, these data suggest that the NURF complex is involved in activation of progesterone target genes, and that direct binding of the NURF complex to DNA is required to accomplish its function in most of the regulated genes.
The genomic regions where PR and NURF colocalized are enriched in Cdk2 and exhibit histone H1 depletion
In order to study the genome-wide effect of NURF recruitment on linker histone dynamics, we selected the defined regions where PR and BPTF are found together (Fig. 6D) and asked whether histone H1 is displaced in these regions following hormone treatment. ChIP experiments performed in 15 genomic regions identified as targets for BPTF and PR by ChIP-seq showed an average of 45% depletion in the histone H1 content in the presence of the hormone compared with control regions where no binding of PR or BPTF is detected (Fig. 6E). The selectivity of the histone H1 displacement was demonstrated by the lack of changes in histone H4 content (Fig. 6E, middle panel). Moreover, in the 15 genomic regions where H1 is displaced, we observed a significant recruitment of Cdk2 after hormone treatment (Fig. 6F). Thus, all of the 15 genomic regions where PR and NURF are localized together after hormone exhibited Cdk2 recruitment and histone H1 depletion.
Discussion
Our results contribute to a better comprehension of the molecular mechanisms of gene induction by describing the very initial steps of hormonal promoter activation. The data reveal an unexpected complexity in the interactions between enzymatic activities implicated in preparing the chromatin for full access of transcription factors. Apart from previously described enzymatic activities (Vicent et al. 2006a), at least four complexes act 1 min after hormone addition. An ATP-dependent chromatin remodeling complex (NURF), a protein kinase complex (Cdk2/CyclinA), a histone lysine demethylase (JARID1B/KDM5B), and a histone lysine methylase (MLL2 or MLL3)-containing complex cooperate in the displacement of histone H1 from the promoter, an important early step in gene induction by progestins.
We showed previously that, in T47D-MTVL cells treated with hormone for 5 min, PR interacts with an exposed HRE on the surface of a nucleosome positioned over the MMTV promoter (Truss et al. 1995) and recruits Brg1/Brm-containing BAF complexes (Vicent et al. 2004). Here we demonstrate that NURF interacts with PR, and that recruitment of the NURF complex in the first minute following hormone addition is a requisite for subsequent binding of BAF and activation of MMTV and other progesterone target genes. We show that NURF is anchored at the promoter of progesterone target genes by an interaction with H3K4me3, likely generated by the MLL2/3 histone lysine methylases of the ASCOM complex. This is reminiscent of the role of hormone-induced acetylation of H3K14 (Vicent et al. 2006a) in anchoring the BAF complex (Vicent et al. 2009). At both phases in activation of the promoter, a histone tail modification stabilizes the binding of an ATP-dependent chromatin remodeling complex to the target promoters.
Another similarity between the two subsequent cycles of promoter chromatin remodeling relates to the role of protein kinases. We found previously that hormone-induced activation of the Src/Ras/Erk cascade leads to phosphorylation of PR at S294 and activation of Msk1, which is targeted to the promoter by PR and phosphorylates H3 at S10, contributing to the displacement of a repressive complex containing HP1γ (Vicent et al. 2006a). Here we show that, prior to this event, 1 min after hormone, PR interacts with a complex of Cdk2 and CyclinA that phosphorylates PR at S400, is recruited to the promoter, and phosphorylates histone H1, leading to its displacement. Thus, there are two similar and consecutive cycles essential for transcriptional activation of hormone-dependent genes, both involving the collaboration between protein kinases, histone-modifying enzymes, and ATP-dependent chromatin remodelers. Each of the remodeling complexes is anchored at the promoter by different epigenetic marks: H3K4me3 established by MLL2/3 anchors NURF, and H3K14ac established by PCAF anchors BAF (Fig. 7). The final output of the first cycle is to decompact the chromatin fiber by displacing histone H1, and the outcome of the second cycle is to open the nucleosome by displacing H2A/H2B dimers.
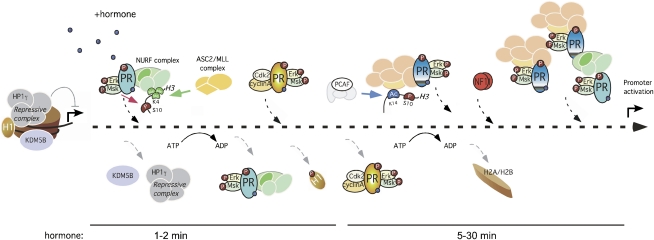
Figure 7.
Model for the initial steps of MMTV promoter activation. Before hormone addition, the MMTV promoter is silent and is associated with repressive complexes that include HP1γ and JARID1B/KDM5B. After 1 min of hormone, the activated complexes of pPR, pErk, and pMsk1, with either the NURF complex or the methyltransferase ASCOM complex, are recruited to the MMTV promoter. The combined action of the ASCOM complex and the displaced KDM5B increases histone H3 in K4 trimethylation, stabilizing NURF at the promoter. Other modifications, such as H3S10phos and H3K14ac, produced likely by Msk and PCAF, could also collaborate in NURF anchoring. NURF remodeling facilitates access of the Cdk2–CyclinA kinase, which phosphorylates histone H1 and promotes its displacement, leading to unfolding of the chromatin fiber. In a second cycle, PR–BAF complexes are recruited along with PR–PCAF complexes that generate H3K14ac and anchor the BAF complex, enabling ATP-dependent H2A/H2B displacement. This nucleosome opening facilitates NF1 binding, generating a stable platform that exposes previously hidden HREs for the recruitment of additional PR and BAF complexes, coactivators, and, eventually, promoter activation.
NURF is required for activation of PR target genes
The chromatin remodeling complex NURF has been shown to be necessary for both transcription activation and repression in vivo (Deuring et al. 2000; Badenhorst et al. 2002; Corona et al. 2007; Bonaldi et al. 2008). Most reports on the role of NURF in gene regulation come from studies in Drosophila, where NURF is involved in the activation of several genes, including the homeotic selector gene engrailed, ultrabithorax, ecdysone-responsive genes, and the roX noncoding RNA (Deuring et al. 2000; Badenhorst et al. 2005; Bai et al. 2007; Bonaldi et al. 2008). These studies were complemented with mechanistic studies using recombinant Drosophila NURF complex (Hamiche et al. 1999; Xiao et al. 2001; Hamiche and Xiao 2004). In contrast, little is known regarding the mechanism of action of NURF in human cells, except for reports on a role in neuronal physiology (Bowser et al. 1995; Mu et al. 1997; Barak et al. 2003). We found that, in T47D-MTVL human breast cancer cells, NURF is essential for efficient hormone-dependent activation of several PR target genes, and is recruited to the target promoters via an interaction with PR. The BAF complex is also recruited to the MMTV promoter within minutes after progestin treatment, but the kinetics of loading of both chromatin remodelers are different. NURF is recruited after 1 min of hormone treatment, while BAF is loaded only after 5 min and its recruitment depends on NURF action. Our findings highlight the notion of transcription initiation as a process involving consecutive cycles of enzymatic chromatin remodeling, where each enzyme complex is necessary at a given time point and catalyzes a particular remodeling step. These results support the existence in T47D-MTVL cells of several pools of PR, associated with the different chromatin remodelers. How the coordinated action of each PR population on target promoters is orchestrated is not well understood, but phosphorylation of the receptor by different kinases and post-translational modifications of nucleosomal histones could provide possible mechanisms.
Local increased H3K4me3, generated by the exchange of KDM5B by ASCOM, anchors NURF at promoters
Although H3K4me3 marks TSSs of virtually all active genes (Santos-Rosa et al. 2002, 2003; Schneider et al. 2004) the role of this modification during MMTV activation has been questioned (Ma et al. 2001; Kinyamu and Archer 2007). We showed that, in T47D-MTVL cells, the MLL2/3-containing complex ASCOM is recruited to a target promoter after 1 min of hormone and increases H3K4me3. Experiments with siRNA knockdown, ChIP, and peptide pull-down assays showed that H3K4me3 is critical for NURF anchoring at the promoter. The very early and transient appearance of the H3K4me3 mark could explain the apparent controversy with previously published studies (Ma et al. 2001; Kinyamu and Archer 2007). We found that the H3K4me3 signal observed at the MMTV promoter is due to the concerted recruitment of the ASCOM complex and the localized displacement of the H3K4me3/2/1 demethylase KDM5B. Knockdown of KDM5B increased the basal and hormone-dependent activity of PR target genes and caused an increase in H3K4me3 levels at the promoters in the absence of hormone.
The molecular mechanism underlying hormone-induced displacement of KDM5B is unclear. It has been reported that KDM5B forms a complex with histone deacetylases (HDACs) (Barrett et al. 2007). We showed previously that an HP1γ-containing complex is bound to the MMTV promoter prior to induction, and is displaced by phosphorylation of H3S10 catalyzed by hormone-activated Msk1 (Vicent et al. 2006a). However, in coimmunoprecipitation experiments, we did not detect an interaction between KDM5B and HP1γ (data not shown). Recently, it has been reported that PARP1 can parylate and inactivate KDM5B catalytic activity (Krishnakumar and Kraus 2010a). Since nuclear receptors are known to activate PARP1 (Kim et al. 2005), it is possible that this pathway participates in the inactivation and displacement of KDM5B following progestin treatment.
The PHD finger present in the BPTF subunit of NURF acts as a highly specialized methyl lysine-binding domain critical for NURF loading (Wysocka et al. 2006). H3S10ph and H3K14ac, two other post-translational modifications present in the MMTV promoter chromatin after hormone addition (Vicent et al. 2006a), increase the binding of the PHD domain to H3K4me3 (Fig. 2G). Binding of BPTF to acetylated lysines could be expected, as the protein contains a bromodomain in its C terminus (Jones et al. 2000), but the interaction with H3 phosphopeptides was not predicted, as BPTF does not encompass a consensus 14-3-3-like domain (Macdonald et al. 2005). Regarding the role of the H3K9me3 signal in NURF recruitment, peptide pull-down experiments showed no interaction of NURF components with the H3K9me3 mark (data not shown). Moreover, either knockdown or inhibition of the methyltransferase G9a increased the basal level of transcription in several target genes without affecting the fold induction after hormone. The same effect was observed when cells were depleted of HP1γ, indicating that the H3K9me3 signal anchors HP1γ at the target chromatin (Vicent et al. 2006a).
The NURF complex is recruited after 1 min of hormone, decreased after 2 min, and is almost undetectable after 5 min (Fig. 4A). How NURF is released from target chromatin is still unknown. Binding of NURF correlates closely with H3K4me3, and therefore a decrease in the trimethylation of H3K4 would explain NURF displacement. It has been proposed that methylation of histone H3R2 by PRMT6 and methylation of H3K4 by MLLs are mutually exclusive (Guccione et al. 2007). Moreover, H3R2 methylation has been reported to block the binding of effectors that harbor methyl-specific binding domains, including PHD domains, chromodomains, and Tudor domains (Hyllus et al. 2007; Bedford and Clarke 2009). Thus, the presence of the H3R2me2 mark could cooperate in erasing the H3K4me3 signal from the promoters and in competing for NURF binding, thus triggering NURF displacement.
Displacement of histone H1 from PR target promoters depends on NURF and Cdk2/CyclinA
MMTV minichromosomes reconstituted with Drosophila embryo extracts were used previously to address the role of histone H1 (Koop et al. 2003). Histone H1 increases nucleosome spacing and compacts the chromatin, hinders access of general transcription factors to the MMTV promoter, and thus inhibits basal transcription (Koop et al. 2003). In the presence of bound PR, H1 is phosphorylated by Cdk2 and subsequently is removed from the promoter upon transcription initiation (Koop et al. 2003). The kinase Cdk2 is known to phosphorylate histone H1 in vivo, resulting in a more open chromatin structure by destabilizing H1–chromatin interactions (Contreras et al. 2003). Histone H1 phosphorylation by Cdk2 has been associated with hormone-dependent transcriptional activation (Bhattacharjee et al. 2001). We found that NURF facilitates the access of Cdk2/CyclinA to target promoter chromatin, and this could explain its role in H1 displacement from the MMTV promoter and from 15 other PR-binding sites that also contain NURF and recruit Cdk2 after hormone treatment. Along with the general effect of Cdk2 inhibition on gene regulation by progestins (R Wright, C Ballaré, and M Beato, unpubl.), these results support a very general role of Cdk2/CyclinA in histone H1 eviction during the initial steps of hormonal chromatin remodeling.
There is evidence for a direct interaction between PR and Cdk2, CyclinA, or cyclinE that could explain how Cdk2/CyclinA is recruited to the target promoters (Faivre et al. 2005; Narayanan et al. 2005). In the T47D-MTVL breast cancer cell line, we found a hormone-independent association of PR with Cdk2 and recruitment of CyclinA to this complex upon hormone addition. Therefore, PR could recruit Cdk2/CyclinA to the target promoter upon hormone addition. We do not know whether NURF and Cdk2/CyclinA form a single ternary complex with PR, or rather are in two different PR-associated complexes. Although by coimmunoprecipitation we detected interaction of PR with both complexes after 1 min of hormone addition (Figs. 1E, 5C), a more in-depth analysis performed at 1-min intervals at 30°C revealed that NURF is recruited before Cdk2/CyclinA (data not shown). These results suggest that NURF recruitment is required for Cdk2/CyclinA loading at target promoters, and support the existence of two independent complexes.
Although H1 displacement takes place locally, it could have a long-range effect on chromatin decompaction, as demonstrated with in vitro assembled condensed chromatin (Routh et al. 2008). Displacement of histone H1 could be a prerequisite for all subsequent steps in remodeling, as SWI/SNF remodeling has been reported to be inhibited by the presence of histone H1 (Horn et al. 2002). A connection between ISWI-containing remodeling machineries and histone H1 dynamics has been reported previously in Drosophila (Corona et al. 2007). In this system, ISWI promotes the association of the linker histone H1 with chromatin (Corona et al. 2007). Along these lines, we cannot discard that NURF is also involved in later steps during hormone induction by helping histone H1 deposition back at the promoter.
How H1 binding is regulated and leads to a more open chromatin structure remains unclear. Some models proposed that Cdk2-dependent H1 phosphorylation leads to the decondensation of chromatin during interphase by disrupting the association of HP1α with the chromatin fiber (Hale et al. 2006). We observed a hormone-dependent displacement of HP1γ from the MMTV promoter without changes in H3K9me3 levels (Vicent et al. 2006a). Whether H1 and Hp1γ are interacting as part of a common repressive complex requires further studies but constitutes an attracting hypothesis. On the other hand, PARP-1 possesses the ability to disrupt chromatin structure by PARylating histones (e.g., H1 and H2B) and a variety of nuclear proteins involved in gene regulation (D'Amours et al. 1999; Krishnakumar and Kraus 2010b). Both PARP-1 and H1 compete for binding to nucleosomes (Kim et al. 2004) and exhibit a reciprocal pattern of binding at actively transcribed promoters: H1 is depleted and PARP-1 is enriched (Krishnakumar et al. 2008). Other post-translational modifications of H1 have been proposed to influence its binding and function. Histone H1 is acetylated at Lys 26 in vivo and can be deacetylated by the NAD+-dependent HDAC SirT1, promoting the formation of repressive heterochromatin (Vaquero et al. 2004). This effect was accompanied by an enrichment of H1 at the promoter, and the spreading of heterochromatin marks like H3K9me3 and H4K20me1 throughout the coding region.
Regarding the NURF-mediated changes in chromatin structure, analysis of nucleosome profiles obtained by MNase digestion before hormone treatment showed a preferential location of nucleosomes overlapping with NURF and PR sites that is less pronounced after hormone activation (data not shown), indicating that chromatin remodeling is involved.
Analysis of the hormone-regulated genes that are affected by depletion of NURF reveals many genes involved in cell cycle and cell proliferation, which could mediate the proliferative response of breast cancer cells to progestins. This may explain the inhibition of cell proliferation in response to progestins that we observed in T47D cells depleted of NURF. A similar inhibition of the proliferative response has been observed in cells depleted of Cdk2 (R Wright, C Ballaré, and M Beato, unpubl.). These results indicate that histone H1 displacement may be a prerequisite for the effects of progestins on cell proliferation, and therefore the enzymes involved in this process would be novel targets for the pharmacological control of breast cancer cell proliferation.
A model of our present view of the initial steps in progesterone activation of the MMTV promoter is shown in Figure 7. Although the different steps of remodeling are depicted as a linear time sequence, we cannot exclude that some of these process occur in parallel and in different time sequences in different target promoters. Our model reflects the average time sequence in the cell population. Briefly, after hormone induction, activated PR carrying Erk and Msk1 binds first to the exposed HRE1 on the surface of the MMTV promoter nucleosome in a process that does not require chromatin remodeling (Pina et al. 1990). Along with the activated PR kinases, the NURF and ASCOM complexes are recruited to the promoter chromatin in one or several complexes. The combined action of ASCOM recruitment and KDM5B displacement enhances H3K4me3 and stabilizes NURF at the promoter. Other modifications, such as H3S10phos and H3K14ac produced by Msk1 and PCAF, could also contribute to NURF anchoring. Once at the promoter, NURF remodels the nucleosome and facilitates the access of PR and the associated Cdk2/CyclinA kinase, which phosphorylates histone H1 and promotes its displacement, contributing to unfolding of the chromatin fiber (Robinson et al. 2006). Although we observed that H3S10 phosphorylation by Msk1 plays a role in HP1γ displacement, we cannot discard that phosphorylation of histone H1 also contributes to this process (Hale et al. 2006). H1-depleted nucleosomes constitute a suitable substrate for recruitment of PR–BAF complexes and further remodeling events catalyzed by BAF and PCAF. H3K14 acetylation by PCAF promotes BAF anchoring (Vicent et al. 2009). BAF mediates ATP-dependent displacement of histones H2A/H2B, and thus facilitates binding of NF1. Bound NF1 stabilizes the open conformation of the H3/H4 tetramer particle that exposes the previously hidden HREs, allowing synergistic binding of further PR–BAF–kinase complexes and PCAF (Vicent et al. 2010).
Finally, given that NURF is also recruited to the promoter 30 min after hormone addition, when no H1 is present, we cannot exclude that NURF catalyzes later steps in chromatin remodeling involving histones or nonhistone chromatin proteins. Indeed, our own results indicate that NURF can act on MMTV minichromosomes lacking histone H1 (Di Croce et al. 1999). In this respect, it remains to be established whether NURF and BAF fulfill partly redundant functions, cooperate on the same promoter, or, rather, are mutually exclusive.
Materials and methods
Cell culture and hormone treatments
T47D-MTVL breast cancer cells carrying one stably integrated copy of the luciferase reporter gene driven by the MMTV promoter (Truss et al. 1995) were routinely grown in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. For the experiments, cells were plated in RPMI medium without phenol red and supplemented with 10% dextran-coated charcoal-treated FBS (DCC/FBS), and, 48 h later, medium was replaced by fresh medium without serum. After 24 h in serum-free conditions, cells were incubated with R5020 (10 nM) for different times at 37°C.
ChIP and re-ChIP assays in cultured cells
ChIP assays were performed as described (Strutt and Paro 1999) using anti-PR (H190), anti-Cdk2 (M2), and anti-HMG2L1 from Santa Cruz Biotechology; anti-H3K4me3, H3K9me3, H3K9ac, and anti-cyclinA from Abcam; anti-ASC-2 and anti-BPTF from Bethyl Laboratories, and anti-SNF2H, anti-H3K4me2, anti-H1 (AE-4) (05-457), anti-H3S10phos (Ab+ 17-685), anti-H3K14ac (07-353), and anti-H3K4me1 from Millipore. Anti-BPTF antibody was a kind gift from R. Shiekhattar. The RSF-1 antibody was a kind gift from Dr. Danny Reinberg. In order to perform ChIPs after short times of induction, cells were incubated during 1, 2, 5, or 30 min with hormone, then the medium was aspirated very quickly and replaced by medium containing 1% formaldehyde. After 10 min of cross-linking at 37°C, the reaction was stop by adding 125 mM glycine. After 5 min at room temperature, the plaques were place on ice and washed twice with cold PBS, and then the cells were collected. Chromatin was prepared following the protocol described above (Strutt and Paro 1999). The efficacy of this protocol was demonstrated by performing kinetics of cross-linking with 1% formaldehyde at 37°C. We found that, already after 1 min of cross-linking, the fold change (t = 0 vs. 1 min) of PR recruitment did not change compared with the standard 10-min cross-linking. Quantification of ChIP was performed by real-time PCR using LightCycler (Roche). The fold enrichment of the target sequence in the immunoprecipitated compared with input (Ref) fractions was calculated using the comparative Ct (the number of cycles required to reach a threshold concentration) method with the equation 2Ct(IP) − Ct(Ref). Each of these values were corrected by the human β-globin gene and were referred to as relative abundance over time 0. Primers sequences are available on request. For re-ChIP assays, immunoprecipitations were washed sequentially as described previously (Shang et al. 2002). Complexes were eluted with 10 mM DTT for 30 min at 37°C, diluted 50 times with dilution buffer, and immunoprecipitated with the indicated antibodies.
RNAi experiments
All siRNAs were transfected into the T47D-MTVL cells using Lipofectamine 2000 (Invitrogen). After 48 h, the medium was replaced by fresh medium without serum. After 1 d in serum-free conditions, cells were incubated with R5020 (10 nM) or vehicle (ethanol) for different times at 37°C. The down-regulation of BPTF, ACF1, KDM5B, HMG2L1, and ASC-2 expression was determined by Western blotting. The depletion of MLL2 and MLL3 (Fig. 2B,E) was estimated by measuring the mRNA levels using real-time PCR. Primer sequences are available on request. Stable KDM5B-depleted T47D-MTVL cells were generated by using lentiviral shRNAs obtained from Sigma (MISSION shRNA lentiviral transduction particles).
RNA extraction and RT–PCR
Total RNA was prepared and cDNA was generated as described previously (Vicent et al. 2006a). Quantification of LUC and GAPDH gene products was performed by real-time PCR. Each value calculated using the standard curve method was corrected by human GAPDH and was expressed as relative RNA abundance over time 0. Primer sequences are available on request.
Coimmunoprecipitation assay
Cells were lysed and cell extracts (2 mg protein) were incubated with protein G/A agarose beads previously coupled with 6 μg of the corresponding antibodies or an unspecific control antibody. The immunoprecipitated proteins were eluted by boiling in SDS sample buffer. Inputs and immunoprecipitations were analyzed by Western blot using BPTF-, Cyclin A-, Cdk2-, and PR-specific antibodies.
Peptide pull-down assays
Nuclear extracts from T47DMTVL breast cancer cells were prepared as described (Dignam et al. 1983). Peptide pull-down assays were performed as described previously (Vermeulen et al. 2007), with the exception of using 100 μg of nuclear extract during incubation of peptide-bound beads. Synthetic biotinylated H3 peptides were kind gifts from M. Vermeulen (University Medical Center, Utrecht, The Netherlands). For Western immunoblotting, antibodies against BPTF, BAF57 (Bethyl Laboratories), TBP (Abcam), and SNF2H (Upstate Biotehnology) were used.
Solexa ChIP-seq analysis
DNA was subjected to deep sequencing using the Solexa Genome Analyzer (Illumina). Single-ended sequences were trimmed to 36 bp and mapped to the human genome assembly hg19 using Bowtie (http//bowtie-bio.sourceforge.net), an ultrafast short-read mapping program (Langmead et al. 2009), keeping only tags that mapped uniquely and with no more than two mismatches.
Peak detection and annotation analysis
The integrated software MACS (Zhang et al. 2008) was used to detect peaks that were enriched from background reads. The algorithm was applied by using the sliding window method to count the reads with default settings and a P-value threshold of 1E-05, and to identify differential binding between the two conditions by treating the T0 of the samples as the control. All of the genome-wide annotations and statistics of protein–DNA interaction patterns from ChIP-seq data were performed with the stand-alone application CEAS (Cis-regulatory Element Annotation System). In the case of the ChIP-seq PR data, the signals of treated and untreated samples were compared using Poisson analysis (P-value < 0.001) (C Ballare´, L Gaveglia, S Althammer, E Eyras, G Castellano, GP Vicent, and M Beato, unpubl.).
Cell proliferation assay
T47DMTVL cells transfected with control or BPTF siRNAs were cultured as described above. Cells (1 × 104) were plated in a 96-well plate in the presence or absence of 10 nM R5020. The cell proliferation ELISA BrdU Colorimetric assay (Roche) was performed according to the manufacturer's instructions. Figure 6C shows the percentage increase of proliferation in the presence versus absence of R5020. The experiments were performed in quintuplicate.
Acknowledgments
We thank Ramin Shiekhattar (The Wistar Institute, USA) for anti-BPTF antibody, Hung-Ying Kao (Case Western Reserve University, USA) for Flag-Cdk2 expression plasmid, Danny Reinberg (New York University, USA) for RSF-1 antibody, and Juan Valcárcel for advice on the manuscript. The experimental work was supported by grants from the Departament d'Innovació Universitat i Empresa (DIUiE), Ministerio de Educación y Ciencia (MEC) BMC 2003-02902, Consolider (CSD2006-00049), Fondo de Investigación Sanitaria (FIS) PI0411605 and CP04/00087, and EU IP HEROIC. G.P.V. was a recipient of a fellowship of the Ramón y Cajal Programme. L.G.′s work was supported by a “La Caixa” fellowship.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.621811.
Supplemental material is available for this article.
References
- Badenhorst P, Voas M, Rebay I, Wu C 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev 16: 3186–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C 2005. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19: 2540–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Larschan E, Kwon SY, Badenhorst P, Kuroda MI 2007. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics 176: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R 2003. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J 22: 6089–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A, Madsen B, Copier J, Lu PJ, Cooper L, Scibetta AG, Burchell J, Taylor-Papadimitriou J 2002. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen?. Int J Cancer 101: 581–588 [DOI] [PubMed] [Google Scholar]
- Barrett A, Santangelo S, Tan K, Catchpole S, Roberts K, Spencer-Dene B, Hall D, Scibetta A, Burchell J, Verdin E, et al. 2007. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer 121: 265–275 [DOI] [PubMed] [Google Scholar]
- Bartsch J, Truss M, Bode J, Beato M 1996. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci 93: 10741–10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG Jr 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 18: 623–635 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee RN, Banks GC, Trotter KW, Lee HL, Archer TK 2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol Cell Biol 21: 5417–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldi T, Straub T, Cox J, Kumar C, Becker PB, Mann M 2008. Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol Cell 31: 762–772 [DOI] [PubMed] [Google Scholar]
- Bowser R, Giambrone A, Davies P 1995. FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain. Dev Neurosci 17: 20–37 [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL 1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res 20: 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT 2003. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 81: 221–227 [DOI] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH 2005. The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620 [DOI] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ 1972. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 148: 558–567 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE 2003. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol 23: 8626–8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW 2004. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW 2007. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5: e232 doi: 10.1371/journal.pbio.0050232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: 249–268 [PMC free article] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- Di Croce L, Koop R, Venditti P, Westphal HM, Nightingale KP, Corona DF, Becker PB, Beato M 1999. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell 4: 45–54 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre E, Skildum A, Pierson-Mullany L, Lange CA 2005. Integration of progesterone receptor mediated rapid signaling and nuclear actions in breast cancer cell models: role of mitogen-activated protein kinases and cell cycle regulators. Steroids 70: 418–426 [DOI] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol 23: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937 [DOI] [PubMed] [Google Scholar]
- Hale TK, Contreras A, Morrison AJ, Herrera RE 2006. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol Cell 22: 693–699 [DOI] [PubMed] [Google Scholar]
- Hamiche A, Xiao H 2004. Methods for analysis of nucleosome sliding by Drosophila NURF. Methods Enzymol 377: 353–363 [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97: 833–842 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9: 263–267 [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21: 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Shimane M 2000. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics 63: 35–39 [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119: 803–814 [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19: 1951–1967 [DOI] [PubMed] [Google Scholar]
- Kinyamu HK, Archer TK 2007. Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol Cell Biol 27: 4891–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y 2007. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Koop R, Di Croce L, Beato M 2003. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J 22: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010a. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39: 736–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010b. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319: 819–821 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Archer TK 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J 17: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci 103: 15392–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee J, Lee SK, Lee JW 2008. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol 22: 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282: 1900–1904 [DOI] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O'Malley BW 2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol 23: 3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chu MJ, Xu J 2007. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol 27: 8073–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, et al. 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 274: 15633–15645 [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11: 1981–1985 [DOI] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, et al. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell 20: 199–211 [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Samuels HH 2005. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev 26: 583–597 [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763 [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437: 436–439 [DOI] [PubMed] [Google Scholar]
- Mu X, Springer JE, Bowser R 1997. FAC1 expression and localization in motor neurons of developing, adult, and amyotrophic lateral sclerosis spinal cord. Exp Neurol 146: 17–24 [DOI] [PubMed] [Google Scholar]
- Narayanan R, Adigun AA, Edwards DP, Weigel NL 2005. Cyclin-dependent kinase activity is required for progesterone receptor function: novel role for cyclin A/Cdk2 as a progesterone receptor coactivator. Mol Cell Biol 25: 264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER 2007. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem 282: 22278–22288 [DOI] [PubMed] [Google Scholar]
- Pierson-Mullany LK, Lange CA 2004. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol Cell Biol 24: 10542–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Bruggemeier U, Beato M 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60: 719–731 [DOI] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6: 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D 2006. EM measurements define the dimensions of the ‘30-nm’ chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci 103: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A, Sandin S, Rhodes D 2008. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci 105: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell 12: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Shang Y, Myers M, Brown M 2002. Formation of the androgen receptor transcription complex. Mol Cell 9: 601–610 [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, et al. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389: 194–198 [DOI] [PubMed] [Google Scholar]
- Strutt H, Paro R 1999. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol 119: 455–467 [DOI] [PubMed] [Google Scholar]
- Trotter KW, Archer TK 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol 24: 3347–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M, Bartsch J, Schelbert A, Hache RJ, Beato M 1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J 14: 1737–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16: 93–105 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M 2004. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell 16: 439–452 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M 2006a. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 24: 367–381 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Zaurin R, Saragueta P, Beato M 2006b. Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways. Ann NY Acad Sci 1089: 59–72 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, Vermeulen M, Mann M, Beato M 2009. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet 5: e1000567 doi: 10.1371/journal.pgen.1000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Zaurin R, Nacht AS, Font-Mateu J, Le Dily F, Beato M 2010. Nuclear factor 1 synergizes with progesterone receptor on the mouse mammary tumor virus promoter wrapped around a histone H3/H4 tetramer by facilitating access to the central hormone-responsive elements. J Biol Chem 285: 2622–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. 2007. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 9: 347–353 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodeling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543 [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25: 801–812 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Yamamoto KR 1984. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell 38: 29–38 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]