Abstract
The Arp2/3 complex nucleates and cross-links actin filaments at the leading edge of motile cells, and its activity is stimulated by C-terminal regions of WASP/Scar proteins, called VCA domains. VCA domains contain a verprolin homology sequence (V) that binds monomeric actin and central (C) and acidic sequences (A) that bind the Arp2/3 complex. Here we show that the C domain binds to monomeric actin with higher affinity (Kd = 10 μM) than to the Arp2/3 complex (Kd > 200 μM). Nuclear magnetic resonance spectroscopy reveals that actin binds to the N-terminal half of the C domain and that both the V and C domains can bind actin independently and simultaneously, indicating that they interact with different sites. Mutation of conserved hydrophobic residues in the actin-binding interface of the C domain disrupts activation of the Arp2/3 complex but does not alter affinity for the complex. By chemical cross-linking the C domain interacts with the p40 subunit of the Arp2/3 complex and, by fluorescence polarization anisotropy, the binding of actin and the Arp2/3 complex are mutually exclusive. Our results indicate that both actin and Arp2/3 binding are important for C domain function but that the C domain does not form a static bridge between the two. We propose a model for activation of the Arp2/3 complex in which the C domain first primes the complex by inducing a necessary conformational change and then initiates nucleus assembly by bringing an actin monomer into proximity of the primed complex.
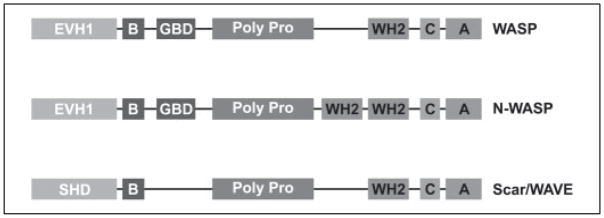
Rapid assembly of actin filaments is required for many basic cellular processes including amoeboid motility, endocytosis, and pathogen invasion (1, 2). One important mediator of actin assembly is the Arp2/3 complex, an essential, evolutionarily conserved, seven-subunit protein complex that nucleates and cross-links new actin filaments. The Arp2/3 complex binds the sides of pre-existing actin filaments and nucleates new filaments branching from the old at a characteristic angle of 70° (3, 4). This activity, called dendritic nucleation (4), generates space-filling networks of cross-linked and entangled actin filaments, the growth of which can generate force and power motility (5). In vitro, three factors cooperate to stimulate the nucleation activity of the Arp2/3 complex: a preexisting actin filament, an actin monomer, and a nucleation-promoting factor (NPF) 3 (6, 7). NPFs include members of the WASP/Scar family of proteins (1, 8). Several WASP/Scar family proteins are regulated by small GTPases and thus serve to link Arp2/3-dependent actin assembly to upstream signals. The region of WASP/Scar family proteins that binds and activates the Arp2/3 complex is called a VCA domain and is usually located at the C terminus. A canonical VCA domain is composed of three sequences motifs: one or two V (verprolin homology) sequences (also known as WASP homology 2 or WH2 domains) that bind actin monomers; a C (central or connecting) sequence that is essential for nucleation and has been shown to interact with the Arp2/3 complex; and an A (acidic) sequence that also binds to the Arp2/3 complex and contributes the majority of the binding energy (1) (Fig. 1).
FIGURE 1. Domain organization of WASP/Scar proteins.
EVH1, Ena/Vasp homology 1 domain; B, basic region; GBD, GTPase-binding domain; Poly Pro, proline-rich region; SHD, Scar homology domain; WH2, verprolin-like or WASP homology 2 domain; C, central or connecting domain; A, acidic domain.
Although the molecular determinants for activation of the Arp2/3 complex have been worked out in some detail, the mechanism of actin nucleation itself is not well understood. The slowest step in spontaneous actin assembly is the formation of a dimer. Conventional actin dimers are extremely unstable and assemble with an equilibrium constant on the order of 4 M (9). On the basis of this fact, Kelleher et al. (10) proposed that the two actin-related subunits of the Arp2/3 complex, Arp2 and Arp3, overcome the kinetic barrier to nucleation by mimicking a conventional actin dimer. In the crystal structure of the complex, Arp2 and Arp3 are in the proper orientation to form an actin-like dimer, but they are separated by a large gap (11) so that formation of a nucleation-competent dimer would require a significant conformational change (1). Goley et al. (12) used fluorescence resonance energy transfer to demonstrate that binding of a VCA domain from WASP induces a conformational change in the complex. The authors (12) also showed that point mutations in the C region that abolish nucleation-promoting activity also abolish the ability to induce the conformational change; they did not test the effect of monomeric actin but found, surprisingly, that filamentous actin had no detectable effect on the conformation of the complex. This result indicates that either (i) filamentous actin does not induce a conformational change in the complex or (ii) that the actin-induced conformational change is different from that induced by VCA and that the fluorescent probes are not positioned to detect it. Electron microscopy studies also support the idea that VCA peptides induce a significant conformational change in the Arp2/3 complex (13, 14) and, consistent with the ability to drive conformational rearrangements, the VCA peptide makes contact with multiple subunits of the Arp2/3 complex (p21, Arp3, Arp2 and p40) (15). Lastly, mathematical modeling of Arp2/3-dependent actin nucleation indicates the presence of a slow, first-order activation step following interaction with monomeric and filamentous actin and a VCA domain (15). This activation step may correspond to a slow conformational rearrangement in the complex.
Previous studies also revealed that actin monomers bound to the VCA domain play an important role in activation of the Arp2/3 complex. In an earlier study (6) we showed that the Arp2 subunit of the complex hydrolyzes ATP during or soon after filament nucleation and that the factors required for dendritic nucleation are also required for ATP hydrolysis. We demonstrated a requirement for monomeric actin by showing that a non-polymerizing, latrunculin-bound actin monomer could substitute for a polymerizable monomer. Marchand et al. (7) demonstrated the importance of monomeric actin by showing that removal of the actin-binding WH2 sequence from a WASP-family VCA domain greatly decreases the efficiency of Arp2/3 activation. These authors (7) also found evidence that the truncated CA domain retained weak monomer binding activity and thus could not conclude that the residual nucleation-promoting activity was monomer-independent.
Here we report a structural and biochemical analysis of the molecular requirements of Arp2/3 activation. By NMR spectroscopy we found that the WH2-actin complex is similar to the previously described ciboulot D1-actin complex (16). This result supports the proposal that the two domains are evolutionarily related (17). In addition, we found that the C domain also binds monomeric actin and that monomer binding plays a role in activating the Arp2/3 complex. This result, together with the previous observation that the C domain binds the Arp2/3 complex, suggests a simple model in which the C domain acts as a “docking ring” that correctly positions an actin monomer onto the actin-related subunits of the complex. To test this model we determined the affinity of the C domain for actin and the Arp2/3 complex, both alone and in combination. We found that the C domain binds much more tightly to actin than to the Arp2/3 complex and that binding of the two is mutually exclusive. In addition, chemical cross-linking revealed that the C domain does not bind Arp2 or Arp3 but contacts the p40 subunit. From these observations, we propose a multistep model for Arp2/3 activation in which the C-domain interacts sequentially with the Arp2/3 complex and the WH2-bound actin monomer. In this model the C domain first binds the Arp2/3 complex and induces a long-lived conformational change. Next, the C domain dissociates and binds an actin monomer, positioning it in the optimal orientation to interact with the “primed” complex.
MATERIALS AND METHODS
Plasmid Construction and Protein Purification
All Scar1 and N-WASP plasmids were constructed by PCR using full-length human Scar1 (gift of Laura Machesky) and rat N-WASP (gift of Wendell Lim). All clones were verified by sequencing. Proteins were expressed in BL21 (pLysS) cells as C-terminal His6, N-terminal GST fusions, or both, where indicated in the text. For uniform labeling with 15N and 2H, BL21 (pLysS) cells freshly transformed with Scar1 VC or VCA expression plasmids were grown in either Silantes [15N,2H]- or [15N]OD2 medium (Cambridge Isotope Laboratories, Cambridge, MA). For residue-specific labeling, cells were grown in M9 minimal medium containing uniformly labeled 15N amino acid (lysine, arginine, alanine, isoleucine, leucine, valine) (Cambridge Isotope Laboratories). Expression of all proteins was induced by the addition of isopropyl 1-thio-β-D-galactopy-ranoside to a final concentration of 0.5 mM. After expression for 3 h, the cells were harvested by centrifugation, resuspended, and disrupted by microfluidization, and supernatants were collected by centrifugation at 10,000 × g for 45 min. His6-tagged proteins were isolated under either native or denaturing conditions on nickel-nitrilotriacetic acid resin. GST fusions were bound to glutathione-agarose beads and cleaved off the column using either thrombin or PreScission protease. All proteins were further purified by chromatography on MonoQ resin (Amersham Biosciences) and verified for molecular weight using MALDI-TOF mass spectrometry. Tag-free peptides were obtained from Synpep Corp. (Alameda, CA) and were >95% pure. Arp2/3 and actin were purified from Acanthamoeba castellani (6, 18) and the actin was pyrene-labeled as described (19).
Fluorescent Labeling
Purified proteins were exchanged into HEK buffer (50 mM KCl, 1 mM EDTA, 10 mM HEPES, pH 7.3) using a NAP-5 column and incubated with a 4–7-fold molar excess of Alexa-488 C5 maleimide (Molecular Probes, Eugene, OR) for 1 h at room temperature and overnight at 4 °C. Samples were spun at 65,000 rpm, and the supernatant was chromatographed on a G-25 column to remove unreacted dye. MALDI-TOF was used to monitor percent labeling.
Actin Polymerization
All pyrene actin polymerization assays were performed with Acanthamoeba actin (6% pyrene labeled) in 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7.0. Ca2+-actin was converted to Mg2+-actin prior to each polymerization reaction by a 2-min incubation with 50 μM MgCl2, 200 μM EGTA. Fluorescence studies were conducted on a K2 Multifrequency Fluorometer (ISS Inc., Champagne, IL). Data were analyzed using KaleidaGraph (Synergy Software, Reading, PA).
Polarization Anisotropy
Fluorescence studies were conducted on a K2 Multifrequency Fluorometer at 25 °C. The samples contained KMEH buffer (10 mM Na-HEPES, pH 7.0, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2). For anisotropy measurements, fixed concentrations of Alexa-488-labeled Scar1 VCA, CA, or C peptides were mixed with various concentrations of either the Arp2/3 complex, latrunculin-stabilized actin, or both, and the data were fitted to a single site-binding isotherm to determine Kd. Alexa-488 was excited with polarized light at 493 nm, and the emitted light was detected at 525 nm. Affinities of unlabeled peptides for either the Arp2/3 complex or actin were determined by competition with Alexa-488-labeled wild-type Scar1 VCA, CA, or C peptides. Anisotropy data were fit to a full solution of the equilibrium competition binding equation using KaleidaGraph (7).
Cross-linking
Proteins were dialyzed into 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.5 mM dithiothreitol, 0.2 mM ATP, and 10 mM imidazole (pH 7.0) at 25 °C to facilitate cross-linking. The indicated protein concentrations were mixed with freshly prepared 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide hydrochloride (EDC) and N-hydroxysuc-cinimide (NHS) for 30 min at room temperature. Samples were methanol/chloroform-precipitated and analyzed by Western blotting with monospecific antibodies against the Arp2/3 complex subunits or by fluorescence imaging of the Alexa-488-labeled C domain.
NMR Spectroscopy
NMR experiments were carried out at 30 °C on Bruker DRX 500- or 600-MHz spectrometers equipped with a triple resonance z axis gradient CryoProbe. Unbound [15N]VCA, residue specific 15N-labeled VC, and [15N,2H]VC samples each contained 400 μM protein in 90% H2O, 10% D2O with modified G-actin buffer (buffer A: 50 mM d6-Na-HEPES (Cambridge Isotope Laboratories), pH 7.5, 100 μM CaCl2, 100 μM ATP, and 0.5 mM Tris(2-carboxyethyl)phosphine). For studies of actin-bound peptides, 5 ml of ~50 μM G-actin was exchanged into 2 mM d6-Na-HEPES, pH 7.5, 10 μM ATP by Nap-5 chromatography and immediately lyophilized. The lyophilized powder was then resus-pended in a solution containing 300 μl of 400 μM peptide in buffer A containing 600 μM latrunculin B (Calbiochem). Samples were spun at 100,000 × g for 30 min resulting in a final actin concentration of 500 μM. Assignments of residues in VC were obtained using a combination of [15N,1H]HSQC (heteronuclear single quantum coherence), [15N,1H]-TROSY-HSQC (transverse relaxation optimized spectroscopy), three-dimensional [15N,1H]NOESY-HSQC, and three-dimensional [15N,1H]TROSY-NOESY-HSQC experiments (nuclear Overhauser effect spectroscopy) (20) on [15N]-VCA, [15N,2H]VCA, residue-specific 15N-labeled VC, and [15N,2H]VC samples bound to monomeric actin. NOESY mixing times were 80 and 120 ms. Data were processed using NMRPipe (21) and analyzed using XEASY (22).
RESULTS
Secondary Structure and Interacting Residues of the VC-Actin Complex
The binding of an actin monomer to a NPF and ultimately to the Arp2/3 complex are essential steps in the activation of the Arp2/3 complex (1). To understand the role of monomeric actin in activating the complex, we sought to define the minimal actin-binding region of the human Scar1 VCA peptide. Using fluorescence polarization anisotropy, we measured the affinities of C-terminal truncation mutants of VCA for monomeric actin (Table 1). Removal of the A domain of Scar1 (VC) or the A domain together with the last five residues of the C domain (VCΔ5, residues 489–538) had no effect on actin binding. Removal of both the A and C sequences (isolated WH2 domain), however, decreased the affinity by 10-fold (3.1 μM versus 0.35 μM, respectively). Therefore, we define the optimal actin-binding region of the human Scar1 VCA domain as VCΔ5.
TABLE 1.
Affinitiies and activities for all Scar1 VCA mutations
| Construct | Kd for actina | Kd for Arp2/3a | Activityb |
|---|---|---|---|
| μM | μM | ||
| Scar VCA | 0.32 ± 0.2 | 0.88 ± 0.2 | 1.0 |
| VC | 0.40 ± 0.1 | >200 | 0.0 |
| VCΔ5 | 0.35 ± 0.1 | ND | ND |
| V | 3.10 ± 0.5 | ND | 0.0 |
| CA | 11.50 ± 2.3 | 0.92 ± 0.1 | 0.08 |
| C | 12.20 ± 2.7 | >200 | 0.0 |
| A | ND | 7.30 ± 1.2 | 0.0 |
| VCA V531A | 0.93 ± 0.1 | 1.30 ± 0.2 | 0.5 |
| VCA V531D | 1.69 ± 0.2 | 1.90 ± 0.1 | 0.1 |
| VCA L535A | 0.09 ± 0.02 | 1.10 ± 0.1 | 0.2 |
| VCA T533D | 0.34 ± 0.2 | 0.74 ± 0.2 | 1.1 |
Affinities were measured by competition with Alexa-488 Scar1 VCA or Alexa-488 Scar1 C using fluorescence polarization anisotropy. Values are listed as ±S.E. for fits to a minimum of 10 data points for each titration derived from three independent experiments. ND, not determined.
Activity was calculated by dividing the maximum number of ends generated for the specified construct by the number of ends generated by wild-type VCA. Activity was measured at maximal concentration for the VCA truncations (see Fig. 3B) and at equimolar amounts for the VCA point mutants (0.5 μM, see Fig. 5). ND, not determined.
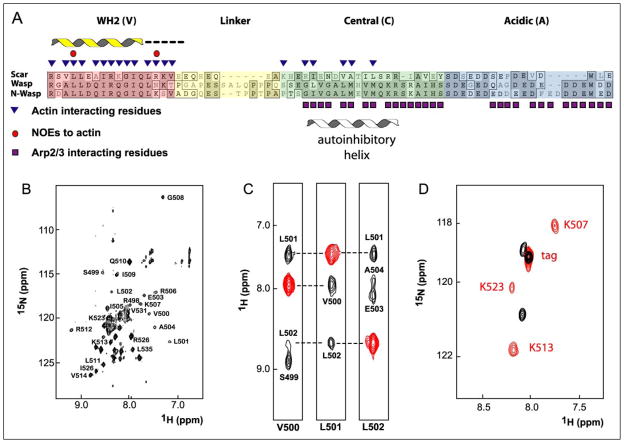
We then used NMR spectroscopy to determine the identity and secondary structure of 15N-labeled residues in Scar1 VC bound to an unlabeled actin monomer. VC was used instead of VCΔ5 because of its higher solubility. First, we used [15N,1H]TROSY-HSQC spectra to look for structural differences between free Scar1 VC and Scar1 VC bound to monomeric actin. Based on chemical shift dispersion, we found that 29 residues in Scar1 VC bind actin (Fig. 2, A and B, supplemental Fig. S1). Changes in the chemical shift of a number of residues became maximal at equimolar amounts of actin and Scar1 VC, consistent with formation of a 1:1 complex.
FIGURE 2. Residues of the VC peptide perturbed upon actin binding and secondary structure of the bound peptide.
A, alignment of human Scar1, WASP, and N-WASP VCA regions. Structural information is displayed above the alignments in B–D. Actin-binding residues were determined by line broadening and NOEs. Secondary structure was determined by the presence and strength of amide-amide NOEs. The helix designates residues with strong amide-amide NOEs, and the dashes denote weak amide-amide NOEs. Arp2/3-interacting residues were determined by line-broadening experiments (24). The autoinhibitory helix denotes the structure of the C domain bound to the GTPase-binding domain of WASP (25). B, 1H-15N TROSY-HSQC spectrum of [15N,2H]Scar1 VC (residues 489 –538) bound to Ca2+-ATP-actin with latrunculin B at 25 °C. The assignment of residues in the bound state is shown. C, selected regions of the three-dimensional [15N,1H]TROSY-NOESY-HSQC spectra of [15N, 2H]VC bound to Ca2+-ATP-actin showing intramolecular amide-amide NOEs. Cross-peaks are shown in red and intramolecular NOEs in black. Connecting residues are denoted. D, overlay of the [15N,1H]HSQC spectra of the free [15N-Lys]VC (black) and the [15N-Lys]VC-actin complex (red).
To better understand the structural changes that occur in VC upon binding actin, we collected [15N,1H]TROSY-NOESY spectra of VC bound to the 43-kDa actin monomer (Fig. 2C). Because of the high correlation time of the actin-VC complex, 100% deuteration of the VC peptide was required to reduce the relaxation time and produce NOEs with sharp line widths, limiting information available to amide-amide intra- and inter-residue amide-Hα NOEs. A combination of TROSY pulse sequences, 100% deuteration, buffer optimization (23), and use of a cryoprobe enabled us to acquire high quality NOESY spectra at protein concentrations of 150–400 μM. From the observed NOEs we determined direct connectivity of a number of residues. Consistent with previous reports (7, 24), our data indicate that the VC domain by itself is unfolded and that about 40% of the VC peptide adopts a helical conformation upon binding actin.
We assigned the NOESY spectra and determined which residues of Scar1 VC contact actin by using residue-specific labeling strategies. Seven separate isotope-labeled samples of Scar1 VC were prepared, each with a different 15N-labeled amino acid: alanine, valine, isoleucine, lysine, arginine, leucine, and glycine. The HSQC spectra of each labeled peptide alone and in complex with actin revealed that many residues undergo significant chemical shift changes and line broadening upon binding actin (Fig. 2D, supplemental Fig. S2). Such changes in the NMR spectra indicate the involvement of these amino acids in the binding interface. We correlated chemical shifts in the bound state with NOEs to partially assign the VC peptide. The entire WH2 domain (residues 498–522) was assigned using sequential NOEs and residue labeling. The presence of strong amide-amide NOEs indicates that the WH2 domain is helical. The intensity of the NOEs decreases dramatically at a position immediately C-terminal to the highly conserved Gly-512 and completely disappears at the C-terminal end of the canonical WH2 domain (Val-514), indicating that this region is either dynamic or in an ordered yet nonhelical conformation (Fig. 2A). We observed two inter-residue amide-aliphatic NOEs between actin and Scar1 VC, one at residue Leu-502 and another at Arg-518. Both residues are highly conserved and are required for actin binding. As NOEs are short-range dipole-dipole interactions limited to distances of up to 5–7 Å, our data indicate that these residues in Scar1 VC are in direct contact with the actin monomer (Fig. 2A).
Consistent with our binding data shown in Table 1, a number of residues in the C domain undergo actin-induced chemical shift changes and line broadening (Fig. 2A). In addition, spectra of the isolated C domain show chemical shift changes and decreases in peak intensity upon binding actin (supplemental Figs. S1 and S2). Unlike in the WH2 region, we detected no amide-amide NOEs in the NOESY spectra of the C domain, either alone or attached to the WH2 domain (Fig. 2A). This does not exclude the possibility that the C domain is helical upon binding to actin, as a number of mechanisms such as dynamics can significantly weaken NOE cross-peaks. In fact, our mutagenesis data (see below) as well as previous studies (24, 25) are consistent with helical structure in the C domain.
Binding and Activation Properties of VCA Truncations
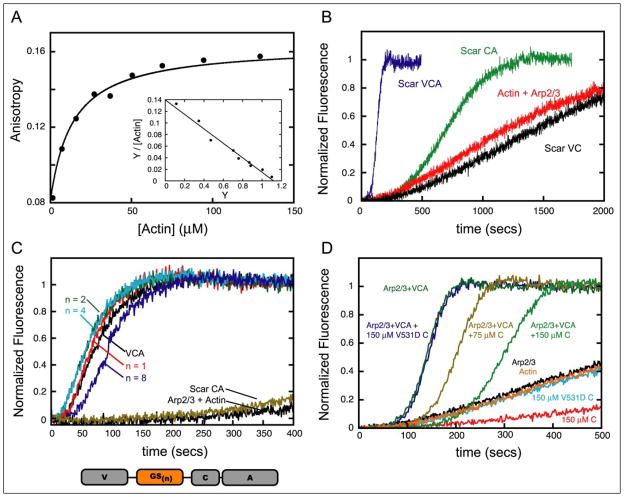
Based on our NMR data it appears that actin contacts residues within the C domain that have previously been shown to be important for activation of the Arp2/3 complex (7, 24, 26). We used polarization anisotropy to confirm that the isolated C domain binds actin directly (Fig. 3A). We first added a cysteine to the C terminus of the C domain and used it to covalently attach the fluorescent dye Alexa-488. Titration with actin caused a saturable increase in the anisotropy of the labeled C domain from 0.08 to 0.16. This increase is consistent with a single actin monomer binding to the C domain (Fig. 3A, inset). Fitting this curve to a single site hyperbola gave a Kd of 13.4 μM. A competition assay for actin between unlabeled and labeled C domain yielded a Kd of 12.2 μM for unlabeled peptide (data not shown), indicating that the dye had no significant effect on actin binding.
FIGURE 3. Binding and activation properties of VCA truncations.
A, C domain binds actin monomers. Conditions: 10 mM HEPES, pH 7.0, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2, 200 μM latrunculin B. Equilibrium binding of the C domain labeled with Alexa-488 (0.2 μM) to monomeric actin measured by fluorescence anisotropy. Curve is best fit of data to binding quadratic (39) yielding equilibrium constant of 13.4 μM (see Table 1). Scatchard plot (inset) indicates a 1:1 stoichiometry. B, time course of pyrene-actin polymerization for VCA truncations. Mg-ATP-actin monomers (4 μM 5% pyrene-labeled) and 10 nM Arp2/3 complex were polymerized in 10 mM HEPES, pH 7.0, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2 at 22 °C, either alone or with 0.5 μM Scar1 VCA (residues 489 –559), 4 μM Scar1 CA (residues 523–559), 4 μM Scar1 VC (residues 489 –543). C, activities of Gly-Ser insertions. Mg-ATP-actin monomers (2 μM 6% pyrene-labeled) and 10 nM Arp2/3 complex were polymerized either alone or with the indicated construct (0.5 μM). Scar1 CA was used at 4 μM. The number of Gly-Ser pairs is indicated by n. The placement of the Gly-Ser pairs in the VCA sequence is illustrated. D, the C domain inhibits spontaneous actin polymerization. Time course of pyrene-actin (4 μM, 5% pyrene-labeled) polymerization in the presence of 150 μM and 75 μM C and C-V531D peptides alone or with 10 nM Arp2/3 complex and 0.25 μM Scar1 VCA peptide.
Previous reports claimed that, under certain conditions, both the VC and CA domains of WASP weakly stimulate nucleation by the Arp2/3 complex (7, 27). Given the fact that both actin and Arp2/3 bind to residues in the C domain, it is possible that these interactions allow the VC and CA peptides to retain some nucleation-promoting activity. To this end, we measured the activities of multiple deletion constructs of Scar1 VCA (Table 1) using pyrene-actin assembly assays. Removal of the WH2 domain from Scar1 VCA (Scar1 CA) significantly reduced the nucleation activity but did not completely abolish it, whereas removal of the acidic domain (Scar1 VC) abolished nucleation activity entirely (Fig. 3B, Table 1). Hufner et al. (27) report that the VC domain of WASP fused to GST weakly promotes nucleation. In the present study, however, we used an untagged, monomeric version of the peptide. We believe the difference in results can be attributed to the dimerization of GST, which has been shown previously to increase artifactually the activity of VCA (28). Therefore, it appears that the CA region retains the minimal activating properties of Scar1 and that the WH2 domain serves to increase the activity of the peptide. To determine the geometric constraints on the ability of the WH2 domain to enhance nucleation by the CA region, we constructed mutant VCA domains with flexible peptide linkers of various lengths inserted between the WH2 domain and the CA region. The linkers had little effect on the kinetics of Arp2/3-mediated actin polymerization, indicating that the effect of the WH2 domain is not sensitive to its distance from or orientation to the CA sequence (Fig. 3C).
The addition of 150 μM C domain peptide (~40:1 ratio of C domain to actin) inhibits spontaneous assembly of actin filaments but does not alter the critical concentration, as judged by pyrene fluorescence (Fig. 3D, data not shown). This agrees with previous data showing that VCA and WH2 peptides both inhibit spontaneous polymerization without sequestering actin monomers (28). Interestingly, the C domain increases the lag time of VCA/Arp2/3-mediated actin polymerization and, qualitatively, appears to decrease the rate of filament generation in a dose-dependent manner. This activity of the C domain, inhibition of spontaneous polymerization, was previously unknown.
We next attempted to understand how the affinity of the V, C, and A regions for actin and Arp2/3 each contribute to nucleation-promoting activity. We measured the affinities of multiple deletion constructs of Scar1 VCA for actin and Arp2/3 using fluorescence anisotropy and competition binding assays (Table 1). Panchal et al. (24) reported that upon titration of a 15N-labeled N-WASP VCA peptide with the Arp2/3 complex, residues in both the C and A domains undergo line broadening, indicative of binding to the complex (Fig. 2A). By polarization anisotropy, we find that the C domain alone binds very weakly to the Arp2/3 complex (Kd>200 μM) (Table 1). By itself, the acidic domain (A) binds the Arp2/3 complex with micromolar affinity. Adding the C sequence (CA) increases the affinity for the complex 8-fold compared with the isolated A sequence. If the increased affinity reflects the C domain binding to the Arp2/3 complex, then the binding energy of this interaction is about 1 kcal/mol, consistent with a Kd of ~184 mM. From our anisotropy data we estimate the Kd of this interaction at ~500 μM. This discrepancy may represent the effect of tethering the two peptides together and/or the effect of a conformational change induced by one peptide on the binding of the other. In our hands, the CA region is the smallest contiguous fragment that 1) binds both actin and the Arp2/3 complex and 2) promotes nucleation activity of the complex.
Actin and Arp2/3 Bind to the C Domain in a Mutually Exclusive Manner
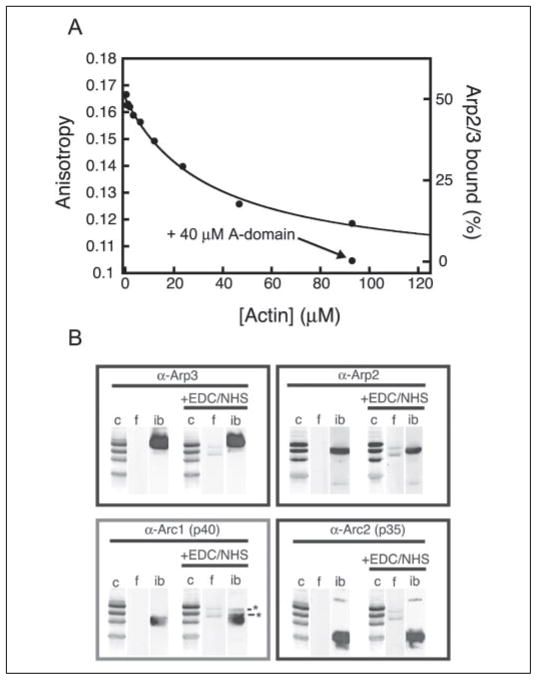
An attractive hypothesis for the role of the C domain in activating the Arp2/3 complex is that it forms a bridge between monomeric actin and the Arp2/3 complex and positions the first monomer of the daughter filament in the proper orientation to interact with the complex. To test this “molecular bridge” hypothesis, we used Alexa-488-labeled CA to monitor whether actin and Arp2/3 can bind simultaneously to the Scar1 C domain (Fig. 4A). We added 1 μM Arp2/3 complex to 20 nM fluorescently labeled CA peptide (~50% bound) and then titrated latrunculin-bound actin monomers to high concentration (100 μM). The addition of monomeric actin decreased anisotropy to a value between that of CA bound to Arp2/3 and CA bound to actin. This suggests that a significant fraction of the Arp2/3-bound CA was released (Fig. 4A). The Arp2/3 complex should still bind to the A domain, albeit with a lower affinity (Table 1), because of its interaction with the highly conserved tryptophan in the A domain. Consistent with this idea, the addition of excess unlabeled A peptide (40 μM) following titration with monomeric actin caused a further saturable decrease in anisotropy to the level of actin plus CA alone. The apparent Kd calculated from the titration with actin is ~10 μM, which agrees well with our direct binding measurements (KD = 12.2 μM) (Table 1). These data are not consistent with the idea that the C domain forms a simple molecular bridge between an actin monomer and the Arp2/3 complex.
FIGURE 4. Interaction of the C domain with Arp2/3 and actin.
A, actin competes with Arp2/3 complex for binding to the C domain. Equilibrium binding of the CA domain labeled with Alexa-488 (0.02 μM) to monomeric actin in the presence of 1 μM Arp2/3 complex measured by fluorescence anisotropy. Anisotropies of different bound states are given on the right side of the graph. At 95 μM actin, 40 μM A domain (544 –559) was added to the sample. B, EDC/NHS cross-linking of Alexa-488-labeled C domain (residues 522–543) to the Arp2/3 complex. Each reaction was immunoblotted with antibodies to the four highest molecular weight subunits of Arp2/3. Coomassie (c), fluorescence (f), and immunoblot (ib) signals for each condition are shown. The only subunit of Arp2/3 that cross-links to the C domain, p40, is shown in the gray box. Asterisks denote the full-length C peptide and its degradation product. This degradation product is missing residues 522–527 of Scar1 as measured by MALDI-TOF spectrometry. Cross-linking reaction conditions: 2 mM EDC/NHS was added to 8 μM Arp2/3 complex alone or with 25 μM Scar1 C for 30 min at room temperature.
C Domain Cross-links to p40 of Arp2/3
To better understand the interaction of the C domain with the Arp2/3 complex, we cross-linked Alexa-488-labeled C domain to the Arp2/3 complex using the zero-length cross-linker EDC in the presence of NHS. Cross-linked species were separated by SDS-PAGE and detected with a fluorescence imager (Fig. 4B). In the presence of the Arp2/3 complex, EDC/NHS produced a single, new, fluorescent, C-domain-containing band of about 45 kDa. Blotting with antibodies against Arp3, Arp2, p35, and p40 indicate that this band represents the C domain cross-linked to the p40 subunit of the complex. The addition of monomeric actin decreased cross-linking to p40, consistent with competition between actin and Arp2/3 for the C domain (supplemental Fig. S3). It is formally possible that binding to filaments or the A peptide could induce a conformational change in the Arp2/3 complex that promotes a novel interaction with the C domain. No new cross-links, however, were formed between the C domain and the Arp2/3 complex in the presence of phalloidin-stabilized actin filaments (with and without latrunculin-actin monomers) or excess A peptide (supplemental Fig. S3, data not shown), suggesting that p40 is the only site at which the C-domain interacts with the activated complex.
Perturbation of C domain Binding to Actin, but Not to Arp2/3, Correlates with Decreased Nucleation-promoting Activity
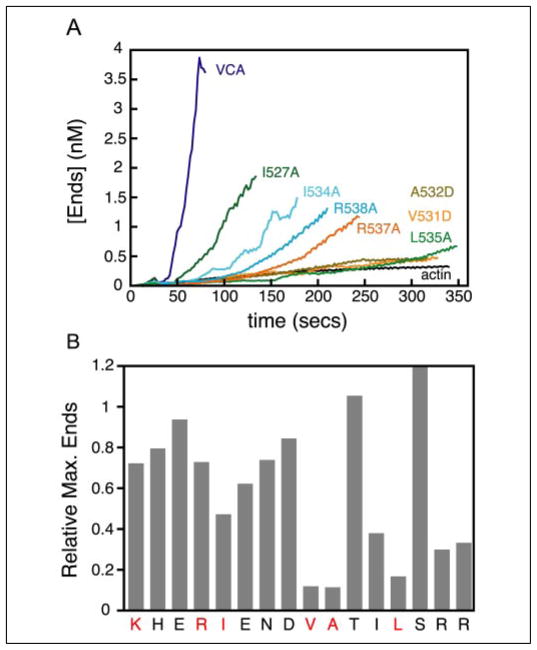
A comparison of the residue-specific data on the binding of Arp2/3, actin, and GTPase-binding domain to the VCA peptide suggests that the binding sites for all three proteins overlap in the C domain (Fig. 2A). To understand the role of each binding event, we performed alanine-scanning mutagenesis on the entire C domain and determined the ability of each mutant to activate the Arp2/3 complex (Fig. 5, Table 1). Several studies report that residues in the C domain are important for the activity of WASP/Scar family proteins (7, 24, 26). We found that every conserved hydrophobic residue in the C domain and both arginines at the C-terminal end are important for NPF activity (Figs. 2A and 5B).
FIGURE 5. Effect of C domain mutations on activation of the Arp2/3 complex.
A, plot of the concentration of ends versus time for each mutant in the C domain of Scar1 VCA-(residues 489 –559) peptides. Curves were generated from time courses of pyrene-actin polymerization for each mutant (0.5 μM) in the presence of 10 nM Arp2/3 complex, 2 μM actin in KMEH buffer at 22 °C. B, relative maximum concentration of ends for each mutant, normalized to wild-type Scar1 VCA. Residues shown to bind actin in Fig. 2A are indicated in red.
Previous studies of similar mutants in the VCA region of N-WASP found no correlation between Arp2/3 activation and affinity for the Arp2/3 complex (24). Because many of the conserved residues in the Scar1 C domain involved in Arp2/3 activation also bind directly to actin monomers, we determined the affinities of mutants for both actin and the Arp2/3 complex (Table 1, supplemental Fig. S4). Perturbing the affinity of the C domain for monomeric actin in either direction significantly decreased nucleation activity. The V531D mutation caused a 5-fold decrease in affinity for actin, whereas, surprisingly, the L535A mutation caused a 3.5-fold increase in affinity for actin. Both mutations produced a 10-fold reduction in the number of filament ends generated in Arp2/3 complex activation assays. This suggests that both increases and decreases in the affinity of the C domain for actin severely hinder nucleation activity. We observed no strong correlation between nucleation-promoting activity and affinity for the Arp2/3 complex, with neither mutation causing more than a 2-fold decrease in affinity for the Arp2/3 complex, which is consistent with previous results (24). In addition, a C domain peptide containing the V531D mutation has no effect on spontaneous polymerization or VCA/Arp2/3-mediated actin polymerization, which further illustrates that this mutation blocks the binding of actin (Fig. 3D).
DISCUSSION
Structure and Function of the WH2 Domain
The WH2 domain was originally identified as a sequence of ~25 residues at the C-terminal end of WASP family proteins that binds actin monomers with micromolar affinity. The WH2 domain binds monomers at the “barbed end” and prevents spontaneous nucleation but does not alter association with the fast growing barbed end of existing filaments. This combination of activities can be used to regulate actin assembly in a variety of ways, and WH2-related sequences are found in many proteins (>30 proteins, SMART Data base (29)) that alter cytoskeletal organization. Understanding the structure and function of this domain alone and in the context of nearby regulatory elements, therefore, will give us insight into several classes of cytoskeletal regulatory mechanism.
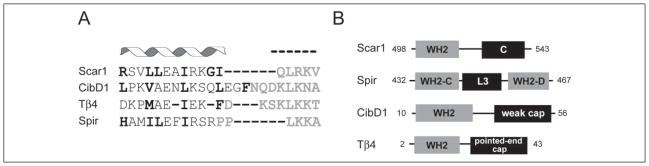
Using NMR, we have shown that the Scar1 WH2 domain by itself is unfolded and that it acquires secondary structure upon binding monomeric actin. This disorder-to-order transition is advantageous in that it provides high specificity at relatively low (micromolar) affinity. Structurally, the Scar1 WH2 domain is defined by an N-terminal helix of 10–16 residues C-terminally capped by 2–5 helix-breaking residues immediately followed by a 4-residue L++φ motif in an extended conformation (where L is a leucine, + is an arginine or lysine, and φ is any hydrophobic residue) (Fig. 2A). The helix is defined by a φφXXφ motif found in many other helical peptides that engage in protein-protein interactions, such as the nuclear hormone co-activator GRIP1 and transcriptional co-activator CBP/p300 (30). The helix-breaking residues are usually glycines (found in all WH2 domains from WASP/Scar1 proteins), prolines, and aspartate. Comparing our data with the recent x-ray crystallographic structure of the ciboulot D1-actin complex and the NMR-derived model of the thymosin β4-actin complex reveals that both domains are structurally homologous to WASP family WH2 domains and use analogous residues to interact with actin (Fig. 6A). In particular we find that Leu-501 and Arg-512 in the Scar1 WH2 domain directly contact the actin monomer (Fig. 2A). The equivalent residues in ciboulot (Val-14 and Lys-31) and thymosin (Met-6 and Lys-18) sit directly in the binding interface in their respective structural models (16, 31, 32). In addition, our NMR data are supported by recently solved atomic structures of actin-bound WH2 domains from WASP and Scar (33).
FIGURE 6. Regions C-terminal to the WH2 domain dictate function.
A, structural alignment of WH2 domains. Scar1 WH2-(498 –5140), ciboulot D1-(10 –33), thymosin β4-(2–20), and Spir C-(432– 448) are shown. Residues directly facing actin are in bold and the extended motif in gray. Actin-binding residues are predicted for Spir. B, residues C-terminal to the WH2 domain dictate the function of the protein. Domain layout of selected WH2 domain-containing proteins: C, central domain that binds to actin and the Arp2/3 complex; L3, linker 3 of Drosophila Spir that can nucleate actin filaments; weak cap, residues that weakly bind the pointed end of actin monomers; pointed-end cap, residues that strongly bind the pointed end of actin monomers.
WASP family proteins contain either one or two WH2 domains, but there is no strong correlation between nucleation-promoting activity and number of WH2 domains present. Rather, it is the sequence on the C-terminal side of the WH2 domain, the C domain, that appears to determine its role in Arp2/3 activation (Fig. 6B). As we have shown, the C domain plays a crucial role in actin nucleation.
Actin and Arp2/3 Binding by the C Domain
Three lines of evidence suggest a critical role for the C domain in the activation of the Arp2/3 complex: 1) the region is highly conserved in all WASP family proteins; 2) mutation of conserved residues decreases nucleation-promoting activity; and 3) in WASP and N-WASP, interaction of the GTPase-binding domain with the C domain strongly inhibits nucleation-promoting activity. Despite its importance, however, the role of the C domain in Arp2/3 activation is not well understood. We found that both actin monomers and the Arp2/3 complex bind to the C domain, and we propose that it may coordinate their interaction.
Previous results indicated that residues in the WASP/Scar family C domain interact directly with and promote a conformational change within the Arp2/3 complex. The crystal structure of the inactive Arp2/3 complex reveals that major conformational rearrangements must occur for Arp2 and Arp3 to form an actin-like dimer capable of nucleating a new filament (11). In lower resolution structural models based on electron microscopy, the Arp2/3 complex appears in at least three conformations: open, intermediate, and closed (13, 14). Binding of full-length WASP promoted the appearance of the closed state of the Arp2/3 complex. Moreover, a FRET (fluorescence resonance energy transfer) assay demonstrated that a wild-type CA peptide from N-WASP induces a conformational change in the Arp2/3 complex that brings the p40 and p21 subunits (and presumably the Arp2 and Arp3 subunits to which they are connected) into closer proximity (12). These authors also found that mutation of a conserved hydrophobic residue in the C domain, L470A (equivalent to V531D in Scar1 (24)), abolishes the ability of the construct to induce a conformational change. This result is consistent with a report that residues in the C domain directly interact with the Arp2/3 complex (24). This C domain-dependent conformational change may correspond to the rate-limiting “activation” step in Arp2/3-dependent nucleation (15).
There is at present no high-resolution structural model for the interaction between VCA domains and the Arp2/3 complex. We showed previously that WASP family CA domains cross-link to multiple subunits of the Arp2/3 complex: p40, Arp2, Arp3, and p21 (15). Here, we found that zero-length chemical cross-linkers cross-link the C domain to the p40 subunit. The p40 subunit contains multiple WD40-like repeats. These repeats are found in many signaling proteins and mediate numerous types of protein-protein interactions, particularly with highly conserved disordered regions. Previously it was shown that within the yeast Arp2/3 complex, the p40 subunit provides most of the binding energy for the CA peptide, and isolated p40 binds VCA with similar affinity to the intact complex (34). These results are not entirely consistent with previous cross-linking studies nor with our data showing that the C domain provides little binding energy for the Arp2/3 complex. Electron microscopic studies of a WASP family protein bound to the Arp2/3 complex suggest that the NPFs contact the junction between p40, Arp2, and Arp3 (13, 14, 35). In addition to these results, Weaver et al. (36) provide evidence that the highly conserved tryptophan found in the C-terminal portion of the A domain of all Arp2/3 activators interacts with the Arp3 subunit. From these results, we propose an orientation for the CA peptide on the Arp2/3 complex (Fig. 7). The N-terminal region interacts specifically with the p40 subunit, and the A domain stretches from this site on p40 to Arp3, where the conserved tryptophan in the acidic domain might fit into a hydrophobic pocket (37). Based on the structure of the actin-bound WH2 domain and weak sequence homology between the WH2 and C domains, Chereau et al. (33) proposed that the C domain interacts with the Arp2 subunit of the complex. Our chemical cross-linking data and evidence cited above, that the C-domain interacts primarily with the p40 subunit of the complex, do not support this model.
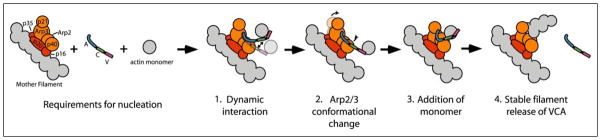
FIGURE 7. Role of the C domain-actin interaction in Arp2/3 activation.
A model of the steps leading to activation of the Arp2/3 is shown. Actin is depicted in gray, WH2 in red, C in green, the A domain in blue, and the Arp2/3 complex in orange and brown. We propose the following steps in the activation of the Arp2/3 complex. 1, the C domain of the VCA peptide binds to the Arp2/3 complex and an actin monomer in a mutually exclusive manner and exists in a dynamic state. Binding of the C domain to the Arp2/3 complex occurs through the p40 subunit, and the A domain is modeled to bind to Arp2 and Arp3 based on previous results (15, 36, 37). The VC region binds to the barbed end of an actin monomer. 2, the CA region induces a conformational change of the Arp2/3 complex bringing p21, Arp3, Arp2, and p40 in closer proximity to each other (12–14). 3, the C domain then binds the actin monomer and, with the WH2 domain, positions it on the Arp2/3 complex. The monomer binds the actin-like Arp2 and Arp3 subunits, forming a nucleus for polymerization and formation of a daughter filament. The addition of the monomer triggers ATP hydrolysis on Arp2. 4, the WH2-actin contact is lost because of filament formation and Arp2 ATP hydrolysis lowers the affinity for the CA region such that VCA is released (6).
Our NMR studies identify residues in the N-terminal half of the C domain that directly contact actin (Figs. 2A, supplemental Fig. S1, Table 1) and demonstrate that the C and WH2 domains bind simultaneously to the same monomer. Furthermore, we find that the C domain inhibits spontaneous nucleation of actin monomers, an activity identical to that of the VCA region and the isolated WH2 (V) domain (28). The WH2 domain sterically blocks the formation of a nucleus by binding to a cleft between subdomains 1 and 3 of actin but allows for barbed-end elongation from preformed filaments. At present, we do not understand why the isolated C domain inhibits spontaneous nucleation. Possibilities include: 1) that it binds to a site on the barbed end that is distinct from that of the WH2 domain; or 2) that it binds a site that interferes with lateral, cross-strand interactions in the filament. We also find that the isolated C domain inhibits VCA/Arp2/3-mediated actin polymerization in a dose-dependent manner. It does not appear that this is caused by directly perturbing the VCA/Arp2/3 interaction, as our results indicate that the C domain interacts very weakly with the Arp2/3 complex (Kd>200 μM) and, at the maximal concentration tested (150 μM), could not act as a competitive inhibitor. The C domain interacts more strongly (>20-fold) with monomeric actin (Kd~10 μM), however, and inhibition may be a consequence of both preventing the interaction of a monomer with the C domain of VCA and the inhibition of spontaneous nucleation. In addition, mutations V531D and L535A, which severely impair the activity of the Scar1 VCA domain, have no effect on affinity for the Arp2/3 complex. Both mutations, however, significantly perturbed actin binding either by strengthening (L535A) or weakening (V531D) it. Furthermore, a C domain peptide containing the V531D mutation inhibited neither spontaneous actin polymerization nor VCA/Arp2/3-mediated actin polymerization. We propose, therefore, that actin binding to the C domain plays a significant role in the first steps of activating the Arp2/3 complex.
Models for Arp2/3 Activation
A realistic model for activation of the Arp2/3 complex must take into account four observations. 1) Binding of the CA region to the Arp2/3 complex induces a conformational change that is probably required for nucleation (12–14). 2) In the presence of the VCA domain, monomeric actin induces a further conformational change that results in hydrolysis of ATP on the Arp2 subunit (6). 3) The C domain binds even more tightly to monomeric actin than to the Arp2/3 complex, and mutations that perturb affinity for actin also perturb Arp2/3 activation. 4) Binding of actin and the Arp2/3 complex to the C domain is mutually exclusive.
The simplest model for Arp2/3 activation is that the C domain acts as a molecular bridge, directly contacting both actin and the Arp2/3 complex and acting as a template for their interaction. This interaction could cause further conformational changes in the complex resulting in ATP hydrolysis by the Arp2 subunit and formation of a stable nucleus. This model is consistent with all available data except that the observation that binding of actin and the Arp2/3 complex to the C domain is mutually exclusive. To account for this fact, we propose that the C domain exists in dynamic equilibrium between binding actin and the Arp2/3 complex (Fig. 7). When bound to the Arp2/3 complex the C domain induces a relatively stable conformational change that brings Arp2 and Arp3 into proximity. The C domain then dissociates, binds to the actin monomer tethered to the WH2 domain, and orients the monomer for proper interaction with Arp2. If the monomer makes productive contact before the complex returns to its inactive state, it triggers creation of a stable heterotrimeric nucleus composed of Arp2, Arp3, and actin and seeds formation of a daughter filament. Following this event, Arp2 ATP hydrolysis releases the CA region from the complex (6). This “dynamic interaction and conformational memory” model would account for the coupling of actin binding and filament nucleating activities as well as the mutually exclusive interaction. The relatively low affinity of the C domain for both actin and the Arp2/3 complex and the fact that strengthening or weakening its interaction with actin inhibits activity suggest that the rate constants that define the C domain interaction with actin and the Arp2/3 complex are tuned to optimal values. The mechanism we put forth here is similar to kinetic proofreading mechanisms that temporally separate the binding step from the activation step using an irreversible reaction such as the hydrolysis of ATP (38). Future experiments detailing the kinetics of the C domain/actin and C domain/Arp2/3 interactions as well as high-resolution structural information of the Arp2/3 complex bound to VCA, F-actin, and monomeric actin are needed to fully resolve the mechanisms of activation of the Arp2/3 complex.
Supplementary Material
Acknowledgments
We are extremely grateful to members of the Mullins, Dötsch, and Vale laboratories for providing helpful discussions, valuable reagents, and technical expertise. We thank Orkun Akin for assistance with MATLAB, Kevin Slep for advice, Brian Kelch for initial experiments, and Brian Volkman for use of NMR facilities.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The abbreviations used are: NPF, nucleation-promoting factor; WASP, Wiskott-Aldrich syndrome protein; Scar, suppressor of cyclic AMP receptor; VCA domain, verprolin homology sequence, central or connecting sequence, and acidic sequence; WH2, WASP homology 2; EDC, 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride; NHS, N-hydroxysulfosuccinimide; HSQC, heteronuclear single quantum coherence; TROSY, transverse relaxation optimized spectroscopy; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; GST, glutathione S-transferase.
Supplemental Material can be found at: http://www.jbc.org/cgi/content/full/M507470200/DC1
This work was supported by National Institutes of Health Grant R01 GM61010.
References
- 1.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Engqvist-Goldstein AE, Drubin DG. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 3.Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 4.Mullins RD, Heuser JA, Pollard TD. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 6.Dayel MJ, Mullins RD. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 8.Gouin E, Welch MD, Cossart P. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Sept D, McCammon JA. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelleher JF, Atkinson SJ, Pollard TD. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 12.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 14.Martin AC, XPX, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch MD, Drubin DG. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Curr Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 16.Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Dide-lot G, Preat T, Knossow M, Guittet E, Carlier MF. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 17.Boquet I, Boujemaa R, Carlier MF, Preat T. Cell. 2000;102:797–808. doi: 10.1016/s0092-8674(00)00068-4. [DOI] [PubMed] [Google Scholar]
- 18.MacLean-Fletcher S, Pollard TD. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 19.Mullins RD. Curr Opin Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- 20.Riek R, Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. J Am Chem Soc. 2002;124:12144–12153. doi: 10.1021/ja026763z. [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 23.Kelly AE, Ou HD, Withers R, Dotsch V. J Am Chem Soc. 2002;124:12013–12019. doi: 10.1021/ja026121b. [DOI] [PubMed] [Google Scholar]
- 24.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 25.Eadie JS, Kim SW, Allen PG, Hutchinson LM, Kantor JD, Zetter BR. J Cell Biochem. 2000;77:277–287. doi: 10.1002/(sici)1097-4644(20000501)77:2<277::aid-jcb10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 26.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 27.Hufner K, Higgs HN, Pollard TD, Jacobi C, Aepfelbacher M, Linder S. J Biol Chem. 2001;276:35761–35767. doi: 10.1074/jbc.M106520200. [DOI] [PubMed] [Google Scholar]
- 28.Higgs HN, Blanchoin L, Pollard TD. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 29.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. Nucleic Acids Res. 2004;32:D142–144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 31.Domanski M, Hertzog M, Coutant J, Gutsche-Perelroizen I, Bontems F, Carlier MF, Guittet E, van Heijenoort C. J Biol Chem. 2004;279:23637–23645. doi: 10.1074/jbc.M311413200. [DOI] [PubMed] [Google Scholar]
- 32.Irobi E, Aguda AH, Larsson M, Guerin C, Yin HL, Burtnick LD, Blanchoin L, Robinson RC. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan F, Egile C, Lipkin T, Li R. J Biol Chem. 2004;279:54629–54636. doi: 10.1074/jbc.M402357200. [DOI] [PubMed] [Google Scholar]
- 35.Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D. PLoS Biol. 2005;3:E383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 37.Beltzner CC, Pollard TD. J Mol Biol. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Hopfield JJ. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.