Abstract
Proteins that are synthesized on cytoplasmic ribosomes but function within plastids must be imported and then targeted to one of six plastid locations. Although multiple systems that target proteins to the thylakoid membranes or thylakoid lumen have been identified, a system that can direct the integration of inner envelope membrane proteins from the stroma has not been previously described. Genetics and localization studies were used to show that plastids contain two different Sec systems with distinct functions. Loss-of-function mutations in components of the previously described thylakoid-localized Sec system, designated as SCY1 (At2g18710), SECA1 (At4g01800), and SECE1 (At4g14870) in Arabidopsis (Arabidopsis thaliana), result in albino seedlings and sucrose-dependent heterotrophic growth. Loss-of-function mutations in components of the second Sec system, designated as SCY2 (At2g31530) and SECA2 (At1g21650) in Arabidopsis, result in arrest at the globular stage and embryo lethality. Promoter-swap experiments provided evidence that SCY1 and SCY2 are functionally nonredundant and perform different roles in the cell. Finally, chloroplast import and fractionation assays and immunogold localization of SCY2-green fluorescent protein fusion proteins in root tissues indicated that SCY2 is part of an envelope-localized Sec system. Our data suggest that SCY2 and SECA2 function in Sec-mediated integration and translocation processes at the inner envelope membrane.
Plastids arose from a primary endosymbiotic event involving a photosynthetic cyanobacterial progenitor and a nonphotosynthetic eukaryotic host (for review, see McFadden, 2001). Over time, many genes were eliminated from the plastid genome and other genes moved from the organelle genome to the nuclear genome (for review, see Bock and Timmis, 2008; Kleine et al., 2009). The corresponding gene products are now synthesized on cytoplasmic ribosomes and are targeted to the plastid by posttranslational mechanisms that involve an N-terminal transit peptide. Although several different import pathways exist, the majority of these proteins are imported into the plastids by the combined action of the TOC complex in the outer plastid envelope and the TIC complex in the inner envelope membrane (for review, see Inaba and Schnell, 2008). Some of the imported proteins are delivered to the inner envelope membrane via a stop-transfer mechanism, which involves lateral diffusion in the plane of the membrane from the TIC complex (Tripp et al., 2007, and refs. therein). Others are delivered to the stroma and, after removal of the transit peptide, many are secondarily targeted to the thylakoid membranes, thylakoid lumen, or the inner envelope membrane (Cline and Dabney-Smith, 2008). The signals and systems involved in targeting to the thylakoid membranes and lumen are relatively well studied and show clear homologies with bacterial transport systems (for review, see Schünemann, 2007; Cline and Dabney-Smith, 2008). Although it has been clearly established that certain inner membrane proteins, most notably TIC21, TIC40, and TIC110, also have soluble stromal intermediates (Li and Schnell, 2006; Tripp et al., 2007; Vojta et al., 2007; Chiu and Li, 2008) and therefore require a postimport pathway for integration, a translocase that mediates insertion into the inner membrane or translocation to the intramembrane space has not been identified (Tripp et al., 2007).
According to the conservative sorting hypothesis (Hartl et al., 1986), proteins that are destined for the inner envelope membrane, which corresponds to the plasma membrane of the original bacterial endosymbiont, should use systems and mechanisms related to those involved in secretion and membrane protein integration in bacteria (for recent review, see Natale et al., 2008; Mandon et al., 2009). In bacteria, most of the exported proteins are translocated by components of the Sec or Tat pathway. The core of the Sec translocon is formed by three gene products, SecY, SecE, and SecG, while SecA, a peripheral protein and ATPase, provides the driving force for translocation. The SecYEG complex is also required for integration of many integral inner membrane proteins. In this case, as the protein is in transit through the SecYEG complex, lateral gates open to allow outward diffusion of the transmembrane helices in the plane of the bilayer. An additional protein called YidC can facilitate this process by interacting with the transmembrane helices. YidC can also act in a Sec-independent fashion to insert a limited number of proteins. The Sec pathway translocates and integrates proteins in an unfolded conformation, by virtue of their interaction with molecular chaperones or because they are cotranslationally translocated. Fully folded proteins that bear a twin Arg in their signal peptides use the Tat (twin Arg) pathway instead (Berks et al., 2003). A complex of TatA, TatB, and TatC forms the translocase for the Tat pathway.
In thylakoids, four different pathways have been described for integration of membrane proteins or translocation to the thylakoid lumen. The Sec pathway, SRP pathway, Tat pathway, and spontaneous pathway each handles a different subset of thylakoid proteins (for recent review, see Cline and Theg, 2007; Schünemann, 2007; Cline and Dabney-Smith, 2008). Biochemical and genetic studies have allowed investigators to identify the components and energy requirements of these systems. The SecYEG translocon is reduced to a complex of SecY and SecE homologs in chloroplasts, and there are two YidC homologs, Alb3 and Alb4. Disruption of the SecY gene in maize (Zea mays) results in pale seedlings and an arrest of seedling growth (Roy and Barkan, 1998). Disruption of the maize SecE gene produces a less severe, pale-green seedling phenotype (Williams-Carrier et al., 2010). The effect of disrupting Alb3 in Arabidopsis (Arabidopsis thaliana) is severe: the plants are albino and show arrested chloroplast development (Sundberg et al., 1997). Disruption of the Alb4 gene has a mild phenotype that appears restricted to nonphotosynthetic proteins (Benz et al., 2009). Disruption of the TatC gene similarly results in albino seedlings and seedling arrest (Motohashi et al., 2001). The characterized chloroplast SRP pathway operates differently from the bacterial and endoplasmic reticulum SRP pathways (Henry et al., 2007). Chloroplast SRP (cpSRP) lacks an RNA component and contains a novel cpSRP43 protein that allows it to function posttranslationally. In the posttranslational mode, the cpSRP pathway employs the Alb3 integrase, is Sec independent, and functions to integrate a single family of membrane proteins, the light-harvesting chlorophyll a/b proteins. Indirect evidence suggests that cpSRP may also function in conjunction with SecY to integrate plastid-encoded thylakoid proteins (Cline and Theg, 2007). Components of the Tat pathway, which was first discovered through work in chloroplasts, include Tha4, Hcf106, and chloroplast TatC (cpTatC; Cline and Theg, 2007). The spontaneous pathway is not thought to involve proteinaceous components in the membrane.
Although much work has been done on these pathways, one fundamental question that has not received much attention relates to the biogenesis of the translocase components. Thylakoids are not permanent features of plastids, and these membranes and associated translocons must be assembled as the thylakoids develop. Each translocon includes one or more subunits that have multiple transmembrane regions; therefore, their assembly depends on a capacity for membrane protein integration in proplastids. Relatively few studies have addressed this question. In bacteria, the homologous subunits are cotranslationally integrated by the Sec machinery (for discussion, see Martin et al., 2009). A recent study of the biogenesis of cpTatC ruled out involvement of the thylakoid Tat, Sec, and SRP systems as well as the YidC homolog Alb3 (Martin et al., 2009). Because cpTatC is not integrated into isolated thylakoids, the spontaneous pathway is also not involved. The authors hypothesized that cpTatC integration, which is Sec dependent in bacteria, may depend on a membrane protein integrase system based in the inner envelope.
We have identified a candidate for this system: an evolutionarily conserved translocase that is associated with the inner envelope. Through investigation of an embryo-lethal mutation in Arabidopsis, we learned that, in addition to the previously described thylakoid-localized SecY (Laidler et al., 1995), a second SecY homolog is encoded in plant genomes. We show that this protein is targeted to plastids, that its function is distinct from that of the previously described SecY homolog, and that it localizes preferentially to the plastid envelope. We have designated this protein as SCY2 (At2g31530) and the previously characterized thylakoid-localized protein as SCY1. A second gene encoding a homolog of SecA, the ATPase driving translocation and integration events, was also identified. We show that this protein (SECA2) is also targeted to plastids and that mutants have the same loss-of-function phenotype as scy2 mutants. Because this translocation/integration system is associated with the envelope, we propose that it may be particularly important for postimport targeting of inner membrane proteins and/or early stages of thylakoid biogenesis.
RESULTS
Disruption of SCY2 Leads to Embryo Lethality
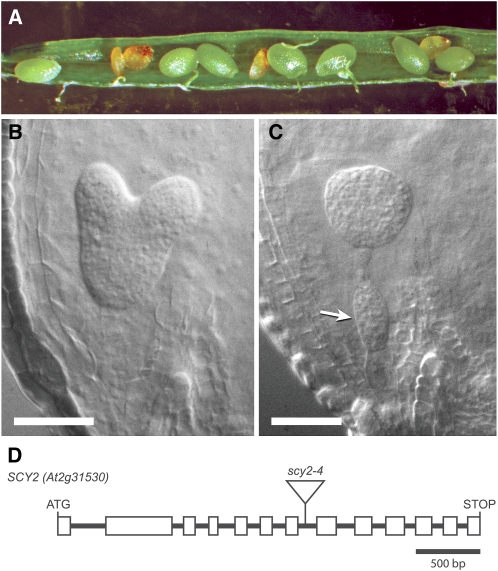
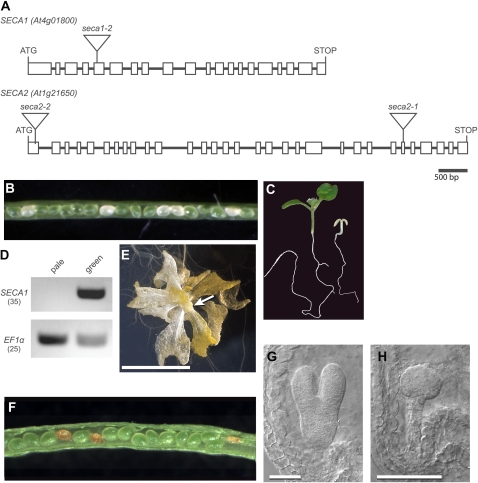
A line showing recessive embryo lethality was identified within a collection of approximately 80 lines generated following Agrobacterium tumefaciens-mediated transformation of wild-type plants (Wassilewskija [Ws] ecotype). Shriveled seed was observed in the seed stock, and kanamycin resistance, associated with the inserted T-DNA, segregated at a ratio of approximately 2:1. When late stage siliques from heterozygous parents were opened, small brown seeds that appeared to be empty, as shown in Figure 1A, were observed at a frequency of 24.3% (n = 596). Earlier stage siliques were opened and cleared, and the embryos inside the seeds were examined. In siliques that contained mostly late heart stage embryos (Fig. 1B), approximately 25% of the seeds contained embryos arrested at the globular stage of embryogenesis, with extra cell divisions apparent in the suspensor (Fig. 1C). Based on the frequency, embryo lethality appeared to be recessive rather than dominant and likely due to disruption of an essential gene. The line was backcrossed to the wild type two times, and cosegregation analysis of a population of over 100 individuals indicated that the embryo-lethal trait was tightly linked to the T-DNA insertion and kanamycin resistance. Sequencing of the flanking DNA revealed that the T-DNA had inserted into the seventh intron of At2g31530, as shown in Figure 1D. A deletion of 21 bp occurred at the insertion site (Supplemental Fig. S1), but the gene was otherwise intact. A clone of At2g31530 genomic sequence (see Fig. 4A, construct 2 below) was introduced by transformation. The genomic clone complemented the mutant phenotype, as evidenced by the identification of individuals that were homozygous for the original T-DNA insertion in At2g31530 in the T2 generation (described below). Three additional alleles in the Columbia background were reported in a large-scale project to identify essential genes in Arabidopsis (www.SeedGenes.org). The At2g31530 locus corresponds to a gene previously identified as emb2289.
Figure 1.
scy2 mutant allele and mutant phenotypes. A, Silique from a SCY2/scy2-4 heterozygous parent showing aborted seeds. B and C, Seeds in younger, developing siliques contain either wild-type-appearing embryos (B) or embryos arrested at the globular stage (C). Extra cell divisions in the suspensor are denoted by the arrow. D, Gene diagram showing introns and exons (boxes) in the open reading frame of SCY2 (At2g31530) and the location of the T-DNA insertion in scy2-4.
Figure 4.
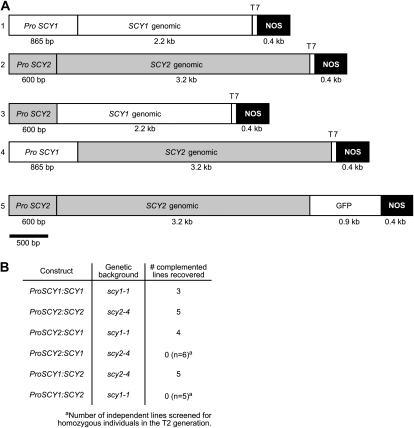
Complementation of scy mutants. A, Constructs used for SCY mutant complementation, promoter-swap experiments, and localization experiments. B, Table indicating the genetic backgrounds and number of independent lines where complementation was confirmed for each construct. For backgrounds where complementation was not observed, the number of lines that were screened is shown.
A cDNA sequence corresponding to the At2g31530 transcript was amplified from light-grown shoots, and the predicted amino acid sequence was used to perform BLAST searches. The databases contain many related sequences from other plants and algae as well as cyanobacteria and other bacteria. The most similar is a predicted protein from Vitis vinifera, which is 75% identical to the predicted mature protein. A SecY sequence from Chlamydomonas reinhardtii is 34% identical, and a Synechococcus SecY sequence is 31% identical. Protein hydrophobicity plots (Supplemental Fig. S2) indicated that the Arabidopsis protein is likely to have 10 transmembrane domains and a topology that is similar to that of SecY from Escherichia coli. The most closely related Arabidopsis protein, which is only 29% identical but has a similar predicted protein topology, is cpSecY (At2g18710), a subunit of a plastid-localized preprotein translocase. Based on the sequence relationships, we renamed the EMB2289 locus (At2g31530) as SCY2 and designated the Ws allele as scy2-4. At2g18710 was designated as the SCY1 locus. In accordance with nomenclature guidelines for Arabidopsis genes, we will refer to the proteins encoded by these loci as SCY2 and SCY1.
Arabidopsis SCY2 Encodes a Membrane-Integrated Plastid Protein
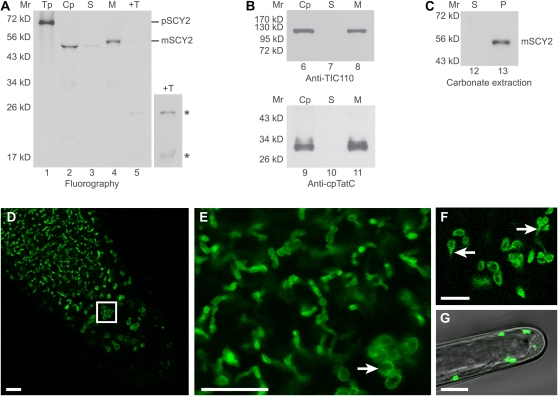
An analysis of the predicted amino acid sequence using TargetP (Emanuelsson et al., 2007) indicated that the N terminus of SCY2 is likely to encode a plastid transit peptide. In order to determine whether SCY2 is capable of localizing to plastids, in vitro translated radiolabeled precursor to Arabidopsis SCY2 (pSCY2) was incubated with isolated pea (Pisum sativum) chloroplasts in an in vitro import assay. pSCY2 was imported into the chloroplasts, processed to a mature 48.3 kD size, and localized to membranes (Fig. 2A, lanes 2 and 4). Immunoblots with antibodies to the inner membrane protein TIC110 and the thylakoid protein cpTatC indicated that the membrane fraction contained both the envelope and the thylakoid membranes (Fig. 2B). Carbonate-extracted membranes retained SCY2, indicating that SCY2 is integrated into the membrane (Fig. 2C). The mature SCY2 protein in whole chloroplasts appears to migrate as a smaller protein than that associated with the membranes (Fig. 2A, compare lanes 2 and 4). However, this is likely to be the result of the highly abundant Rubisco large subunit present in chloroplasts that distorts the migration of other proteins in this region of the gel, resulting in a concave moon band shape. Protease treatment of membranes produced two degradation products that measured 24.1 and 18.2 kD in size (Fig. 2A, +T). Degradation products are expected from protease treatment of membrane-integrated proteins that possess both exposed and membrane-protected regions (Mori et al., 2001). Together, these results suggest that SCY2 is a membrane-integrated plastid protein.
Figure 2.
Arabidopsis SCY2 is a membrane-integrated plastid protein. A to C, In vitro import assays. A, Radiolabeled precursor to SCY2 (pSCY2) was incubated with isolated pea chloroplasts in an in vitro import reaction for 30 min. Chloroplasts were treated with 100 μg mL−1 thermolysin, repurified, washed, lysed, and fractionated into stroma and membranes. Translation product (Tp) equivalent to 5% of the assay, chloroplasts (Cp), stroma (S), membranes (M), and thermolysin-treated membranes (+T) were analyzed using SDS-PAGE and fluorography. A longer exposure (4×) of thermolysin-treated membranes is shown at right to visualize mature SCY2 degradation products, indicated by asterisks. B, Chloroplasts, stroma, and membranes were also analyzed by SDS-PAGE and immunodetection with antibodies to TIC110 (applied at 1:5,000) or cpTatC (applied at 1:20,000). C, Membranes from the SCY2 import reaction were subjected to alkaline carbonate extraction. The supernatant (S) and membrane pellet (P) were analyzed using SDS-PAGE and fluorography. The positions of molecular mass markers (Mr) are indicated at the left of each gel or blot image. D to G, Localization of the SCY2-GFP fusion protein in root tips (D–F) and root hairs (G) by confocal laser scanning microscopy. The boxed area in D indicates the amyloplasts in the columella of the root tip. E and F are higher magnification views showing plastids of different morphology. Amyloplasts in the columella cells are indicated by the white arrow in E, and stromules are indicated by the white arrows in F. Bars = 10 μm.
To confirm that SCY2 is plastid localized, a construct encoding a SCY2-GFP translational fusion was generated (see Fig. 4A, construct 5 below) and introduced into Arabidopsis plants. The construct included 600 bp of sequence upstream of the ATG and the complete SCY2 coding sequence minus the last codon and stop codon. This was introduced into SCY2/scy2-4 plants, and the T2 generation was screened for plants that were homozygous for scy2-4. Several individuals were identified, indicating that SCY2-GFP complements the scy2-4 mutation. Based on this, we suggest that the distribution of the GFP signal is likely to represent the distribution of functional SCY2. GFP signal, although rather weak, could be readily detected in tissues with nongreen plastids, such as the root tip (Fig. 2, D–F), root hairs (Fig. 2G), and dark-grown hypocotyls (data not shown). In the root tips, GFP signal was associated with organelles of diverse size and shape (Fig. 2, D–F). These correspond to the different types of plastids that have been previously described in roots, including statoliths in the columella cells of root cap (Fig. 2D, boxed area) and other types of amyloplasts. In the amyloplasts, where the center is occupied by one or more large starch grains, the GFP signal appeared as a ring or halo, indicating that the signal was confined to the periphery of these organelles (Fig. 2, E and F). This was less obvious in plastids in regions more distal to the tip, which tended to have elongated profiles and appeared to be interconnected, forming a network throughout the cells (Fig. 2E). Stromules, which are thin envelope-bound extensions of stroma (Köhler and Hanson, 2000), were also visible (Fig. 2F), suggesting that the GFP signal was associated with the envelope membranes. Unfortunately, we could not extend the in vivo observations to tissues with green plastids, because the intense autofluorescence of chlorophyll masked the weaker signal from the GFP fusion protein. Microarray analyses indicate that SCY2 is constitutively expressed at a low level in most Arabidopsis tissues (www.genevestigator.com, Arabidopsis eFP Browser [Winter et al., 2007]). This, combined with the in vitro localization results with chloroplasts, suggests that SCY2 localizes to a variety of plastid types, including chloroplasts.
Disruption of SCY1 Leads to Seedling Lethality
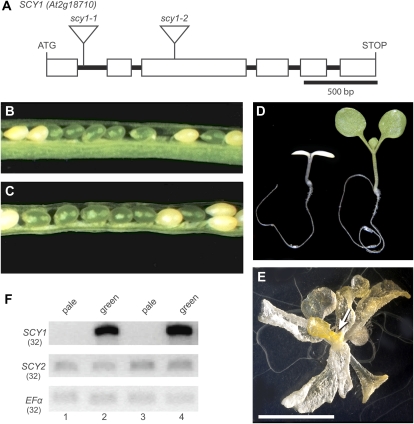
The embryo-lethal phenotype associated with scy2 mutations indicated that SCY2 is an essential protein, yet the Arabidopsis genome contains another locus, SCY1 (At2g18710), that encodes a plastid-localized SecY homolog. To investigate the relationship between the functions of the gene products encoded by SCY1 and SCY2, scy1 mutants were isolated and characterized. Two potential loss-of-function T-DNA alleles were identified in the Ws background. As shown in Figure 3A, scy1-1 has a T-DNA insert in the first intron and scy1-2 has a T-DNA insert in the third exon.
Figure 3.
scy1 mutant alleles and mutant phenotypes. A, Gene diagram showing introns and exons (boxes) in the open reading frame of SCY1 (At2g18710) and the location of T-DNA insertions in scy1-1 and scy1-2. B and C, Opened siliques showing white seeds in scy1-1 (B) and scy1-2 (C). D, Pale homozygous scy1-1 mutant seedling and green sibling. E, scy1-1 plant grown for 5 weeks on 1% Suc. The arrow indicates floral buds. Bar = 1 cm. F, RT-PCR analyses. cDNA prepared from RNA isolated from either pale seedlings or green siblings was amplified with primers flanking the T-DNA insertion sites. Lanes 1 and 2, scy1-1; lanes 3 and 4, scy1-2. The number of PCR cycles is indicated by parentheses. Full-length SCY1 transcripts do not accumulate in the pale seedlings.
Analysis of phenotypic changes in the scy1 mutants indicated that SCY1 is also an essential gene. When seeds from a heterozygous parent were sown on Suc-containing medium, small pale seedlings appeared at a frequency of 20.6% (n = 1,672) for the scy1-1 line and 25.8% (n = 1,540) for the scy1-2 line. The pale seedlings were identified as homozygous scy-1 and scy1-2 plants through PCR-based genotyping (data not shown). In addition, when maturing siliques were opened, 25.52% (n = 239) and 22.49% (n = 209) of the seeds were pale for the scy1-1 line (Fig. 3B) and the scy1-2 line (Fig. 3C), respectively. Because Arabidopsis seeds reflect the color of the embryos inside them, this suggests that SCY1 is required for normal greening during embryogenesis. Assembly of chlorophyll-protein complexes does not appear to be essential for embryo viability, because most of the pale embryos completed germination and survived to the seedling stage. They remained approximately the same size as their green siblings until 4 d after germination. By 5 d after germination, their smaller size became apparent. Wild-type seedlings produced true leaves by day 8 after germination, while the pale seedlings initially showed no signs of apical growth (Fig. 3D). However, if the pale seedlings were cultured for extended periods in the presence of 1% Suc, they grew slowly and produced lateral organs in a spiral phyllotaxy. Unlike normal leaves, these were translucent and yellow in color (Fig. 3E). The plants eventually formed pale floral buds but were infertile, at least when grown on plates. No full-length SCY1 transcripts could be detected in the pale seedlings by reverse transcription (RT)-PCR (Fig. 3F), indicating that the phenotypic changes in scy1-1 and scy1-2 reflect complete loss of function of SCY1.
SCY1 and SCY2 Have Distinct Activities
Disruptions in SCY1 and SCY2 genes exhibit mutant phenotypes at different developmental stages. To examine whether differences in expression, rather than differences in function, are the basis of genetic nonredundancy, we performed a promoter-swap experiment. Primers used in these experiments for genotyping scy1 and scy2 alleles in the presence of various transgenes are listed in Supplemental Table S1, and a summary of the results is presented in Figure 4B. First, we determined that the SCY1 genomic sequence that included 800 bp of SCY1 upstream sequence (Fig. 4A, construct 1, ProSCY1:SCY1) was sufficient to rescue scy1-1 mutations. Three lines showing full complementation of the seedling-arrest mutant phenotype were recovered. For each line, plants that carried the ProSCY1:SCY1 transgene and were homozygous for scy1-1 were identified in the T2 generation and their genotype was confirmed in the T3 generation. Although a few pale plants were observed in the population, most of the plants were indistinguishable from the wild type, as shown in Supplemental Figure S3. Second, we determined that the SCY2 genomic sequence that included 600 bp of SCY2 upstream sequence (Fig. 4A, construct 2, ProSCY2:SCY2) was sufficient to rescue scy2-4 mutations. Five lines showing full complementation of the embryo-lethal trait were recovered. Plants that carry the ProSCY2:SCY2 transgene and are homozygous for scy2-4 were indistinguishable from the wild type, as shown in Supplemental Figure S3. The siliques of individual plants showed either no embryo lethality or up to 25% embryo lethality, which likely reflects the dosage and segregation of the transgene. Surviving plants in the following generation were all homozygous for scy2-4. Third, we generated two promoter-swap constructs, ProSCY2:SCY1 (Fig. 4A, construct 3) and ProSCY1:SCY2 (Fig. 4A, construct 4). These were introduced into genetic backgrounds that contained either scy1-1 or scy2-4 alleles. Four lines of scy1-1 homozygous plants carrying the ProSCY2:SCY1 construct were recovered. Some of the plants were indistinguishable from the wild type (Supplemental Fig. S3), but most were lighter green, possibly because the SCY2 promoter is not as strong as the SCY1 promoter in shoot tissues. Five lines of scy2-4 homozygous plants carrying the ProSCY1:SCY2 construct were recovered. These plants were visually indistinguishable from the wild type, as shown in Supplemental Figure S3. In contrast, when the ProSCY2:SCY1 construct was introduced into backgrounds with scy2-4 mutations, no homozygous individuals were identified in the T2 generation of six independent lines. Similarly, when the ProSCY1:SCY2 construct was introduced into backgrounds with scy1-1 mutations, only heterozygous individuals were viable in the T2 generation of five independent lines. Based on these results, we conclude that the coding sequence, rather than the upstream regulatory regions, is the primary determinant of the ability to complement. This indicates that SCY1 and SCY2 proteins cannot substitute for each other but, rather, play different essential roles in plant cells.
SCY2 Is Localized in Envelope Membranes
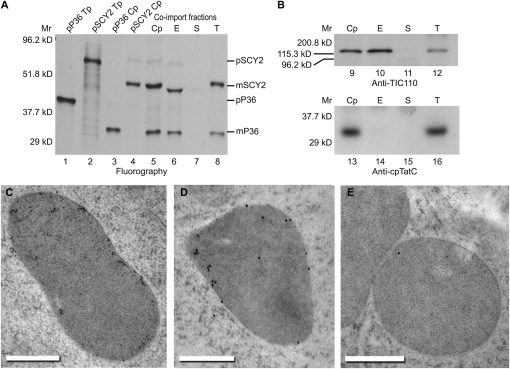
SCY1 has previously been localized to the thylakoid membranes (Schuenemann et al., 1999). Given that SCY2 is required prior to thylakoid development in embryos and accumulates in nongreen plastids after germination, we considered the possibility that SCY2 localizes to a different membrane, possibly the plastid envelope. In vitro import assays were performed with pea chloroplasts, followed by membrane fractionation (Fig. 5, A and B). SCY2 distribution was compared with that of a radiolabeled inner envelope phosphate translocator (P36), the precursor of which was coimported with pSCY2. Processed forms of both proteins can be detected in both the envelope and thylakoid fractions. Fractionation quality was judged by the relative amounts of envelope and thylakoid marker proteins (TIC110 and cpTatC) that could be found in each respective fraction by immunoblotting (Fig. 5B). The envelope fraction is free of thylakoid contamination, because cpTatC was only found in the thylakoid fraction. However, fractionation was not complete, because a small amount of TIC110 was found in the thylakoid fraction (Fig. 5B). Imported radiolabeled P36 was also found in the thylakoid fraction, further indicating that the thylakoid fraction was partly contaminated with envelope membranes (Fig. 5A). Envelope membrane contamination of thylakoids is common, especially when fractionation is conducted on chloroplasts recovered from protein import assays. Therefore, in order to eliminate envelope vesicles from the thylakoid fraction, thylakoid membranes recovered from a SCY2 import assay were sequentially washed by pelleting through a 10% Percoll cushion as described by Rawyler et al. (1992). Even after all detectable TIC110 was washed from the thylakoid fraction, a small but significant amount of SCY2 remained (Supplemental Fig. S4). These experiments suggest that imported SCY2 localizes predominantly, although perhaps not exclusively, to envelope membranes.
Figure 5.
SCY2 localizes to the envelope membranes. A, Import assays followed by membrane fractionation. Radiolabeled precursors to P36 (pP36) and SCY2 (pSCY2) were each incubated with isolated pea chloroplasts in in vitro import reactions for 15 min. Both precursors were also coimported into isolated chloroplasts for 15 min. Chloroplasts from all three import reactions were repurified and washed. Chloroplasts that underwent coimport were lysed and fractionated (see “Materials and Methods”). Translation products (Tp) equivalent to 5% of each assay, chloroplasts (Cp), envelope membranes (E), stroma (S), and thylakoid membranes (T) were analyzed by SDS-PAGE and fluorography. B, Immunoblots of chloroplasts and fractions from pSCY2/pP36 coimport probed with antibodies to TIC110 (applied at 1:5,000) or cpTatC (applied at 1:20,000). C to E, Immunogold localization of GFP. Thin sections of plastids in root cells expressing SCY2-GFP (C and D) or wild-type root cells (E) were incubated with GFP antibodies, applied at 1:10 (C) or 1:30 (D and E), and goat anti-rabbit secondary antibodies conjugated to 10-nm gold (C) or 15-nm gold (D and E). The gold label is found preferentially at the periphery of the plastids. Bars = 500 nm.
To examine SCY2 distribution in vivo, root tips from one line of seedlings used for GFP imaging were subjected to high-pressure freezing, freeze substitution, and immunogold labeling with antibodies to GFP. Gold particles were preferentially associated with the plastids in the root cells of plants expressing the GFP fusion protein (Fig. 5, C and D). Few gold particles were observed in association with other components of the cytoplasm or with the plastids of wild-type plants (Fig. 5E). Labeling of plastids was also not seen in controls where the secondary antibody was used alone (data not shown). This indicates that the labeling is specific and confirms our identification of the GFP-labeled organelles viewed by confocal microscopy as plastids. As Figure 5, C and D, show, the gold particles are found predominantly at the periphery of the plastids. Given the dimensions of antibody molecules, any gold particles within 35 nm of the periphery can reflect envelope localization (Rohde et al., 1991). Some of the gold particles were more distantly and centrally located, but these are less frequently observed. We conclude that in the plastids of roots, in cells where SCY2 is normally expressed, SCY2 is associated primarily with the envelope membranes.
Arabidopsis Has Two SecA Proteins with Distinct Functions
If SCY2 functions as the translocon for a canonical Sec system, it should work in concert with a SecA protein, the ATPase that threads the polypeptide through the SecY channel. The Arabidopsis genome contains two loci that encode SecA proteins, At4g01800 and At1g21650. We initiated a study of the functions of these proteins by identifying T-DNA mutant alleles in the SALK collection and performing a phenotypic screen. We identified one allele of At4g01800 (SALK_063371) that is a candidate loss-of-function allele. The T-DNA inserted in the fifth exon in this line (Fig. 6A). An analysis of this allele and the phenotype of albino or glassy yellow1 (agy1) mutant plants carrying a second allele was recently published (Liu et al., 2010). The essential parts of our analysis, which confirm their observations, are presented here. When siliques developing on plants heterozygous for the SALK allele were opened, pale seeds (Fig. 6B) were observed at a frequency of 26.0% (n = 642). When seeds from these parents were sown on plates containing Suc, 24.7% of the seedlings (n = 473) were also pale (Fig. 6C). PCR genotyping showed that these are homozygous for the mutation, and they also fail to produce full-length transcripts, as shown by RT-PCR (Fig. 6D). Like scy1 mutants, when grown for extended periods on 1% Suc, the pale seedlings produce yellow and translucent (glassy) lateral organs (Fig. 6E), as reported previously (Liu et al., 2010). SecA proteins usually partition between soluble and peripheral membrane phases, with membrane association due to specific interaction with SecY (for recent review, see Natale et al., 2008). The similarity in the mutant phenotypes is consistent with the hypothesis that this protein functions with SCY1. Therefore, we refer to the product of the At4g01800 (AGY1) locus by the registered gene product symbol SECA1 and designate the SALK T-DNA allele as seca1-2.
Figure 6.
seca mutant alleles and mutant phenotypes. A, Gene diagrams of SECA1 (At4g01800) and SECA2 (At1g21650) showing introns and exons (boxes) in the open reading frames and the location of T-DNA insertions in the mutants. B, Opened silique of a SECA1/seca1-2 heterozygous plant showing white seeds. C, Pale seca1-2 homozygous mutant seedling and green sibling. D, RT-PCR analysis. cDNA prepared from RNA isolated from either pale seedlings or green siblings was amplified with primers flanking the seca1-2 T-DNA insertion site or primers specific for EF1α (loading control). The number of PCR cycles is indicated by parentheses. Full-length SECA1 transcripts do not accumulate in the pale seedlings. E, A seca1-2 plant grown for 5 weeks on 1% Suc. The arrow indicates floral buds. Bar = 1 cm. F, Opened silique of a SECA2-1/seca2-1 heterozygous plant showing aborted seeds. G and H, Seeds in younger, developing siliques contain either wild-type-appearing embryos (G) or embryos arrested at the globular stage (H). Bars = 50 μm.
We identified one allele of At1g21650 (SALK_014008) as a potential loss-of-function allele. The T-DNA inserted into the 26th exon in this line (Fig. 6A). A total of 24.5% of the seeds (n = 931) in the siliques of heterozygous parents abort (Fig. 6F), and analysis of cleared siliques indicated that the embryos arrest at the globular stage of development (Fig. 6, G and H). A cosegregation analysis (n = 33) performed after five backcrosses to Columbia wild type showed that the embryo-lethal trait is closely linked to the insertion. Eighteen plants in the population were heterozygous for the mutant allele, and only these plants produced siliques with aborted embryos. We subsequently identified a second potential loss-of-function allele with a T-DNA insertion in the first exon (Fig. 6A) in the Versailles collection (FLAG_315D07; Ws ecotype). Heterozygous plants carrying this T-DNA insertion also produced seeds with embryos arrested at the globular stage (data not shown), and the embryo-lethal trait also cosegregated with the T-DNA (n = 22). Based on the similarity to the mutant phenotype of SCY2, we have designated the At1g21650 locus as SECA2 and the T-DNA alleles as seca2-1 (SALK_014008) and seca2-2 (FLAG 315D07). Although many genes, when mutated, produce embryo-lethal phenotypes, the fact that both scy2 and seca2 mutations produce arrested globular stage embryos is consistent with the hypothesis that SECA2 functions with SCY2.
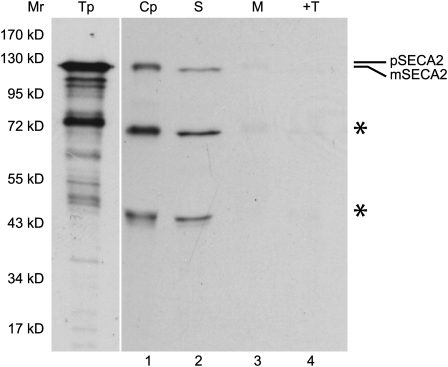
SECA2 has not been characterized previously. We confirmed that it is likely to be directed to plastids through an import assay. A full-length cDNA clone was isolated and sequenced (GenBank accession no. GU289737) and subsequently used for in vitro transcription and translation. The sequence of this clone included 21 extra nucleotides in exon 28 relative to accession NM_102014, suggesting that alternative splicing of the transcript may occur. The predicted protein is only 46% identical to SECA1 at the amino acid level but contains all of the signature domains for SecA proteins. In vitro coupled transcription and translation produced the predicted precursor to Arabidopsis SECA2 (pSECA2) at 119.7 kD. When incubated with isolated pea chloroplasts, the precursor was processed to a mature size of 116 kD and localized to the stroma (Fig. 7). In vitro translation also produced several smaller translation products. In vitro translation of very large polypeptides frequently yields carboxyl-truncated proteins due to premature termination. This appears to be the case for SECA2, as two smaller processed proteins were also recovered with chloroplasts and localized to the stroma (Fig. 7).
Figure 7.
Arabidopsis SECA2 is a plastid-localized protein. Radiolabeled precursor to SECA2 (pSECA2), obtained by a coupled transcription and translation reaction, was incubated with isolated chloroplasts in an in vitro import reaction for 45 min. Chloroplasts were treated with 100 μg mL−1 thermolysin, repurified, washed, lysed, and fractionated into stroma and membranes. Translation product (Tp) equivalent to 0.8% of the assay, chloroplasts (Cp), stroma (S), membranes (M), and thermolysin-treated membranes (+T) were analyzed using SDS-PAGE and fluorography. Smaller translation products, likely to be carboxyl-truncated pSECA2 resulting from premature termination, also appeared to be imported and processed, as indicated by asterisks. Translation products that were exposed for one-fourth the length of time of chloroplasts and chloroplast fractions are shown. The positions of molecular mass markers (Mr) are shown at left.
SecE Is Required for Greening of Seedlings
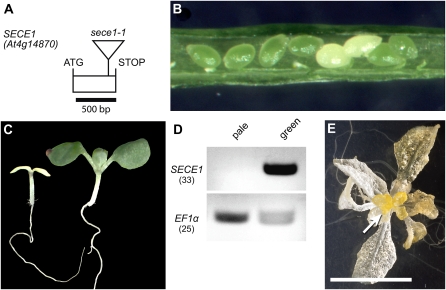
SecY proteins have been shown to physically interact with SecE proteins to form the membrane-associated portion of the translocon. Only one plastid-localized SecE homolog, encoded by At4g14870, has been reported thus far in Arabidopsis (Schuenemann et al., 1999). We identified one potential loss-of-function allele in the Versailles collection of T-DNA mutants. In the FLAG 583E05 line, the T-DNA inserted into the single exon that constitutes the coding region, disrupting the SecE domain (Fig. 8A). When siliques of plants heterozygous for this mutation were opened, they were found to contain pale seeds (Fig. 8B) at a frequency of 24.7% (n = 482). When seeds from these parents were sown, 21.6% (n = 707) of the seedlings were pale, as shown in Figure 8C, and these were homozygous for the mutation, as confirmed by PCR genotyping. RT-PCR analysis indicated that SecE transcripts fail to accumulate in the pale seedlings (Fig. 8D). As with the scy1 and seca1 mutants, the pale seedlings produce translucent and yellow leaf-like lateral organs and floral buds with extended culture in the presence of 1% Suc (Fig. 8E). A cosegregation analysis (n = 124) performed after one backcross to the Ws wild type showed that the production of white seeds is closely linked to the insertion. Fifty-seven plants in the population were heterozygous for the mutant allele, and only these plants produced siliques that contained approximately 25% white seeds. Genomic sequence corresponding to the At4g14870 locus was introduced into heterozygous plants by transformation. Individuals that contained the introduced sequence and were homozygous for the original T-DNA insertion in At4g14870 were identified by PCR genotyping in the T1 generation. These individuals were viable and green; therefore, we conclude that the disruption of At4g14870 was responsible for the albino phenotype. The similarity of the mutant phenotype to the phenotype of scy1 and seca1 mutants is consistent with the hypothesis that the gene product functions in complexes with SCY1 and SECA1. Consequently, we have designated the At4g14870 locus as SECE1 and the T-DNA allele as sece1-1.
Figure 8.
sece1 mutant allele and mutant phenotypes. A, Gene diagram of SECE1 showing the single exon in the open reading frame and the location of the T-DNA insertion in the sece1-1 mutant. B, Opened silique of a SECE1/sece1-1 heterozygous plant showing white seeds. C, Pale sece1-1 homozygous mutant seedling and green sibling. D, RT-PCR analysis. cDNA prepared from RNA isolated from either pale seedlings or green siblings was amplified with primers flanking the sece1-1 T-DNA insertion site or primers specific for EF1α. The number of PCR cycles is indicated by parentheses. Full-length SECE1 transcripts do not accumulate in the pale seedlings. E, sece1-1 plant grown for 5 weeks on 1% Suc. The arrow indicates floral buds. Bar = 1 cm.
DISCUSSION
Dual Sec Systems in Plastids Reflect an Ancient Divergence
The genes that encode plastid-localized SecY, SecA, and SecE that function in protein translocation events in the thylakoid membranes have been described previously (Voelker et al., 1997; Roy and Barkan, 1998), and the functions of the proteins have been biochemically characterized (Nakai et al., 1994; Yuan et al., 1994; Schuenemann et al., 1999). We have identified and characterized two additional loci in the Arabidopsis genome that encode plastid-localized homologs of SecY and SecA, designated as SCY2 and SECA2. Other plants and green algae such as Chlamydomonas also contain two SCY genes and two SECA genes. In each organism, one of the SCY sequences is more closely related to SCY1 and the other to SCY2, while one of the SECA sequences is more closely related to SECA1 and the other to SECA2. Therefore, there are two distinct lineages in each family. These may reflect an ancient divergence that predated the emergence of the plant kingdom, because each protein is more similar to bacterial forms of the protein than they are to the other SCY or SECA protein. Subfunctionalization presumably led to the establishment of two essential Sec systems in plastids. Other organisms that separately evolved dual Sec systems and contain two SecY/SecA pairs include certain gram-positive bacteria. These bacteria contain one essential Sec system and an accessory Sec system specific for a small subset of exported proteins (Rigel and Braunstein, 2008).
SCY1 and SCY2 Perform Different Functions
SECA1 and SCY1 have been shown to function in thylakoid transport of lumenal and some membrane proteins through genetic (Voelker et al., 1997; Roy and Barkan, 1998) and biochemical (Nakai et al., 1994; Yuan et al., 1994; Mori et al., 1999; Schuenemann et al., 1999) investigations. Based on their characteristics in in vitro transport assays, they appear to operate very similarly to the E. coli Sec translocase. In mature chloroplasts, SCY1 is localized strictly to the thylakoids and cannot be detected in the envelope membranes even with extended exposure of the immunoblots (Fincher et al., 2003). The phenotype of mutations in SCY1, SECA1, and SECE1 in maize (Voelker et al., 1997; Roy and Barkan, 1998; Williams-Carrier et al., 2010) is impairment in thylakoid biogenesis and, for SCY1, subsequent seedling lethality. Similarly, we show here that mutations in Arabidopsis SCY1, SECA1, and SECE1 yield chlorotic phenotypes. The pale-seedling phenotype exhibited by these components is similar to that resulting from the disruption of genes for other thylakoid translocases such as Alb3 and TatC. These considerations suggest a SECA1/SCY1 translocation function restricted to thylakoid biogenesis.
At present, experimental evidence that SCY2 and SECA2 function in protein translocation is lacking. Nevertheless, sequence similarity to well-characterized Sec proteins and conservation of signature sequence elements strongly suggest that they do function in protein transport. For example, Arg-357, Pro-358, Gly-359, and Thr-362 in loop 8 of E. coli SecY have been identified as functionally important for translocation (Mori and Ito, 2001). All are completely conserved in SCY1, and all except Arg-357, which has a conservative change to a Lys residue, are conserved in SCY2. Residues that give rise to suppressor mutations in transmembrane 7 of E. coli SecY are also largely conserved in SCY1 and SCY2 (Osborne and Silhavy, 1993). In addition, a critical Tyr residue and L-X-X-Y motif are found in the C termini of both SCY1 and SCY2. These residues contact SecA during the process of translocation in bacteria (Mori and Ito, 2006). The SecA2 protein contains the highly conserved motor and preprotein-binding domains that are signatures of the SecA family of proteins. These considerations suggest that both SCY2 and SECA2 are functional for protein transport.
The difference between the loss-of-function phenotypes establishes that SCY1 and SCY2, as well as SECA1 and SECA2, are genetically nonredundant. Whereas mutations in SCY1 and SECA1 give rise to chlorotic seedlings, mutations in either SCY2 or SECA2 arrest embryo development at the globular stage. The possibility existed that the different phenotypes resulted from different expression patterns. However, the promoter-swap experiments reported here demonstrated that the differing phenotypes were not due to different expression patterns but rather were specifically linked to the coding regions of the two genes. An analogous situation exists for the Toc159 family of proteins that serve as preprotein receptors for the plastid import apparatus. TOC159 is most abundant in green tissues, while other members of the same family, TOC120 and TOC132, are constitutively expressed at low levels (Kubis et al., 2004). However genetic and biochemical studies have shown that the TOC159 coding sequence specifies recognition of chloroplast proteins, whereas the TOC132/120 coding sequences specify recognition of “housekeeping” proteins that accumulate in most plastid types (Ivanova et al., 2004).
The correspondence between the mutant phenotypes of SCY1 and SECA1 and SCY2 and SECA2 suggests that each SECA protein works preferentially or exclusively with one SCY protein (i.e. they form a coupled system). If this were not the case and either SECA could work with either SCY, we would have expected to see genetic redundancy, and a double mutant would be needed in order to see defects. Instead, the single mutants have strong defective phenotypes, and they are both essential genes. Because many proteins that will be integrated or translocated through a Sec system interact with the SECA component first, this suggests that specific targeting to the respective SCY is effected by differential binding to either SECA1 or SECA2. A similar situation is found for the accessory Sec system in Streptococcus gordonii, which is responsible for exporting GspB, a cell surface protein that promotes adherence to platelets (for review, see Rigel and Braunstein, 2008). Individual disruption of either SecA2 or SecY2 eliminates the export of GspB.
Role of SECE
SecE is an essential component of the Sec systems in bacteria and in thylakoids. In E. coli, SecE has three transmembrane domains, but only one of these is essential for function (Schatz et al., 1991). In E. coli, SecE has been shown to interact with the hinge region of SecY and is thought to act as a “molecular clamp,” contributing to the stability of the translocation complex (for review, see Natale et al., 2008). In the thylakoid membranes of plastids, SecE includes a single transmembrane domain and has been shown to interact with SCY1 (Schuenemann et al., 1999). A single SecE homolog has been identified to date in plants (Schuenemann et al., 1999; Fröderberg et al., 2001; Williams-Carrier et al., 2010). If this protein were essential for both SCY1- and SCY2-containing complexes, the expected mutant phenotype would be embryo lethality. Instead, plants that are homozygous for sece1-1 mutations are albino seedlings with Suc-dependent development. The similarity in mutant phenotypes suggests that SECE1 plays an essential role only in SCY1-containing complexes, suggesting that another protein serves the SecE function for SCY2. The accessory Sec system of S. gordonii is encoded in an operon that contains SecA2, SecY2, and several accessory factors that are essential for export. One of these factors is distantly related to SecE, with a similarity of only 52% to the B. subtilis protein (Rigel and Braunstein, 2008). A BLAST search with default parameters failed to identify any SecE proteins in the database, suggesting that a second SecE in Arabidopsis might not be recognized as such. The large collections of Arabidopsis embryo-lethal mutants might be a good starting point for a search for a second plastid-localized protein with SecE activity.
What Does the SCY2 System Do?
Until bona fide SECA2/SCY2 substrates are identified, the actual function of SCY2 in plastid biogenesis will be unclear. However, several characteristics of SCY2 are relevant for hypotheses regarding substrates. The first is our observation that GFP-tagged SCY2 is associated with the envelope in root plastids and SCY2 localizes preferentially to the envelope membranes in an in vitro import assay. The in vitro assay also suggested that some SCY2 localized to thylakoids. Localization of the endogenous protein will be essential to determine if SCY2 is actually present in thylakoid membranes. The chloroplast import assay correctly localizes envelope, thylakoid, and stromal proteins of chloroplasts. However, it may not report correctly for proteins that function early during plastid development or that redistribute during development, such as the plastid signal peptidase Plsp1, which appears to change localization from envelope to thylakoids during development (Inoue et al., 2005; Shipman and Inoue, 2009). Unfortunately, the immunocytochemistry study could not be extended to other tissues, because the GFP antibodies label green plastids in wild-type samples nonspecifically (i.e. when GFP is absent). Regardless of whether SCY2 redistributes to thylakoid membranes in chloroplasts, the scy2-4 embryos arrest at the globular stage, before the proliferation of the thylakoid membranes (Mansfield and Briarty, 1991), which starts at the heart stage. By this reasoning, the most likely site of SCY2 function at this critical stage would be the envelope. The fact that SecY is found in both the cell and thylakoid membranes of cyanobacteria, and in the envelope and thylakoid membranes of cyanelles (Nakai et al., 1993; Yusa et al., 2008), implies a need for an envelope-based Sec system.
Most proteins of the thylakoid membrane are localized by a process called conservative sorting, wherein the Toc/Tic import system delivers the proteins to the stromal compartment, which is the ancestral endosymbiont cytoplasm. From there, they follow ancestral targeting and translocation pathways conserved from the endosymbionts. The efficiency of conservative sorting from an evolutionary standpoint is that the intraplastid trafficking machinery for conserved proteins was already in place and coevolved with its substrate proteins, eliminating the need for new translocation mechanisms. By this reasoning, SCY2 is likely to be situated in the inner envelope membrane facing the stroma and to transport proteins from the stroma into the interenvelope space or insert them into the inner envelope membrane. Several nucleus-encoded envelope proteins are candidate SCY2 substrates. TIC40 and TIC110 are transiently present in the stroma as soluble intermediates and integrated into the inner envelope membrane, possibly by a conservative sorting pathway of unknown identity (Li and Schnell, 2006; Tripp et al., 2007). Although soluble intermediates of TIC21 and TIC110 accumulate in null Tic40 mutants, TIC40 is thought to play an accessory, rather than a central, role, because translocation is slowed but not blocked (Chiu and Li, 2008). Null Tic40 homozygotes are viable and have chlorotic phenotypes (Chou et al., 2003; Kovacheva et al., 2005). On the other hand, Tic110 homozygotes arrest at the globular stage (Inaba et al., 2005; Kovacheva et al., 2005), just as scy2 and seca2 mutants do. Several other inner envelope proteins have embryo-defective mutant phenotypes (Kobayashi et al., 2007), although the route they take to the envelope is not known. One other intriguing possibility is that SCY2 may function in the integration of multispanning thylakoid translocase subunits. Thylakoids are thought to initiate through the budding or invagination of the inner envelope membrane (for discussion, see von Wettstein, 2001; Vothknecht and Westhoff, 2001), and translocases that mediate the expansion and population of proteins of the system may themselves need to be initially inserted in the envelope (for discussion, see Cline, 2003). This has been proposed for the cpTatC subunit based on experimental observations that are inconsistent with integration by any of the known thylakoid translocase systems (Martin et al., 2009). The cpTat complex assembles in scy1 and alb3 mutants, and biochemical inhibitors of cpTat and SRP were without effect on cpTatC localization. In E. coli, TatC insertion is Sec dependent and likely occurs cotranslationally (Yi et al., 2003). If the mechanism and machinery are conserved in plastids, it is possible that SCY2 acting at the envelope could mediate a pseudo-cotranslational integration of cpTatC, wherein import across Toc and Tic and integration via SCY2 would be coupled. Pseudo-cotranslational integration appears to occur for TIC40 in vitro, where the TIC40 N terminus inserted and was processed while the C terminus was still in the import channel (Li and Schnell, 2006). If cpTatC is initially inserted in the inner envelope membrane, this would necessitate envelope-to-thylakoid relocation by a membrane-flow transport system. Evidence that a system of this kind exists in chloroplasts has been presented by other investigators (Hoober et al., 1991; Morré et al., 1991; Westphal et al., 2001).
Sodium azide, which has been shown to inhibit SecA function, was previously used to identify substrates of the SECA1/SCY1 system in pea chloroplasts (Yuan et al., 1994) and in principle could be used to verify usage of the SECA2/SCY2 system. However, sodium azide is an unreliable reporter of Sec usage for two reasons. First, not all SecA proteins are sensitive to azide (e.g. the SECA1 of spinach [Spinacia oleracea] chloroplasts; Berghöfer et al., 1995). Second, SECA2 may not be required for some of the proposed membrane protein substrates of SCY2 (i.e. cpTatC, TIC40, and TIC110) because of the short translocated loops of these proteins. In E. coli, integration of membrane proteins with transported loops of 30 residues or less does not require SecA (Andersson and von Heijne, 1993; Deitermann et al., 2005). Thus, more sophisticated in vivo analyses (Voelker and Barkan, 1995) or reconstituted integration assays, like the system described by Li and Schnell (2006), will be required for the identification of SCY2 substrates.
In conclusion, we suggest that SECA2/SCY2 must be considered top candidates for a role as mediators of the postimport or conservative sorting pathway for inner membrane proteins in plastids. The duplication and subfunctionalization of Sec systems in plastids or the bacterial ancestors of plastids appears to have been an ancient event and may have been associated with the differentiation of two membrane systems, the envelope and the thylakoids.
MATERIALS AND METHODS
Identification of T-DNA Insertional Alleles
The scy2-4 allele was isolated following the transformation of Arabidopsis (Arabidopsis thaliana Ws ecotype) wild-type plants with a construct consisting of the cauliflower mosaic virus 35S promoter fused to AGL15 antisense sequence. DNA flanking the insertion was amplified from kanamycin-resistant plants through tail PCR (Liu and Whittier, 1995). Plants carrying scy1 mutant alleles were identified by screening the T-DNA populations (Ws ecotype) at the University of Wisconsin-Madison Arabidopsis knockout facility (Sussman et al., 2000). The scy1-1 and scy1-2 alleles were identified in the α (kanamycin-resistant) population using oligonucleotides annealing to the T-DNA left border (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) and regions upstream (scy1-2, 5′-TCTCCTCCTTCTTTGGCATGATTATATTT-3′) or downstream (scy1-1, 5′-CCCCATGTGAGATGCTTATACATAGAAAA-3′) of SCY1.
Plants carrying the seca1-2 (SALK 063371) and seca2-1 (SALK 014008) mutant alleles were isolated from the SALK insertional mutant population (Alonso et al., 2003) by screening seed stocks obtained from the Arabidopsis Biological Resource Center at Ohio State University. The seed stocks from which the seca2-2 and sece1-1 mutants were isolated were part of the INRA-Versailles collection (Ws-4 ecotype) and correspond to FLAG_315D07 and FLAG_583E05, respectively. Each mutant was backcrossed to the wild type at least three times and maintained as a heterozygous line. Plants were grown at 22°C with 16-h days and 8-h nights.
Plant Genotyping
Plants carrying mutant alleles were identified through PCR-based genotyping. DNA was isolated from seedlings or leaf tissue as described previously (Adamczyk et al., 2007). PCR amplification was performed with ExTaq polymerase (TaKaRa Bio) and gene-specific and/or T-DNA-specific primers (Supplemental Table S1). For plants carrying transgenes, primers were used that could differentiate between the introduced sequences and the endogenous loci.
RNA Isolation and RT-PCR Analyses
RNA was extracted from seedlings using the phenol extraction procedure described previously (Lehti-Shiu et al., 2005). For RT-PCR analyses, cDNA was synthesized from 1 μg of RNA using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. To amplify EF1α transcripts, oligonucleotide 279 (5′-GTTTCACATCAACATTGTGGTCATTGG-3′) and oligonucleotide 280 (5′-GAGTACTTGGGGGTAGTGGCATCC-3′) and either 18 or 25 cycles (as indicated) were used. To amplify SCY1 transcripts, oligonucleotide 188 (5′-GGTAAGCGAAGTTTCCTCTTACTCT-3′) and oligonucleotide 561 (5′-ATTAACCCCGCCGAGTGGTATGTA-3′) and 30 cycles were used. To amplify SECA1 transcripts, oligonucleotide 1,069 (5′-TGGTGAAGGGAAAACGCTTGTTGCT-3′) and oligonucleotide 1,070 (5′-TGCCCACTGTTCACGGGGAT-3′) and 33 cycles were used. To amplify SECE1 transcripts, oligonucleotide 1,065 (5′-CACTAACCGCACAATTCTCG-3′) and oligonucleotide 1,066 (5′-AGTCTTGAACACCTCTTCCG-3′) and 33 cycles were used.
Generation of Transgenic Plants
Constructs used for scy1 and scy2 mutant complementation and localization experiments are shown in Figure 4A. All of the constructs were transferred into a modified pPZP221 transformation vector (Hajdukiewicz et al., 1994) containing the NOS terminator. The ProSCY2:SCY2 construct consisted of 600 bp of sequence upstream of ATG, the full coding sequence of SCY2 (minus the final 3 bp) with all of the introns, and a C-terminal extension encoding a T7 epitope tag (Novagen). For the ProSCY2:SCY1 construct, SCY1 genomic sequence was amplified using an oligonucleotide that would introduce an EcoRI site after the ATG. This was inserted behind the SCY2 upstream sequence using the EcoRI site in that position in SCY2. The rest of the construct included the full coding sequence of SCY1 (minus the final 3 bp) with all of the introns and a C-terminal extension encoding a T7 epitope tag. To generate the ProSCY1:SCY1 construct, SCY2 upstream sequence was removed from the ProSCY2:SCY1 construct and replaced with 800 bp of SCY1 upstream sequence amplified using an oligonucleotide that would add an EcoRI site in the appropriate position after the ATG. The ProSCY2:SCY2-GFP construct included 3.8 kb of SCY2 genomic sequence (600 bp upstream of the ATG plus the coding sequence) fused in frame with GFP. The SECE1 construct for mutant complementation consisted of a 1.4-kb BamHI fragment that included 388 bp of upstream sequence, the full coding sequence, and 455 bp of downstream sequence. Further details of construct generation are available upon request.
Constructs were introduced into plants using the floral dip method (Clough and Bent, 1998) and Agrobacterium tumefaciens GV3101. Recipient plants were heterozygous for scy1-1, scy2-4, sece1-1, or wild-type Ws. Transgenic progeny were selected on medium supplemented with 100 μg mL−1 gentamycin.
Light and Confocal Microscopy
Developing embryos were visualized in lightly fixed, cleared ovules as described previously (Lehti-Shiu et al., 2005).
For confocal microscopy, 5- to 7-d-old seedlings in the T2 generation of multiple lines carrying the ProSCY2:SCY2-GFP construct were removed from agar plates and placed under a coverslip. Confocal fluorescence images were collected using a Zeiss LSM 510 META confocal laser scanning microscope. Samples were viewed using a 40× C-Apochromat water-immersion objective. For detecting GFP, an excitation wavelength of 488 nm was used, and emission was collected with bandpass 500 to 550 IR. Images were also gathered in a spectral series to confirm that the emission was at the expected wavelengths for GFP and not due to chlorophyll autofluorescence. Images were edited using the LSM Image browser (http://www.zeiss.com/lsm) and Adobe Photoshop 6.0.
High-Pressure Freezing and Immunolabeling
Root tips were collected from transgenic and wild-type 7-d-old seedlings and processed as described by Otegui et al. (2006). Samples were subjected to high-pressure freezing (Baltec HPM 010) and freeze substitution at −90°C in 0.1% (w/v) uranyl acetate, 0.2% (w/v) glutaraldehyde in acetone. After 5 d, the samples were warmed to −60°C and infiltrated gradually with Lowicryl HM20 resin (Electron Microscopy Sciences) over 3 to 4 d. Finally, the samples were polymerized at low temperature with UV light. Thin sections (80 nm) were mounted on Formvar-coated nickel grids and incubated in 10% (w/v) nonfat milk in phosphate-buffered saline (PBS) with 0.1% Tween 20 for 10 min at room temperature, prior to incubation for 1 h in either 1:10 or 1:30 dilutions of anti-GFP rabbit polyclonal antibodies (Torrey Pines Biolabs) in the PBS-Tween 20 buffer. Sections were then rinsed for 30 s with PBS with 0.5% Tween 20 and incubated for 1 h in a 1:10 dilution of goat anti-rabbit antibodies conjugated with 10-nm gold or 15-nm gold (Electron Microscopy Sciences). After a final 30-s rinse in PBS with 0.5% Tween 20 and a rinse with deionized water, specimens were observed using a transmission electron microscope (Philips CM-120; FEI Co.). Controls included incubations of wild-type tissues with the same antibodies and incubations where the primary antibodies were omitted.
Cloning of cDNAs and Construction of Precursors
A DNA fragment encoding the precursor to SCY2 was amplified using Pfu Turbo DNA polymerase (Stratagene) and gene-specific primers flanked by restriction sites from cDNA generated from light-grown shoot RNA. The oligonucleotide 5′-TTGGATCCCTCCTCATCCTCTCGATGAAT-3′ (oligonucleotide 697), which added a BamHI site before the ATG, was used in combination with 5′-TTGGTACCTAAGTTCTTCTGTTTCTCCAATCTCC-3′ (oligonucleotide 698), which added a KpnI site 28 bp after the stop codon. The resulting fragment was cloned into pGEM-T (Promega) for sequence analysis. It was excised and subcloned into pGEM3Zf+ for in vitro transcription and translation.
DNA fragments encoding two overlapping halves of the precursor to SECA2 (pSECA2) were amplified from Arabidopsis cDNA generated from 7-d-old Columbia seedling RNA. The first half was amplified using 5′-ACCCCATTCTTTCAGCAGTTC-3′ (oligonucleotide 934) and 5′-AACCCGGGAAATGGGTTCTGTTTCTAATCTTGTGA-3′ (oligonucleotide 856), which introduced a SmaI site upstream of the ATG. The second half was amplified using 5′-CAGGCTGTGGAGGCTAAAGA-3′ (oligonucleotide 933) and 5′-TTGGTACCTCAGTTTTTCCCCTCAGGTAA-3′ (oligonucleotide 857), which introduced a KpnI site downstream of the stop codon. The two fragments were cloned into pGEM-T, and clones with the same orientation were subsequently fused together using an internal SalI site. For import assays, the full-length cDNA sequence was reamplified using 5′-TTGGTACCATGGGTTCTGTTTCTAATCTTGTGA-3′ (oligonucleotide 1,005), which changed the SmaI site to a KpnI site, and 5′-TTGGTACCTCAGTTTTTCCCCTCAGGTAA-3′ (oligonucleotide 857), cloned into pGEM-T, and then subcloned as a KpnI-KpnI fragment behind the SP6 promoter in pGEM-3Z. Orientation was confirmed by restriction digests and sequencing.
Chloroplast Protein Import and Fractionation
In vitro translated precursor proteins were produced by coupled SP6 transcription translation (Promega) in the presence of [3H]Leu (Perkin-Elmer). Translation products were diluted with 1 volume of 60 mm Leu in 2× import buffer (IB; 1× = 50 mm HEPES/KOH, pH 8.0, and 0.33 m sorbitol) prior to use unless otherwise indicated. Intact chloroplasts were isolated from 9- to 10-d-old pea seedlings (Pisum sativum ‘Laxton’s Progress 9 Improved’) and were resuspended in IB at 1 mg mL−1 chlorophyll (Cline, 1986). Radiolabeled precursor proteins were incubated with isolated chloroplasts (0.33 mg mL−1 chlorophyll), 5 mm MgATP, and IB in 120 μE of light in a 25°C water bath for the times specified in the figure legends. After import, samples were treated with 100 μg mL−1 thermolysin on ice for 30 min. Proteolysis was stopped by the addition of 0.5 m EDTA to a final concentration of 10 mm, and chloroplasts were reisolated by centrifugation through 35% Percoll and 5 mm EDTA in IB. Intact chloroplasts were washed, resuspended to equal concentrations of chlorophyll, and then analyzed using SDS-PAGE. Alternatively, chloroplasts were lysed hypotonically by resuspension in 10 mm HEPES/KOH, pH 8.0, and incubation on ice for 10 min. Lysed chloroplasts were analyzed using SDS-PAGE and/or fractionated into membranes and stroma by differential centrifugation (12,000g, 10 min, 4°C). Membranes were resuspended in IB, analyzed using SDS-PAGE, and/or combined with 100 μg mL−1 thermolysin on ice for 30 min. Proteolysis was stopped by the addition of 10 mm EDTA before analysis by SDS-PAGE and fluorography.
Chloroplasts were also fractionated into envelope, stroma, and thylakoids by differential centrifugation as follows. Chloroplasts were lysed by resuspension to 0.5 mg chlorophyll mL−1 in 10 mm HEPES-KOH, pH 8.0, for 10 min on ice, and the lysate was centrifuged at 3,850g for 25 s in a swinging-bucket microfuge at 2°C. The clear supernatant (supernatant 1) was transferred to a separate tube. The remaining partially pelleted chloroplasts were resuspended in 1 mL of 10 mm HEPES-KOH, pH 8.0, and centrifuged at 3,300g for 8 min. The supernatant (supernatant 2) was transferred to a separate tube. Both supernatants were centrifuged at 150,000g for 30 min to pellet the envelope membranes. The stromal extract is the supernatant of ultracentrifugation of supernatant 1. The pellets that resulted from ultracentrifugation were combined. The envelope and thylakoid pellets were resuspended to the original lysate volume in IB before analysis by SDS-PAGE and fluorography.
Electrophoresis
SCY2- and SECA2-containing samples were incubated in sample buffer (0.1 m Tris-HCl, pH 6.8, 8 m urea, 5% SDS, 20% glycerol, and 10% β-mercaptoethanol) for 1 h at room temperature before electrophoresis. Gels were processed for fluorography as described previously (Cline, 1986; Cline and Mori, 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GU289737 (SECA2 cDNA). The Arabidopsis Genome Initiative locus identifiers for the Arabidopsis genes are as follows: SCY1 (At2g18710), SCY2 (At2g31530), SECA1 (At4g01800), SECA2 (At1g21650), and SECE1 (At4g14870).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of the transgene and insertion site in the scy2-4 mutant.
Supplemental Figure S2. Hydrophobicity plots of SCY1, SCY2, and Synechocystis SecY.
Supplemental Figure S3. Phenotype of scy mutant plants complemented with various transgenes.
Supplemental Figure S4. A small amount of SCY2 remains associated with thylakoid fractions devoid of envelope membranes.
Supplemental Table S1. Primers used for PCR genotyping.
Supplementary Material
Acknowledgments
We thank Gabriele Monshausen and Sarah Swanson for help with confocal microscopy; Marisa Otegui, Christoph Spitzer, and Christine Ondzighi for help with sample preparation for electron microscopy; Claudia Lipke and Rafael Buono for plant photography; Kandis Elliot for help with figure preparation; Guang Wu and Chieh-Ting Wang for experimental assistance; and numerous undergraduate students for genotyping. Seeds were provided by the Arabidopsis Biological Resource Center and INRA-Versailles. The confocal imaging was performed at the Plant Imaging Center, Department of Botany, University of Wisconsin-Madison.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson H, von Heijne G. (1993) Sec dependent and Sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J 12: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M, Bals T, Gügel IL, Piotrowski M, Kuhn A, Schünemann D, Soll J, Ankele E. (2009) Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol Plant 2: 1410–1424 [DOI] [PubMed] [Google Scholar]
- Berghöfer J, Karnauchov I, Herrmann RG, Klösgen RB. (1995) Isolation and characterization of a cDNA encoding the SecA protein from spinach chloroplasts: evidence for azide resistance of Sec-dependent protein translocation across thylakoid membranes in spinach. J Biol Chem 270: 18341–18346 [DOI] [PubMed] [Google Scholar]
- Berks BC, Palmer T, Sargent F. (2003) The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol 47: 187–254 [DOI] [PubMed] [Google Scholar]
- Bock R, Timmis JN. (2008) Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Li HM. (2008) Tic40 is important for reinsertion of proteins from the chloroplast stroma into the inner membrane. Plant J 56: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM. (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. (1986) Import of proteins into chloroplasts: membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J Biol Chem 261: 14804–14810 [PubMed] [Google Scholar]
- Cline K. (2003) Biogenesis of green plant thylakoid membranes. Green B, Parson W, , Advances in Photosynthesis and Respiration, Vol 13. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 353–372 [Google Scholar]
- Cline K, Dabney-Smith C. (2008) Plastid protein import and sorting: different paths to the same compartments. Curr Opin Plant Biol 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Mori H. (2001) Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol 154: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Theg SM. (2007) The Sec and Tat protein translocation pathways in chloroplasts. Dalbey R, Koehler C, Tamanoi F, , Molecular Machines Involved in Protein Transport across Cellular Membranes, Vol 25. Elsevier, London, pp 463–492 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deitermann S, Sprie GS, Koch HG. (2005) A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J Biol Chem 280: 39077–39085 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fincher V, Dabney-Smith C, Cline K. (2003) Functional assembly of thylakoid ΔpH-dependent/Tat protein transport pathway components in vitro. Eur J Biochem 270: 4930–4941 [DOI] [PubMed] [Google Scholar]
- Fröderberg L, Röhl T, van Wijk KJ, de Gier JW. (2001) Complementation of bacterial SecE by a chloroplastic homologue. FEBS Lett 498: 52–56 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Schmidt B, Wachter E, Weiss H, Neupert W. (1986) Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47: 939–951 [DOI] [PubMed] [Google Scholar]
- Henry R, Goforth RL, Schuenemann D. (2007) Chloroplast SRP/FtsY and Alb3 in protein integration into the thylakoid membrane. Dalbey RE, Koehler C, Tamanoi F, , Molecular Machines Involved in Protein Transport across Cellular Membranes, Vol 25. Elsevier, London, pp 493–521 [Google Scholar]
- Hoober JK, Boyd CO, Paavola LG. (1991) Origin of thylakoid membranes in Chlamydomonas reinhardtii y-1 at 38°C. Plant Physiol 96: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ. (2005) Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17: 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Schnell DJ. (2008) Protein trafficking to plastids: one theme, many variations. Biochem J 413: 15–28 [DOI] [PubMed] [Google Scholar]
- Inoue K, Baldwin AJ, Shipman RL, Matsui K, Theg SM, Ohme-Takagi M. (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J Cell Biol 171: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ. (2004) Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell 15: 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. (2009) DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. (2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler RH, Hanson MR. (2000) Plastid tubules of higher plants are tissue-specific and developmentally regulated. J Cell Sci 113: 81–89 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, et al. (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidler V, Chaddock AM, Knott TG, Walker D, Robinson C. (1995) A SecY homolog in Arabidopsis thaliana: sequence of a full-length cDNA clone and import of the precursor protein into chloroplasts. J Biol Chem 270: 17664–17667 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE. (2005) Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol Biol 58: 89–107 [DOI] [PubMed] [Google Scholar]
- Li M, Schnell DJ. (2006) Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J Cell Biol 175: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Gong Q, Ma Y, Li P, Li J, Yang S, Yuan L, Yu Y, Pan D, Xu F, et al. (2010) cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J Exp Bot 61: 1655–1669 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. (2009) Translocation of proteins through the Sec61 and SecYEG channels. Curr Opin Cell Biol 21: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S, Briarty L. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Martin JR, Harwood JH, McCaffery M, Fernandez DE, Cline K. (2009) Localization and integration of thylakoid protein translocase subunit cpTatC. Plant J 58: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. (2001) Primary and secondary endosymbiosis and the origin of plastids. J Phycol 37: 951–959 [Google Scholar]
- Mori H, Ito K. (2001) An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci USA 98: 5128–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K. (2006) Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA 103: 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Cline K. (2001) Chloroplast TatC plays a direct role in thylakoid ΔpH-dependent protein transport. FEBS Lett 501: 65–68 [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K. (1999) Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol 146: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré DJ, Selldén G, Sundqvist C, Sandelius AS. (1991) Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol 97: 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi R, Nagata N, Ito T, Takahashi S, Hobo T, Yoshida S, Shinozaki K. (2001) An essential role of a TatC homologue of a ΔpH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 10499–10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M, Goto A, Nohara T, Sugita D, Endo T. (1994) Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein transport. J Biol Chem 269: 31338–31341 [PubMed] [Google Scholar]
- Nakai M, Sugita D, Omata T, Endo T. (1993) Sec-Y protein is localized in both the cytoplasmic and thylakoid membranes in the cyanobacterium Synechococcus PCC7942. Biochem Biophys Res Commun 193: 228–234 [DOI] [PubMed] [Google Scholar]
- Natale P, Brüser T, Driessen AJM. (2008) Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane: Distinct translocases and mechanisms. Biochim Biophys Acta 1778: 1735–1756 [DOI] [PubMed] [Google Scholar]
- Osborne RS, Silhavy TJ. (1993) PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J 12: 3391–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA. (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawyler A, Meylan M, Siegenthaler PA. (1992) Galactolipid export from envelope to thylakoid membranes in intact chloroplasts. I. Characterization and involvement in thylakoid lipid asymmetry. Biochim Biophys Acta 1104: 331–341 [DOI] [PubMed] [Google Scholar]
- Rigel NW, Braunstein M. (2008) A new twist on an old pathway: accessory Sec [corrected] systems. Mol Microbiol 69: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Lim F, Wallace JC. (1991) Electron microscopic localization of pyruvate carboxylase in rat liver and Saccharomyces cerevisiae by immunogold procedures. Arch Biochem Biophys 290: 197–201 [DOI] [PubMed] [Google Scholar]
- Roy LM, Barkan A. (1998) A SecY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J Cell Biol 141: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J. (1991) One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J 10: 1749–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann D, Amin P, Hartmann E, Hoffman NE. (1999) Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J Biol Chem 274: 12177–12182 [DOI] [PubMed] [Google Scholar]
- Schünemann D. (2007) Mechanisms of protein import into thylakoids of chloroplasts. Biol Chem 388: 907–915 [DOI] [PubMed] [Google Scholar]
- Shipman RL, Inoue K. (2009) Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett 583: 938–942 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp J, Inoue K, Keegstra K, Froehlich JE. (2007) A novel serine/proline-rich domain in combination with a transmembrane domain is required for the insertion of AtTic40 into the inner envelope membrane of chloroplasts. Plant J 52: 824–838 [DOI] [PubMed] [Google Scholar]
- Voelker R, Barkan A. (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 14: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Mendel-Hartvig J, Barkan A. (1997) Transposon-disruption of a maize nuclear gene, tha1, encoding a chloroplast SecA homologue: in vivo role of cp-SecA in thylakoid protein targeting. Genetics 145: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta L, Soll J, Bölter B. (2007) Requirements for a conservative protein translocation pathway in chloroplasts. FEBS Lett 581: 2621–2624 [DOI] [PubMed] [Google Scholar]
- von Wettstein D. (2001) Discovery of a protein required for photosynthetic membrane assembly. Proc Natl Acad Sci USA 98: 3633–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht UC, Westhoff P. (2001) Biogenesis and origin of thylakoid membranes. Biochim Biophys Acta 1541: 91–101 [DOI] [PubMed] [Google Scholar]
- Westphal S, Soll J, Vothknecht UC. (2001) A vesicle transport system inside chloroplasts. FEBS Lett 506: 257–261 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A. (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. (2003) YidC is strictly required for membrane insertion of subunits a and c of the F1F0ATP synthase and SecE of the SecYEG translocase. Biochemistry 42: 10537–10544 [DOI] [PubMed] [Google Scholar]
- Yuan J, Henry R, McCaffery M, Cline K. (1994) SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science 266: 796–798 [DOI] [PubMed] [Google Scholar]
- Yusa F, Steiner JM, Löffelhardt W. (2008) Evolutionary conservation of dual Sec translocases in the cyanelles of Cyanophora paradoxa. BMC Evol Biol 8: 304–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.