Abstract
Several members of the Kruppel-like factor (KLF) family of transcription factors play important roles in differentiation, survival, and trafficking of blood and immune cell types. We demonstrate in this study that hematopoietic cells from KLF4−/− fetal livers (FL) contained normal numbers of functional hematopoietic progenitor cells, were radioprotective, and performed as well as KLF4+/+ cells in competitive repopulation assays. However, hematopoietic “KLF4−/− chimeras” generated by transplantation of KLF4−/− fetal livers cells into lethally irradiated wild-type mice completely lacked circulating inflammatory (CD115+Gr1+) monocytes, and had reduced numbers of resident (CD115+Gr1−) monocytes. Although the numbers and function of peritoneal macrophages were normal in KLF4−/− chimeras, bone marrow monocytic cells from KLF4−/− chimeras expressed lower levels of key trafficking molecules and were more apoptotic. Thus, our in vivo loss-of-function studies demonstrate that KLF4, previously shown to mediate proinflammatory signaling in human macrophages in vitro, is essential for differentiation of mouse inflammatory monocytes, and is involved in the differentiation of resident monocytes. In addition, inducible expression of KLF4 in the HL60 human acute myeloid leukemia cell line stimulated monocytic differentiation and enhanced 12-O-tetradecanoylphorbol 13-acetate induced macrophage differentiation, but blocked all-trans-retinoic acid induced granulocytic differentiation of HL60 cells. The inflammation-selective effects of loss-of-KLF4 and the gain-of-KLF4-induced monocytic differentiation in HL60 cells identify KLF4 as a key regulator of monocytic differentiation and a potential target for translational immune modulation.
Although members of the Sp/Kruppel-like factor (KLF)3 family of transcription factors, KLF1–17 and Sp1–8 in mammals, share a high degree of sequence homology in their zinc-finger regions, their non-DNA binding portions are diverse in composition and function (1–4). Several of the 17 mammalian KLFs, including KLF1, KLF2, KLF3, KLF6, KLF11, and KLF13, have been shown to play crucial roles in hematopoiesis and immunity (5–13). In addition to regulating quiescence in T lymphocytes, KLF2 (LKLF; KLF4’s closest family member) regulates trafficking of T lymphocytes by regulating the expression of key trafficking molecules, including L-Selectin (CD62L) and β7 integrin (3, 4, 14). KLF4 (also known as gut KLF (15)) itself down-modulates expression of the CD11d myeloid-associated molecule (16) and mediates proinflammatory signaling in human macrophages (17). KLF4 expression is induced in response to IFN-γ, LPS, or TNF-α in human macrophages, and decreases in response to TGF-β1. KLF4 binds to and induces the iNos promoter, and KLF4 knockdown diminishes LPS or IFN-γ induction of iNOS. Recently, Feinberg et al. demonstrated that KLF4 is highly expressed in the human monocyte/macrophage lineage and ectopic expression of KLF4 induces monocytic differentiation of HL60 cells (18). KLF4 was demonstrated to be a downstream target of the Ets transcription factor PU.1 and a direct transcriptional regulator of CD14. The authors also showed that common myeloid progenitors and hematopoietic stem-progenitor cells (HSPCs) from KLF4−/− mice produced fewer monocytic cells and increased granulocytic cells in clonogenic assays (18). Although these studies strongly implicate KLF4 as a regulator of macrophage differentiation and activation, detailed in vivo studies have been difficult due to nonhematopoietic lethality of complete KLF4 deficiency (KLF4−/−) shortly after birth (19).
Monocytes are the circulating precursors of tissue macrophages and dendritic cells. Monocyte-macrophage development in the mouse is thought to occur in ordered progression, starting from a CD115+Ly6C+CCR2+ precursor in the bone marrow (BM), which gives rise to three distinct subpopulations of circulating blood monocytes: 1) Ly6ChighCD62L+CCR2+ “inflammatory” monocytes, which are short-lived and migrate to sites of inflammation; 2) Ly6ClowCD62L−CCR2− “resident” monocytes, which remain in the circulation longer than their Ly6Chigh counterparts and are thought to generate and replace resident dendritic cells and macrophages; and 3) a small population of Ly6Cmid cells, which may represent either a functionally distinct population of monocytes or simply the transition from Ly6Chigh to Ly6Clow. All three monocyte subpopulations have the ability to differentiate into macrophages or dendritic cells in vitro (20).
KLF4 is expressed at high levels in mouse and human embryonic stem (ES) cells; and recently, expression of KLF4, along with three other transcription factors (Oct3/4, Sox2, and c-Myc), was found to be sufficient to induce developmental reprogramming of mature mouse fibroblasts to reacquire key properties of pluripotent ES cells (21–25). KLF4 mediates the binding of Oct3/4 and Sox2 to the mouse Lefty1 proximal promoter (26). To date, however, Lefty1 is the only known ES-specific KLF4 target, and the mechanism by which KLF4 participates in reprogramming of mouse fibroblasts remains cryptic.
We previously identified KLF4 among transcripts over-expressed in human hemopoietic stem cell (HSC)-enriched populations (CD34+/[CD38/Lin]low), as compared with hematopoietic progenitor (HPC)-enriched populations (CD34+/[CD38/Lin]high) (27). Based on the expression pattern of KLF4 in HSPCs and human monocyte-macrophages, and its close homology to KLF2, we hypothesized that loss of KLF4 would disrupt HSPC and/or monocyte-macrophage function. We report here that despite high levels of KLF4 expression in HSPCs, key in vitro and in vivo hematopoietic functions of these cells were not affected by loss of KLF4. However, hematopoietic “KLF4−/− chimeras” generated by transplantation of KLF4−/− fetal liver (FL) cells into lethally irradiated congenic wild-type (wt) mice, had fewer monocytic cells in their BMs, lower numbers of resident monocytes (CD115+Gr1−) in their blood, and completely lacked inflammatory monocytes (CD115+Gr1+) in their blood and spleens. Furthermore, BM monocytic cells from KLF4−/− chimeras expressed lower levels of key trafficking molecules and were more apoptotic. KLF4−/− chimeras had normal numbers of peritoneal exudate (PE) macrophages, and KLF4−/− macrophages were functionally intact in their iNos generation and expressed TNF-α in response to activation. Finally, conditional KLF4 expression in human HL60 leukemia cells induced features of monocytic differentiation and enhanced 12-O-tetradecanoylphorbol 13-acetate (TPA) induced macrophage differentiation, but blocked all-trans-retinoic acid (RA) induced granulocytic differentiation of HL60 cells. These data identify KLF4 as a key in vivo regulator of mammalian monocyte differentiation.
Materials and Methods
Mice, FL transplantation, competitive repopulation, and peritoneal macrophages
Mouse BM cells were harvested from 6 to 10 wk old C57BL/6 (CD45.2+) and C57BL/6-Ly5.2 (CD45.1+) mice by flushing femurs and tibias. FL cells were isolated from embryonic (postcoital) day (E) 14.5 KLF4−/− (19) and wt littermates (on a C57BL/6 background, provided by Dr. Julie Segre, National Institutes of Health, Bethesda, MD) and genotyped by PCR. To generate hematopoietic chimeras, 1–2 × 106 KLF4−/− or wt FL cells (CD45.2+) were transplanted i.v. into 6–8-wk-old lethally (1000 cGy) irradiated CD45.1+ hosts. For competitive repopulation experiments, 5 × 105 FL cells (CD45.2+) plus 5 × 105 wt BM cells (CD45.1+) were co-transplanted into lethally irradiated CD45.1+ hosts.
Peritoneal macrophages were isolated 8–12 wk after FL transplant by flushing the peritoneal cavity with 10 ml PBS. For inflammation experiments, 0.75 ml 4% brewer’s thioglycolate (Sigma-Aldrich) was injected into the peritoneum 3 days before peritoneal macrophage harvest. Three × 105 macrophages were plated in each well of a 96-well flat-bottom plate and allowed to adhere overnight. Then, wells were washed three times with fresh medium to remove nonadherent cells. All studies performed in mice were approved by the Johns Hopkins University Animal Care and Use Committee.
Cell lines and culture
HL60 (American Type Culture Collection) human leukemia cells and primary mouse peritoneal macrophages were cultured in RPMI 1640 (Invitrogen Life Technologies) supplemented with 10% FBS (FBS; Gemini Bio-Products), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen Life Technologies). Stable cell lines transduced to express a KLF4-estrogen receptor (ER) fusion and GFP (see below) were enriched by FACS-sorting for GFP+ cells, followed by limiting dilution cloning. At least three clones were examined for each construct. In brief, 4-hydroxy-tamoxifen (4HT; in 100% ethanol) was added to cell cultures to achieve a final concentration of 200 nM. TPA and all-trans-RA were purchased from Sigma-Aldrich and resuspended in DMSO or ethanol, respectively. For hematopoietic colony-forming cell (CFC) assays, 5 × 104 mouse FL or BM cells were plated in methylcellulose medium (Stem Cell Technologies) supplemented with 50 ng/ml recombinant mouse stem cell factor (SCF), 10 ng/ml recombinant mouse IL-3, 10 ng/ml recombinant human IL-6, and 3 U/ml recombinant human erythropoietin (all cytokines from PeproTech). Monocyte-macrophage-specific CFC assays were performed as above, except medium was supplemented only with 50 ng/ml SCF and 10 ng/ml recombinant mouse monocyte-CSF (M-CSF). Colonies were counted 7 days after plating.
Plasmid constructs and dual promoter lentiviral vectors
The human KLF4 open reading frame was generated by RT-PCR, and cloned into an intermediate vector downstream of an elongation factor 1α promoter. A modified ligand-binding domain construct of the mouse ER, which is selectively responsive to 4HT (28), was fused to the 3′ terminus of KLF4 (KLF4-ER). The elongation factor 1α α-KLF4-ER cassette was then subcloned into the FUGW (29) lentiviral plasmid (provided by Dr. David Baltimore, California Institute of Technology, Pasadena, CA). All clones were sequence verified. Lentiviral packaging and transductions were conducted as described previously (30, 31).
Flow cytometric analysis and cell purification
The following FITC-, PE-, PerCP-Cy5.5-, or allophycocyanin-coupled mAbs against mouse leukocyte differentiation Ags were obtained from BD Biosciences: CD3, CD4, CD8, CD45R/B220, CD45.1, CD45.2, CD69, NK1.1, and Gr1 (recognizes Ly6C and Ly6G). CD115 and F4/80 mAbs were purchased from eBioscience. Blood samples were lysed with RBC lysis buffer (eBioscience), and then cells were blocked with Fc Block (BD Biosciences), stained at 4°C with optimal amounts of fluorochrome-coupled mAbs diluted in staining medium (PBS containing 2.5% FBS) for 30 min, then washed. At least 20,000 events were acquired per immunostain on a FACSort or FACSCalibur (BD Biosciences) flow cytometer. Similarly, levels of competitive repopulation were measured using CD45.1-PE and CD45.2-FITC mAbs (BD Biosciences). Percent engraftment of donor CD45.2+ FL cells was calculated as: 100 × (number of CD45.2+ cells)/(number of CD45.1+ cells + number of CD45.2+ cells). For cell fractionation, whole BM or splenocyte suspensions were RBC lysed and stained with mAbs specific for T cells, B cells, NK cells, erythroid progenitors, and monocytes, then enriched by cell sorting on a FACSVantage or FACSAria (BD Biosciences) flow cytometer. CD115+ cells were enriched, for Western blotting, using anti-biotin MicroBeads (Miltenyi Biotec) according to the manufacturer’s protocol. Enrichment by positive selection was verified (>40% CD115+) by flow cytometry.
Control or genetically modified HL60 cells were stained using propidium iodide (PI; Sigma-Aldrich) at selected time points after addition of 4HT or vehicle control (32). DNA content analysis was performed using the Dean-Jett-Fox model in the FlowJo FACS analysis software (Tree Star).
Macrophage activation and Griess assay
Adherent peritoneal macrophages (described above) were treated with various concentrations of LPS (Sigma-Aldrich) at 37°C for 24 h. The generation of nitrite was used as a measure of NO release from peritoneal macrophages; nitrite concentrations in supernatants were measured by the Griess reaction as described by the manufacturer’s protocol (Promega). RNA was harvested from cells 24 h after treatment with LPS.
qRT-PCR amplification and Western blots
RNA was isolated using the RNAeasy kit (Qiagen), and Superscript III (Invitrogen Life Technologies) was used to make cDNA, according to the manufacturer’s protocols. Real-time quantitative-RT-PCR (qRT-PCR) was conducted as described previously (27). Unless stated otherwise, qRT-PCRs were normalized with hydroxymethylbilane synthase expression. Sequences for all primers used for qRT-PCR are available upon request.
Western blots were performed using standard procedures with the following modifications. Cells were cultured in complete medium supplemented with MG132 (30 μM; Sigma-Aldrich) for 5 h before harvesting (33). Cells were lysed in RIPA buffer (Sigma-Aldrich) supplemented with complete mini tablets (Roche) and lysates were quantified and loaded immediately. Polyclonal Abs recognizing KLF4 and GAPDH were purchased from Santa Cruz Biotechnology and Abcam, respectively. Densitometry was performed on a Bio-Rad GS-800 Calibrated Densitometer using Quantity One program from Bio-Rad.
Results
KLF4 was expressed in HSCs and monocyte-macrophages
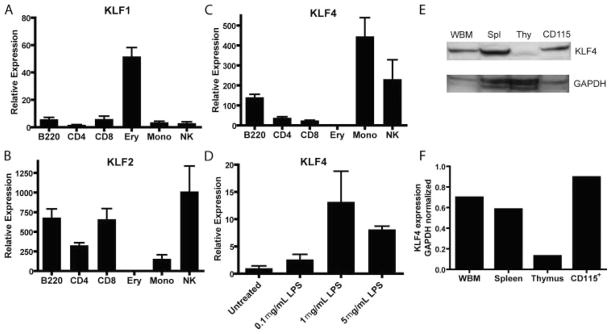
We previously reported KLF4 among the transcripts over-expressed in human HSC-enriched populations (CD34+/[CD38/Lin]low), as compared with HPC-enriched populations (CD34+/[CD38/Lin]high) (27), and similar results have been found in mice (34). To further characterize the hematopoietic expression of KLF4 in mouse cells, we isolated several types of committed blood and immune cells from wt C57BL/6 mice, and we measured the expression of KLF4, and for comparison, its two closest family members, KLF1 and KLF2 (Fig. 1, A–C). As anticipated (35), KLF1 was highly expressed only in mouse BM erythroid cells. KLF2 was expressed in all mouse lymphoid populations, but at low levels in monocytic and erythroid BM cells. KLF4 was expressed most highly in mouse BM monocytic cells (CD45+CD115+Gr1+), consistent with previous microarray experiments in humans, which found the highest levels of KLF4 transcripts in CD14+ monocytes (36). We found intermediate levels of KLF4 in NK cells and B cells and low levels in T cells and erythroid BM cells. We confirmed that KLF4 protein was expressed in cell lysates from whole BM, spleen, thymus, and CD115+ purified cells (Fig. 1, E and F). We next measured the expression of KLF4 in activated peritoneal macrophages. As has been demonstrated in human cells (17), activation of mouse macrophages using LPS induced KLF4 expression (Fig. 1D).
FIGURE 1.
KLF4 was expressed at high levels in BM monocytic cells and activated macrophages. A–C, Committed B cells (CD45+B220+), CD4 T cells (CD45+CD3+CD4+), CD8 T cells (CD45+CD3+CD8+), and NK cells (CD45+NK1.1+) were FACS-sorted from C57BL/6 splenocytes. Erythroid (CD45−Ter119+) and monocytic cells (CD45+CD115+Gr1dimF4/80+) were FACS-sorted from BM. RNA was harvested, and qRT-PCR was performed in triplicate and normalized to hydroxymethylbilane synthase. D, Peritoneal macrophages were exposed to LPS for 24 h. Data are representative of n = 3 experiments, and error bars represent SEM. E, Western blots of cell lysates from whole BM, spleen, thymus, and purified CD115+ BM monocytic cells. Cells were cultured for 5 h in the presence of the proteasome inhibitor MG132 before harvesting for Western blot. F, Densitometric analysis of blot in E; KLF4 expression was normalized to GAPDH expression in each cell type.
KLF4−/− FLs contained normal numbers of functional HPCs and HSCs
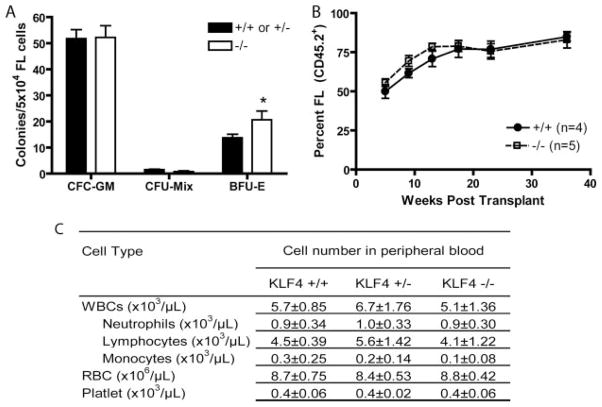
KLF4−/− mice die shortly after birth due to an epithelial defect (19), but have been reported to have blood. We found that FLs from E14.5 KLF4−/− embryos had similar numbers of total cells as wt embryos (data not shown). FL cells from KLF4−/− embryos formed similar numbers of in vitro granulocyte/monocyte (CFC-GM) and granulocyte/monocyte/erythroid (CFC-Mix) colonies, and slightly more erythroid (BFU-E) colonies (Fig. 2A). To examine the ability of FL cells from wt and KLF4−/− embryos to form monocytic/macrophage (CFC-M) colonies specifically, we cultured FL cells in methylcellulose containing only M-CSF and SCF; there were no significant differences in the numbers of CFC-M from wt or KLF4−/− embryos (data not shown). In several experiments, we observed only slight differences between KLF4+/+ and KLF4+/− mice, so we have grouped these in certain (indicated) experiments, such as Fig. 2A.
FIGURE 2.
Loss of KLF4 did not affect HPC or HSC function. A, Five × 104 E14.5 FL cells were plated in methylcellulose CFC assay cultures containing hematopoietic cytokines (see Materials and Methods). Colonies were counted 7 days after plating. Data is representative of n = 6 for KLF4+/+ or KLF4+/− and n = 3 for KLF4−/− FL cells. In several experiments, we observed only a slight difference between KLF4+/+ and KLF4+/− chimeras, so we have grouped these in several (indicated) experiments. Error bars, SEM; *, p = 0.031. B, Competitive repopulation assay comparing donor cell repopulating capacity of 1) KLF4+/+ CD45.2+ FL vs CD45.1+KLF4+/+ BM (●, solid line) to that of 2) CD45.2+KLF4−/− FL vs CD45.1+KLF4+/+ BM (□, dotted line). Percents FL-derived (CD45.2+) granulocytes are plotted over 40 wk. C, Complete blood cell counts were performed on blood from KLF4+/+ (n = 5), KLF4+/− (n = 5), and KLF4−/− (n = 10) chimeras. Before performing complete blood cell counts, donor chimerism of >95% was confirmed by flow cytometry.
To examine the in vivo hematopoietic capacity of KLF4−/− cells, we performed transplants of wt or KLF4−/− FL cells into lethally irradiated congenic adult recipient mice. KLF4−/− cells provided radioprotection, and complete blood cell counts performed 6 and 10 wk after transplant showed no significant differences between mice transplanted with wt or KLF4−/− FL cells (Fig. 2C). To assess competitive engraftment and repopulation ability, equal numbers of wt or KLF4−/− FL donor cells (CD45.2+) were cotransplanted with congenic (CD45.1+) wt BM donor cells into lethally irradiated congenic (CD45.1+) adult recipient mice, and FL cell-derived (CD45.2+) engraftment levels were measured over a 40 wk time course. We observed no significant differences in engraftment/repopulation by KLF4−/− compared with wt FL cells (Fig. 2B).
KLF4 was essential for normal late monocytic differentiation in vivo
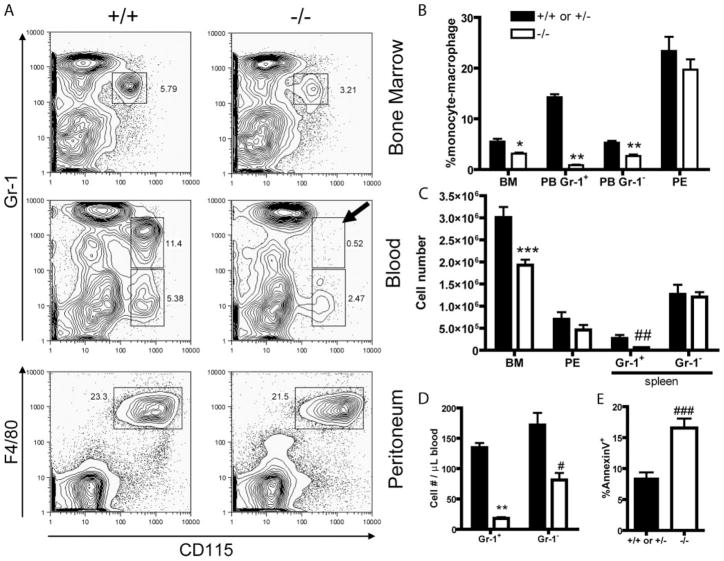
Because we found high levels of KLF4 in BM monocytic cells, we next examined the BM, PB, spleen, and peritoneal fluid of KLF4+/+ or +/− and KLF4−/− chimeras for monocyte-macrophages (Fig. 3). KLF4−/− chimeras had a 43% reduction in the number of donor-derived (CD45.2+) BM monocytic cells. Peripheral blood from KLF4−/− chimeras had a 53% reduction in the number of donor-derived resident monocytes (CD115+Gr1−) and was essentially void of donor-derived inflammatory monocytes (CD115+Gr1+; Fig. 3A middle right panel with arrow). Spleens from KLF4−/− chimeras had essentially no donor-derived inflammatory monocytes, but the numbers of donor-derived resident monocytes were unaffected. The numbers of donor-derived peritoneal macrophages were unaffected by loss of KLF4. Examination of other cell lineages showed no significant differences.
FIGURE 3.
Loss of KLF4 disrupted late monocytic differentiation. A, Representative FACS plots of BM, blood, and peritoneal exudates (PE) isolated from KLF4+/+ or KLF4+/− and KLF4−/− chimeras 10 wk after transplant. Cells were stained with indicated mAbs. Cells are morphologically gated on total viable cells except for peripheral blood, which is gated based on monocytic light scattering. In all plots, cells are gated on CD45.2+ cells (donor-derived). Arrow added to emphasize lack of inflammatory monocytes in KLF4−/− chimeras’ blood. B, Percent donor-derived monocytic cells (CD45.2+CD115+Gr1+ or CD45.2+CD115+Gr1−) or macrophages (CD45.2+CD115+F4/80+) found in BM, PB, and PE from KLF4+/+ or KLF4+/− and KLF4−/− chimeras. The graphs shown are the combined results of ≥ three independent FL transplants with n ≥ 5 in all experiments. * and **, p = 0.0009 and p < 0.0001, respectively. C, Similar to B except total number of cells is plotted. *** and ## indicate p = 0.0012 and p = 0.0129, respectively. D, Numbers of Gr1+ and Gr1− monocytes in peripheral blood from KLF4+/+ or KLF4+/− and KLF4−/− chimeras. Total white blood cell count × percent monocytes by flow cytometry determined the total numbers of monocytes. ** and #, p < 0.0001 and p = 0.0016, respectively. E, Percent Annexin V-positive BM monocytic cells (CD45.2+CD115+Gr1+AnnexinV+). ###, p = 0.0008.
We next sought to understand the cause of the decreased numbers of monocytic cells in the KLF4−/− chimeras. Because KLF4−/− FL cells showed no defect in colony formation in vitro, we examined donor-derived monocytic cells from KLF4−/− chimeras to see whether loss of KLF4 affected the survival of these cells in vivo. Although there were no differences in the numbers of Annexin V-positive granulocytic or lymphoid cells, there was a 2-fold increase in the numbers of Annexin V-positive monocytic cells in the BMs of KLF4−/− chimeras, suggesting that KLF4 may be involved in the survival of monocytic progenitors.
Loss of KLF4 disrupted expression of key trafficking molecules
Recent reports demonstrated that the closely related KLF2 transcription factor regulates several critical trafficking molecules in T lymphocytes (3, 4). Therefore, we measured the expression of several molecules (previously shown to be regulated by KLF2) in KLF4−/− monocytic cells. Donor-derived BM monocytic cells from KLF4−/− chimeras expressed lower levels of β7 integrin and the inflammatory monocyte marker CD62L, but expressed slightly higher levels of F4/80, a late monocyte-macrophage differentiation marker (Fig. 4A); there was no effect on the level of CD69. Consistent with our flow cytometric measurements, BM monocytic cells from KLF4−/− chimeras had lower levels of β7 integrin and CD62L mRNA by qRT-PCR (Fig. 4C); levels of CCR2 were reduced 6-fold in the in KLF4−/− monocytic cells.
FIGURE 4.
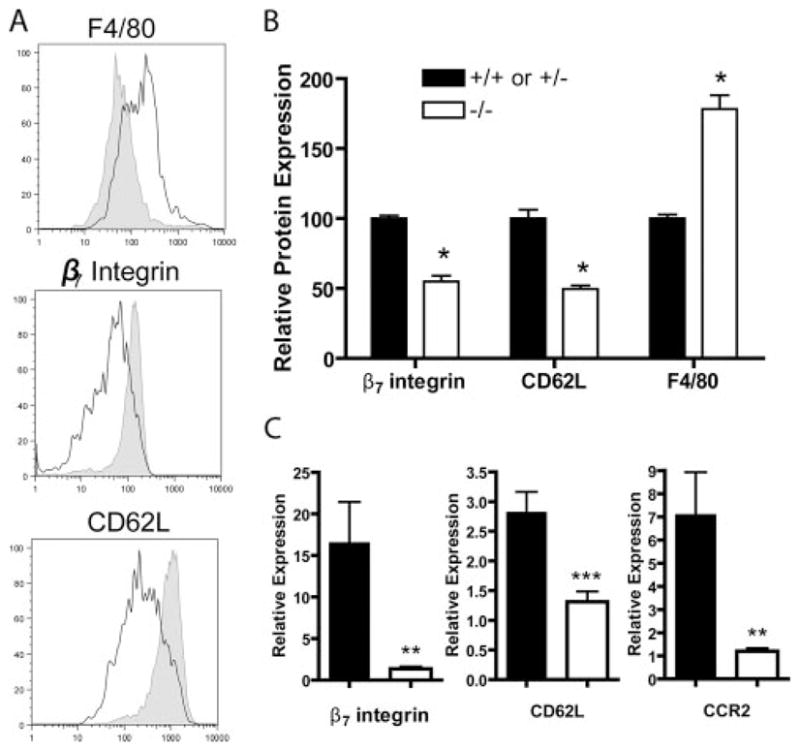
BM monocytic cells from KLF4−/− chimeras had dysregulated differentiation and cell surface molecules. A, Representative histograms of CD45.2+CD115+Gr1+ BM monocytic cells from KLF4+/+ (shaded) and KLF4−/− (empty) chimeras. B, Normalized mean fluorescence intensity of indicated molecules from CD115+Gr1+CD45.2+ BM monocytic cells. n = 8 from ≥ three independent FL transplants from KLF4+/+ or KLF4+/− and KLF4−/− chimeras; *, p < 0.0001. C, qRT-PCR of mRNA from KLF4+/+ or KLF4+/− or KLF4−/− BM monocytes. n = 3 from three independent FL transplants; ** and ***, p = 0.03 and p = 0.02, respectively.
KLF4−/− macrophages became activated in response to LPS
Previous reports have shown that KLF4 is an important mediator of proinflammatory signaling in human macrophages in vitro (17). Thioglycolate-induced peritoneal macrophages from KLF4+/+ and KLF4−/− chimeras became larger and contained cytoplasmic granules after culture in the presence of LPS, indicating similar activation. In addition, we found no significant reduction between KLF4+/+ and KLF4−/− macrophages in nitrite production (Fig. 5A) or TNF-α mRNA up-regulation.
FIGURE 5.
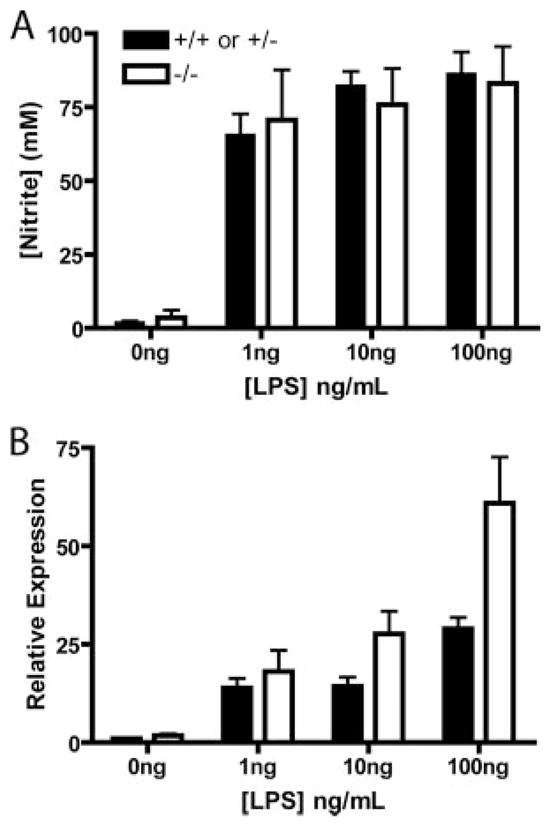
Loss of KLF4 did not reduce iNos activity or TNF-α mRNA levels. Thioglycolate-stimulated peritoneal macrophages were harvested from KLF4+/+ or KLF4+/− and KLF4−/− chimeras and allowed to adhere to tissue culture plates overnight. Cells were washed three times and medium was added with the indicated amounts of LPS. A, After 24 h of culture in the presence of LPS, Greiss assays were performed on supernatants from each well and cells were harvested for RNA analysis. B, TNF-α mRNA levels, measured by qRT-PCR (n = 2).
Induction of KLF4 expression in HL60 cells stimulated monocytic differentiation, as well as p21-associated proliferative arrest
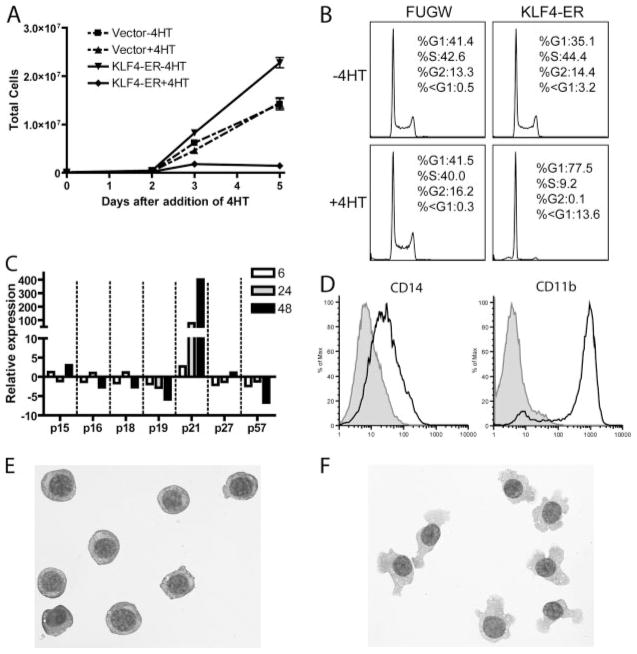
To determine whether KLF4 is sufficient to regulate components of monocyte-macrophage differentiation, we examined the effects of enforced KLF4 expression on the HL60 human leukemia cell line, as a model of granulocytic or monocyte-macrophage differentiation. We were unable to obtain an HL60 cell line which stably expressed KLF4, suggesting that constitutive KLF4 expression was lethal or growth inhibitory. Therefore, we used an approach that had been described previously for KLF1 (37), in which KLF4 is fused to a tamoxifen-sensitive mutant of the estrogen receptor. The KLF4-ER fusion protein was subcloned into a lentivirus and used to transduce HL60 cells (see Materials and Methods). Because it had been demonstrated that KLF4 up-regulates p21 and down-regulates proliferation in other cell types (38), we selected and cellularly subcloned KLF4-ER-transduced HL60 clones which, in the absence of 4HT, expanded at rates similar to those of untransduced cells, to bias toward minimally leaky cell lines. Upon addition of 4HT to these selected KLF4-ER-transduced HL60 sub-cloned cell lines, proliferation slowed dramatically (Fig. 6A). PI staining 48 h after the addition of 4HT (but not vehicle controls) showed that KLF4-ER-induced cells arrested markedly in G1 phase, as compared with transduction controls; the minor subdiploid population of PI-stained cells and absence of Annexin V-positive cells (data not shown) indicated that KLF4 expression did not greatly enhance apoptosis (Fig. 6B). Similar to previous reports in other cell types (38), we found that KLF4-induced cell cycle arrest was associated with increased levels of p21 (Fig. 6C), but not other cell cycle inhibitors. To assess the effects of induced KLF4 on monocytic differentiation, we immunostained KLF4-ER-transduced cells 48 h after addition of 4HT. The 4HT-treated cells (but not vehicle or transduction controls) expressed higher levels of the CD14 and CD11b monocytic markers (Fig. 6D), were non-adherent, and had monocytic morphology (Fig. 6, E and F).
FIGURE 6.
Induced expression of KLF4 in HL60 cells induced proliferative arrest, p21, and features of monocytic differentiation. In brief, 105 FUGW-transduced and KLF4-ER-transduced HL60 cells were plated, and viable cells were counted each day using trypan blue. 4HT or ethanol was added to each well on day 0, and cells were split 1/2 when they reached 106 cells/ml. Shown are representative results from one of three independent clones. B, Day 2 HL60 cells were stained for DNA content with PI. C, Results are from a single experiment in which KLF4-ER-transduced HL60 cells were treated with 4HT and harvested at 6, 24, and 48 h after treatment. qRT-PCR results were normalized to KLF4-ER cells treated with ethanol. D, KLF4-ER-transduced HL60 cells were treated with ethanol (shaded) or 4HT (empty) for 48 h and then stained with the indicated mAbs. E and F, Cytospins of KLF4-ER-transduced HL60 cells cultured for 5 days in the presence of vehicle (ethanol; E) or 4HT (F).
KLF4 expression enhanced macrophage differentiation and blocked granulocytic differentiation of HL60 cells
We next examined the effects of conditional KLF4 expression on HL60 cells induced to differentiate into granulocytes or macrophages. HL60 cells differentiate into macrophages or granulocytes when cultured with TPA or RA, respectively. KLF4-ER transduced HL60 cells differentiated into monocytes in the presence of 4HT and macrophages in the presence of TPA (Fig. 7A), as expected. When we cultured KLF4-ER transduced HL60 cells in the presence of both TPA and 4HT, we observed a much more robust macrophage differentiation, with essentially every cell present in the culture contributing to a monolayer of macrophages (Fig. 7A, lower right). To assess granulocytic differentiation of HL60 cells, we measured the surface expression of CD66. When cultured in the presence of RA, a portion of the HL60 cells differentiate into granulocytes and up-regulate CD66. In contrast to the enhanced macrophage differentiation, granulocytic differentiation was blocked when KLF4-ER transduced cells were cultured with 4HT and RA (Fig. 7B). Interestingly, KLF4 induced monocytic differentiation was completely blocked when RA was present in the culture (Fig. 7B, lower right).
FIGURE 7.
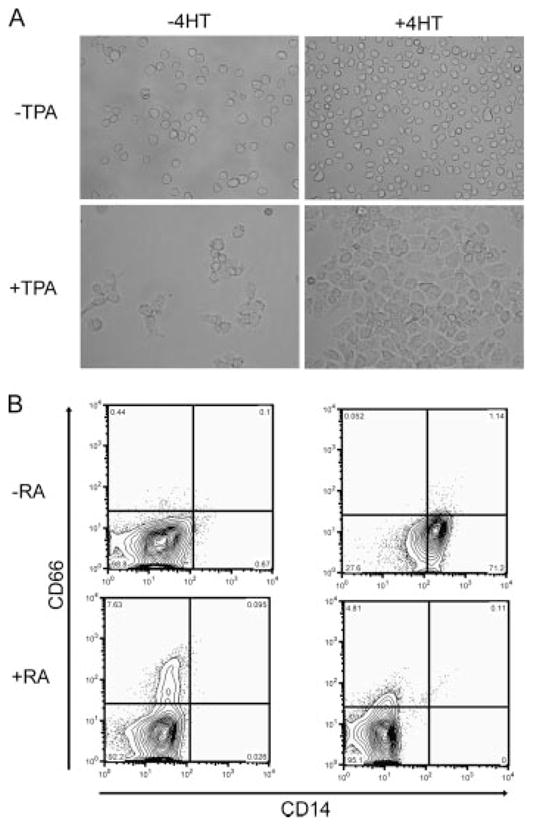
KLF4 expression enhanced macrophage differentiation and blocked granulocytic differentiation. A, HL60 cells were cultured in the presence of TPA (50 ng/ml) and/or 4HT for 48 h. Photomicrographs were taken at 60× magnification and are representative of three independent experiments. B, HL60 cells were cultured for 5 days in the presence or absence of RA (1 μM) and/or 4HT. Cells were immunostained with CD66 and CD14. The plots shown are representative of three independent experiments.
Discussion
We found that induced KLF4 expression stimulated monocytic differentiation in HL60 cells (Fig. 6) and acted synergistically with TPA to promote macrophage differentiation, but blocked RA granulocytic differentiation of HL60 cells, extending previous in vitro studies in human cells (18, 38–40). Our loss-of-function studies in mice now define KLF4 as an essential regulator of in vivo mammalian monocytic differentiation/survival. In the investigations herein, loss of KLF4 disrupted normal in vivo monocytic differentiation. Wt mice transplanted with KLF4 deficient cells (“KLF4−/− chimeras”) had decreased numbers of resident monocytes in blood and completely lacked inflammatory monocytes in blood and spleen. In addition, monocytic cells from KLF4−/− chimeras had abnormal characteristics, with increased F4/80 and decreased β7 integrin, CD62L (a marker of inflammatory monocytes), and CCR2 expression. Despite these qualitative abnormalities, the reduced numbers of resident monocytes, and the absence of inflammatory monocytes, numbers of peritoneal macrophages were not affected by loss of KLF4.
Results from studies in human macrophages identified KLF4 as a potential mediator of proinflammatory signaling (17). Similar to human cells, mouse macrophages up-regulated KLF4 upon activation with LPS (Fig. 1D). We examined the function of KLF4−/− macrophages to see whether loss of KLF4 prevented these cells from responding to LPS. We found that, upon activation, KLF4−/− macrophages expressed iNos similarly to wt macrophages and expressed even higher levels of TNF-α. Thus, KLF4 is dispensable for macrophage activation.
Our observation that the BMs of KLF4−/− chimeras contained about half the normal numbers of CD115+Gr1+ (resident) monocytic precursor cells suggested that KLF4 is important for the survival or production of these cells. An antiapoptotic role for KLF4 has been described previously in which KLF4 prevents apoptosis by up-regulating p21 and inhibiting expression of BAX (40). In support of a survival defect, we found that KLF4−/− chimeras’ BM monocytic cells were twice as apoptotic as KLF4+/+ cells. In addition, whereas the majority of HL60 cells were killed by treatment with TPA, induction of KLF4 rescued almost all of the cells and promoted macrophage differentiation, suggesting that KLF4 may enhance the survival of these cells. Because absence of KLF4 did not affect CFC-M or other HSPCs, our data do not support a defect early in monocyte development. A caveat is that it is possible that such an effect was masked because in vitro cultures do not correctly mimic in vivo development. Indeed, previous reports have shown that Ly6C (detected in our experiments using Gr1 mAb), a characteristic of monocytic precursors and inflammatory monocytes, is rapidly down-regulated upon in vitro culture (41). Taken together, these data suggest that KLF4 may be important for the in vivo differentiation or survival of inflammatory monocytes.
Previous reports have found high levels of KLF4 in HSC-enriched populations (27, 34) and have identified the ability of KLF4 to participate in reprogramming of embryonic pluripotency (21, 42, 43). Because KLF4 regulates p21 expression (Fig. 6C) (38), we initially hypothesized that KLF4 might regulate quiescence of HSCs by inducing p21, as has been demonstrated for deficiency of p21, per se (44). However, we showed herein, that despite high levels of KLF4 expression, HSPC functions were unaffected by loss of KLF4. KLF4−/− cells performed equally to wt cells in competitive HSC repopulation assays, as well as other HSPC assays (Fig. 2). Thus, the apparently dispensable role of KLF4 in stem cell biology remains unclear; it remains possible that some compensatory mechanism is responsible for this lack of phenotype in HSPCs. Furthermore, the ectopic expression of KLF4 has strikingly different effects in HSPCs, monocytes, and fibroblasts.
Current models of monocytic maturation propose that resident monocytes (Ly6C−) develop from inflammatory monocytes (Ly6C+). This is based on the observation that, following ablation of circulating monocytes, inflammatory monocytes are detected first, followed by resident monocytes (20). Thus, we were surprised to find that KLF4−/− chimeras completely lacked inflammatory monocytes, but retained (a reduced number of) resident monocytes. Our results might fit the current model if KLF4−/− inflammatory monocytes are extremely short-lived during their development to resident monocytes. We also found that KLF4−/− monocytic precursors expressed lower levels of CD62L and CCR2, suggesting that they already may be expressing a resident monocytic phenotype. Taken together, these data suggest the possibility of a common precursor that gives rise to both resident and inflammatory monocytes, in which KLF4 is necessary for inflammatory monocyte differentiation.
Monocytic cells play integral roles in innate and adaptive immunity, and recruitment of monocytes to sites of injury or inflammation is crucial in the pathogenesis of inflammatory diseases such as rheumatoid arthritis and atherosclerosis (45, 46). Previous studies have demonstrated that depletion of monocytic cells is beneficial in the prevention of RA and atherosclerosis in mouse models (47, 48). The inflammatory monocyte-selective role of KLF4 makes it an attractive potential target for investigation and treatment of inflammation and inflammatory diseases.
Footnotes
This research was supported in part by a Fellow Award from the National Foundation for Cancer Research and National Institutes of Health Grant CA070970.
Abbreviations used in this paper: KLF, Kruppel-like factor; HSPC, hematopoietic stem-progenitor cell; BM, bone marrow; ES, embryonic stem; HSC, hemopoietic stem cell; HPC, hematopoietic progenitor; FL, fetal liver; wt, wild type; PE, peritoneal exudate; TPA, 12-O-tetradecanoylphorbol 13-acetate; RA, retinoic acid; ER, estrogen receptor; 4HT, 4-hydroxy-tamoxifen; CFC, colony-forming cell; SCF, stem cell factor; M-CSF, monocyte-CSF; PI, propidium iodide; qRT-PCR, quantitative-RT-PCR.
Disclosures
The Johns Hopkins University holds patents on CD34 monoclonal Abs and inventions related to stem cells. C.I.C. is entitled to a share of the sales royalty received by the University under licensing agreements between the Johns Hopkins University, Becton Dickinson Corporation, and Baxter HealthCare Corporation. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- 1.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 2.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 4.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 5.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 6.Perkins AC, Yang H, Crossley M, Fujiwara Y, Orkin SH. Deficiency of the CACCC-element binding protein, BKLF, leads to a progressive myeloproliferateve disease and impaired expression of SHP-1. Blood. 1997;90:575. [Google Scholar]
- 7.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 9.Pilon AM, Nilson DG, Zhou D, Sangerman J, Townes TM, Bodine DM, Gallagher PG. Alterations in expression and chromatin configuration of the alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol Cell Biol. 2006;26:4368–4377. doi: 10.1128/MCB.02216-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu P, Morris PE, Haar JL, Wani MA, Lingrel JB, Gaensler KM, Lloyd JA. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood. 2005;106:2566–2571. doi: 10.1182/blood-2005-02-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, McPherson L, Feng D, Song A, Dong C, Lyu SC, Zhou L, Shi X, Ahn YT, Wang D, Clayberger C, Krensky AM. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178:5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emery DW, Gavriilidis G, Asano H, Stamatoyannopoulos G. The transcription factor KLF11 can induce gamma-globin gene expression in the setting of in vivo adult erythropoiesis. J Cell Biochem. 2007;100:1045–1055. doi: 10.1002/jcb.21093. [DOI] [PubMed] [Google Scholar]
- 13.Mitsuma A, Asano H, Kinoshita T, Murate T, Saito H, Stamatoyannopoulos G, Naoe T. Transcriptional regulation of FKLF-2 (KLF13) gene in erythroid cells. Biochim Biophys Acta. 2005;1727:125–133. doi: 10.1016/j.bbaexp.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 15.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noti JD, Johnson AK, Dillon JD. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem. 2005;280:3449–3457. doi: 10.1074/jbc.M412627200. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 20.Sunderkotter C, Nikolic T, Dillon MJ, Van Rooijen N, Stehling M, Drevets DA, Leenen PJ. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 23.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 24.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, Niwa H. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgantas RW, III, Tanadve V, Malehorn M, Heimfeld S, Chen C, Carr L, Martinez-Murillo F, Riggins G, Kowalski J, Civin CI. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 28.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Zhan X, D’Costa J, Tanavde VM, Ye Z, Peng T, Malehorn MT, Yang X, Civin CI, Cheng L. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Alder JK, Chun JH, Friedman AD, Heimfeld S, Cheng L, Civin CI. HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells. 2006;24:876–888. doi: 10.1634/stemcells.2005-0598. [DOI] [PubMed] [Google Scholar]
- 32.Cao W, Britos-Bray M, Claxton DF, Kelley CA, Speck NA, Liu PP, Friedman AD. CBF β-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 33.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 34.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 35.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coghill E, Eccleston S, Fox V, Cerruti L, Brown C, Cunningham J, Jane S, Perkins A. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood. 2001;97:1861–1868. doi: 10.1182/blood.v97.6.1861. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26:2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bruijn MF, Slieker WA, van der Loo JC, Voerman JS, van Ewijk W, Leenen PJ. Distinct mouse bone marrow macrophage precursors identified by differential expression of ER-MP12 and ER-MP20 antigens. Eur J Immunol. 1994;24:2279–2284. doi: 10.1002/eji.1830241003. [DOI] [PubMed] [Google Scholar]
- 42.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 43.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:260–262. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 44.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 45.Szekanecz Z, Koch AE. Macrophages and their products in rheumatoid arthritis. Curr Opin Rheumatol. 2007;19:289–295. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- 46.Bobryshev YV. Monocyte recruitment and foam cell formation in atherosclerosis. Micron. 2006;37:208–222. doi: 10.1016/j.micron.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Solomon S, Rajasekaran N, Jeisy-Walder E, Snapper SB, Illges H. A crucial role for macrophages in the pathology of K/B × N serum-induced arthritis. Eur J Immunol. 2005;35:3064–3073. doi: 10.1002/eji.200526167. [DOI] [PubMed] [Google Scholar]
- 48.Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein E. Proc Natl Acad Sci USA. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]