Abstract
The targets of broadly cross-neutralizing (BCN) antibodies are of great interest in the HIV vaccine field. We have identified a subtype C HIV-1-superinfected individual, CAP256, with high-level BCN activity, and characterized the antibody specificity mediating breadth. CAP256 developed potent BCN activity peaking at 3 years postinfection, neutralizing 32 (76%) of 42 heterologous viruses, with titers of antibodies against some viruses exceeding 1:10,000. CAP256 showed a subtype bias, preferentially neutralizing subtype C and A viruses over subtype B viruses. CAP256 BCN serum targeted a quaternary epitope which included the V1V2 region. Further mapping identified residues F159, N160, L165, R166, D167, K169, and K171 (forming the FN/LRD-K-K motif) in the V2 region as crucial to the CAP256 epitope. However, the fine specificity of the BCN response varied over time and, while consistently dependent on R166 and K169, became gradually less dependent on D167 and K171, possibly contributing to the incremental increase in breadth over 4 years. The presence of an intact FN/LRD-K-K motif in heterologous viruses was associated with sensitivity, although the length of the adjacent V1 loop modulated the degree of sensitivity, with a shorter V1 region significantly associated with higher titers. Repair of the FN/LRD-K-K motif in resistant heterologous viruses conferred sensitivity, with titers sometimes exceeding 1:10,000. Comparison of the CAP256 epitope with that of the PG9/PG16 monoclonal antibodies suggested that these epitopes overlapped, adding to the mounting evidence that this may represent a common neutralization target that should be further investigated as a potential vaccine candidate.
The development of an HIV-1 vaccine is widely assumed to require both a cellular and a neutralizing antibody (nAb) component. Despite substantial efforts to produce a vaccine immunogen capable of inducing broadly nAb responses, little success has been achieved (16, 18, 29). The inability to design an appropriate antigen is hindered by the enormous antigenic variability, as well as a myriad of masking mechanisms, exhibited by the envelope glycoprotein (5, 19). The identification of new nAb targets on the HIV-1 envelope, as well as unraveling how antibodies to these targets develop in natural infection, is therefore a high priority for HIV vaccinology.
The majority of individuals mount a potent nAb response within the first few months of HIV-1 infection that largely targets variable regions of the envelope glycoprotein and is very type specific (4, 14, 20, 33, 45). However, only a subset of infected people (20 to 30%) develop broadly cross-neutralizing (BCN) antibodies effective against diverse heterologous viruses (23, 35, 38, 39). The concerted efforts by numerous groups to understand the polyclonal specificities of these BCN plasmas have defined the contributions of antibodies against known targets such as the CD4 binding site, CD4-induced epitopes, and the membrane-proximal region (MPER), as well as unknown targets on gp120 and gp41 (6, 11, 23, 35, 38, 39). Identification of the targets of such BCN antibodies is crucial, as these epitopes likely represent novel vaccine targets that may be more amenable to vaccine design than current targets, many of which have been defined on the basis of the specificities of a few well-characterized monoclonal antibodies (MAbs) (e.g., 4E10, 2F5, IgG1b12, and 2G12) (5).
The existence of quaternary neutralizing epitopes (QNEs), those that are present only within the context of a native trimeric envelope “spike,” was first demonstrated by the isolation of MAb 2909, which binds an epitope encompassing parts of the V2 and V3 loops but does not bind monomeric gp120 (10, 17). Similar neutralizing MAbs from rhesus macaques infected with simian/human immunodeficiency virus SF162P4 (2.2G, 2.3E, and 2.5B) target the same overall QNE, though each possesses distinct amino acid and glycan requirements (34). Although these antibodies are very potent, they are also extremely type specific, neutralizing only SF162. However, the isolation of MAbs PG9 and PG16, somatic variants isolated from an HIV-1 subtype A-infected individual, which recognize QNEs and show substantial breadth and potency across subtypes, confirmed the notion that BCN antibodies against these types of epitopes exist and possibly define a valuable new vaccine target (43).
The epitope recognized by PG9/PG16 is dependent on the N160 glycosylation motif in V2, as well as other residues in the stem of V1/V2, V2, and V3 (with PG16 showing much greater dependence on residues in V3 than PG9) (43). PG9/PG16-like antibodies are increasingly being defined as those that are not generally adsorbed by gp120 and are sensitive to mutations at N160 and to treatment with kifunensine (a mannose analogue that inhibits α-mannosidases in the type 1 endoplasmic reticulum and the Golgi apparatus) (7, 44). The binding of such antibodies to a region largely described as highly variable confirms the fact that within V2 there are regions and residues that are highly conserved, even between different subtypes (48). Crystallographic and structure-function analyses of PG16 identified extensive affinity maturation and an unusually long complementarity-determining region (CDR) H3 (28 residues) comprising nearly half of the CDR surface and forming a stable, axe-shaped subdomain that protrudes from the antibody as critical to its broad neutralization capacity (27, 28). This raises the questions of whether such antibodies would be hard to elicit in natural infection and whether they would require several years of antigen exposure to develop (27, 28).
The prevalence of BCN antibodies targeting QNEs is unknown. The fact that several studies mapping polyclonal sera (using reagents that are often based on monomeric gp120 and peptides) have been unable to account for the total neutralizing capacity of BCN plasma suggests that many BCN antibodies recognizing QNEs may have gone undetected. Binley et al. (1), using a subset of 8 of 24 subtype B and C sera, showed that anti-gp120 antibodies accounted for a significant but variable fraction of neutralization, depending on the isolate against which adsorbed serum was tested. Similarly Walker et al. (44) reported the possibility of BCN antibodies against QNEs (on the basis of a lack of adsorption using monomeric gp120) in 8 of 18 individuals with BCN activity, with some of these nAbs better adsorbed by a YU-2 gp140-foldon trimer. In the Centre for the AIDS Programme of Research in South Africa (CAPRISA) Acute Infection cohort of subtype C-infected individuals, 4/7 BCN individuals from a total of 40 subjects appear to have antibodies recognizing QNEs (data not shown). These data suggest that these types of antibodies may occur quite frequently.
Here we describe the development of V2-dependent QNE antibodies in CAP256, a subtype C-infected (and later superinfected) individual in the CAPRISA Acute Infection cohort (42) who developed unusually high-titer nAbs directed primarily at subtype C and A viruses. We have mapped the precise residues crucial for the formation of this V2-dependent quaternary BCN epitope and the variation in the fine specificity of the BCN response that occurred over 4 years of infection. These data point to this region of V2 as being sufficiently conserved and immunogenic to elicit BCN antibodies.
MATERIALS AND METHODS
CAPRISA participant CAP256.
CAP256 was enrolled in the CAPRISA Acute Infection study (42) that was established in 2004 in Durban and Vulindlela, South Africa. The CAPRISA Acute Infection study was reviewed and approved by the research ethics committees of the University of KwaZulu-Natal (E013/04), the University of Cape Town (025/2004), and the University of the Witwatersrand (MM040202). All participants provided written informed consent for study participation.
Cell lines.
The JC53bl-13 cell line, engineered by J. Kappes and X. Wu, was obtained from the NIH AIDS Research and Reference Reagent Program. 293T cells were obtained from George Shaw (University of Alabama, Birmingham). Both cell lines were cultured in Dulbecco's modified Eagle medium (Gibco BRL Life Technologies) containing 10% heat-inactivated fetal bovine serum and 50 μg/ml gentamicin (Sigma). Cell monolayers were disrupted at confluence by treatment with 0.25% trypsin in 1 mM EDTA.
Neutralization assays.
Env-pseudotyped viruses were obtained by cotransfecting the Env plasmid with pSG3ΔEnv (45) using Fugene transfection reagent (Roche) as previously described (14). Neutralization was measured as described previously (14) by a reduction in luciferase gene expression after single-round infection of JC53bl-13 cells with Env-pseudotyped viruses. Titers were calculated as the reciprocal plasma dilutions causing a 50% reduction in the number of relative light units (ID50).
rgp120 production and purification.
The consensus C (ConC) plasmid pConCgp120-opt (catalog no. 11405), obtained through the AIDS Research and Reference Reagent Program from Beatrice Hahn, was transfected into 293T cells using TransIT-LT1 (Mirus, Pittsburgh, PA). The supernatant was collected after 4 days for purification of recombinant gp120 (rgp120). gp120 was isolated using Galanthus nivalis lectin agarose matrix (Sigma-Aldrich, St. Louis, MO) and eluted with 0.5 M methyl-α-d-mannopyranoside (Sigma). The remaining supernatant was passed over the agarose matrix a further three times before ConC rgp120 was concentrated using an Amicon Centricon filter with a molecular weight cutoff of 30,000. The purified rgp20 was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Serum adsorption of anti-gp120 antibodies from CAP256 plasma.
Purified rgp120 was coupled to cyanogen bromide-activated Sepharose 4B (Pharmacia Biotech, Uppsala, Sweden) overnight at 4°C before washing away of unbound protein with phosphate-buffered saline (PBS) containing 0.5 M NaCl. Beads were blocked with 1 M ethanolamine (pH 8.0) for 2 h and washed three times, alternating between 0.1 M CH3COONa (pH 4.0)-0.5 M NaCl and 0.1 M Tris (pH 8.0)-0.5 M NaCl, before a final wash with PBS. Coated beads were used to adsorb anti-gp120 antibodies as follows. CAP256 plasma was diluted 1:10 in PBS, and 100 μl was added to the coated beads and incubated overnight at 4°C. Adsorbed plasma was removed and stored. Beads were washed twice with 1 ml PBS, which was also collected and combined with the flowthrough. Fetal bovine serum (400 μl) was added before concentration of the adsorbed plasma back down to 500 μl using an Amicon Ultra 4 filter unit (molecular weight cutoff, 30,000). Anti-gp120 binding capacity was measured by enzyme-linked immunosorbent assay (ELISA) as described below. Neutralization assays using adsorbed material were performed in the Pinter lab with U87 cells as described previously (17).
gp120 binding ELISA.
ConC rgp120 (100 ng/well), JRFL rgp120 (50 ng/well), and IIIB lysate (200 ng/well; ENI Diagnostic, Inc., Silver Spring, MD) was used to coat 96-well ELISA plates in sodium bicarbonate buffer, pH 9.8, overnight at 4°C. After blocking with 2% Blotto in PBS (pH 7.4) at 37°C for 30 min, serial dilutions of adsorbed CAP256 plasma (and unadsorbed plasma) were added to the plate. After incubation for 60 min at 37°C, plates were washed and bound antibodies were detected using a 1:2,000 dilution of alkaline-phosphate conjugated anti-human IgG (Zymax), followed by addition of substrate (1 mg/ml p-nitrophenol in diethanolamine buffer [pH 9.8]). Absorbance (405 nm) measurements were taken after 30 min using a Spectra SLT ELISA plate reader (TECAN Instruments, Research Triangle Park, NC).
Generation of chimeras and mutant envelopes.
Chimeric V1V2 Envs were created using an overlapping PCR strategy with the inserts and flanking regions amplified in separate reactions. After linkage, the 3-kb chimeric PCR fragments generated using primers EnvAdir and EnvM (9) were cloned into pCDNA 3.1 (directional; Invitrogen) and screened for function as previously described (12). In some cases, small regions of V2 were exchanged by the insertion of synthetic oligonucleotides at unique restriction sites engineered into the V2 region. Chimerism was confirmed by sequence analysis. Site-directed mutagenesis was performed using the Stratagene QuikChange II kit (Stratagene).
RESULTS
Development of high-titer cross-nAbs in CAP256 that preferentially neutralize subtype C viruses.
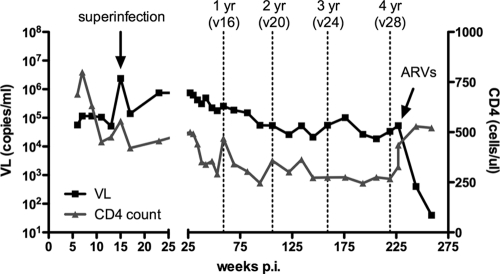
CAP256, a participant in the CAPRISA Acute Infection cohort, was infected with an HIV-1 subtype C virus in July 2005 and subsequently superinfected 13 to 15 weeks later with an unrelated second subtype C virus (data not shown). She had a median viral load of 105,000 RNA copies/ml over 4.5 years of infection, peaking at 2,390,000 RNA copies/ml at the time of superinfection, with steadily declining CD4 counts (Fig. 1). In November 2009, she was diagnosed with and treated for extrapulmonary tuberculosis and started on antiretroviral (ARV) therapy in January 2010, 4 years 7 months postinfection (p.i.). She was one of 7 women from this cohort of 40 who at 3 years of infection showed a high level of cross-neutralizing antibody activity (data not shown).
FIG. 1.
CAP256 viral loads and CD4+ T cell counts over 4 years of infection. Key time points for neutralization assays at 1, 2, 3, and 4 years p.i. are indicated with dotted lines. The peak viral load (VL) at the point of superinfection and the time point at which CAP256 was started on ARV therapy are shown by arrows. v16, visit 16, etc.
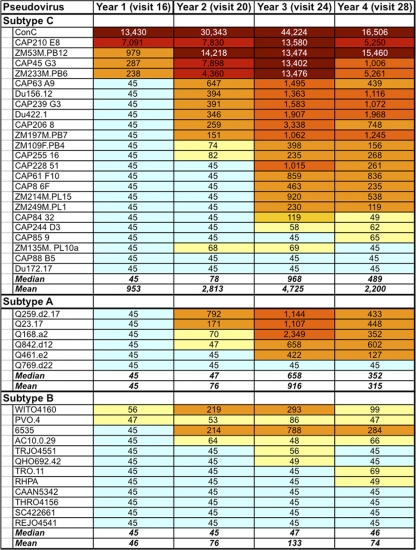
The development of neutralization breadth in CAP256 was assessed by testing serum/plasma at 1 (visit 16), 2 (visit 20), 3 (visit 24), and 4 (visit 28) years p.i. (prior to her initiation on ARV therapy) against a panel of 42 heterologous pseudoviruses (Fig. 2). This included pseudoviruses from the tier 2 subtype A, B, and C panels (21, 22) and pseudoviruses derived from the CAPRISA Acute Infection cohort (14). CAP256 developed substantial neutralization breadth over time, with cross-nAbs increasing incrementally for the first 3 years. Thus, heterologous neutralization was detectable as early as 1 year but was limited to 7/42 (17%) viruses, increasing to 22/42 (52%) viruses at 2 years, and peaking at 3 years with activity against 32/42 (76%) viruses. Although nAb titers overall declined slightly at 4 years of infection, the percentage of viruses neutralized remained static (32/42, 76%). CAP256 showed a distinct subtype bias, preferentially neutralizing subtype C and A viruses (P = 0.0011 and P = 0.0268, respectively) over subtype B viruses. At the peak of BCN activity, measured at 3 years p.i., 88% (21/24) of subtype C viruses were neutralized at a median ID50 titer of 1:968. Unusually high titers against some subtype C viruses were observed, with titers against ConC exceeding 1:10,000 and even >1:44,000. Subtype A viruses were also frequently neutralized, with 5/6 viruses (83%) neutralized at a median titer of 1:658 at 3 years p.i. Titers against subtype A viruses, however, did not reach the levels observed against many subtype C viruses, with a measured maximum titer of antibody against Q168 of 1:2,349. In stark contrast to both subtypes A and C, subtype B viruses were neutralized far less frequently (only 6/12 viruses at 3 years p.i.), and in many cases, the nAb titers were marginal, with a median titer of only 1:47 and a maximum of 1:788 against 6535, which is a tier 1B virus that is relatively neutralization sensitive (37). Therefore, CAP256 developed potent cross-nAbs that preferentially neutralized viruses of subtypes A and C, with activity accumulating gradually and peaking at 3 years of infection.
FIG. 2.
Development of cross-nAbs against heterologous viruses of subtypes A, B, and C in CAP256 over 1 to 4 years of infection. Titers shown are ID50.
CAP256 BCN antibodies were not adsorbed by gp120.
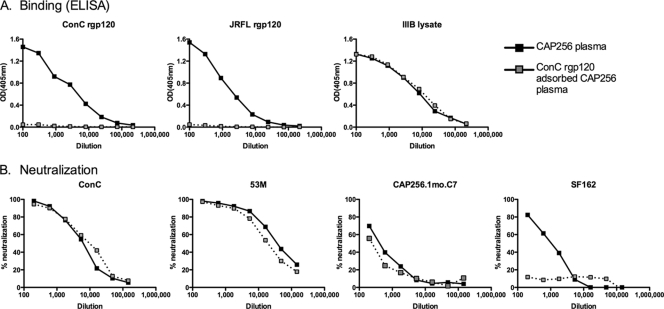
To assess whether the epitopes recognized by CAP256 plasma antibodies at 3.25 years (visit 25) were present on the monomeric gp120 structure, we used rgp120 from the ConC envelope, which was highly sensitive to these antibodies. Adsorption of CAP256 plasma using gp120 bound to Sepharose beads resulted in a significant decrease in the levels of gp120 binding antibody in the flowthrough as measured by ELISA against ConC gp120 and JR-FL gp120 (Fig. 3 A). Binding of adsorbed plasma to the IIIB viral lysate, consisting mostly of viral core proteins, was unaffected, as expected. However, gp120 depletion had no effect on the neutralization activity of this plasma sample against heterologous viruses ConC and ZM53M or the CAP256 autologous enrolment virus (CAP256.1mo.C7) (Fig. 3B). ConC gp120-adsorbed plasma did, however, show reduced neutralization activity against SF162, reflecting the adsorption of antibodies that are able to neutralize this tier 1 virus (Fig. 3B). These data suggested that the BCN antibodies in CAP256 recognized quaternary epitopes dependent on the trimeric form of the envelope glycoprotein, which could not be depleted with gp120.
FIG. 3.
Effect of rgp120 adsorption on CAP256 neutralization. (A) Binding of CAP256 plasma and rgp120-adsorbed plasma on binding to ConC rgp120, JRFL rgp120, and IIIB viral lysate measured by ELISA (optical density [OD] at 405 nm). (B) Neutralization by CAP256 plasma and ConC rgp120-adsorbed CAP256 plasma of ConC, ZM53M, autologous virus CAP256.1mo.C7, and SF162.
The QNE in CAP256 involves the V1V2 region.
We have previously described the construction of variable-loop chimeric viruses used to map the specificities of antibodies mediating autologous neutralization (25, 26). Here we have made use of chimeras consisting of CAP45 (neutralized by CAP256 at high titer) and CAP84 (not neutralized at 2 years p.i., Fig. 2) to map heterologous neutralization. Chimeras that contained the V1V2, C3V4, V4, and V5 regions of CAP45 in the CAP84-resistant backbone were used to localize the region targeted by the QNE antibody in CAP256. Figure 4 shows that plasma from CAP256 at 2 years p.i. (visit 20) neutralized CAP45 at an ID50 of 1:7,898 but failed to neutralize CAP84 (ID50 of less than 1:45). Transfer of the CAP45 V1V2 region into CAP84 to produce the 84-45-84 V1V2 region chimera resulted in neutralization sensitivity, reflected in a titer of 1:5,075 (Fig. 4). In contrast, all of the other chimeras (C3V4, V4, and V5) were completely resistant to neutralization (titers of <1:45). This suggested that heterologous neutralization of the CAP45 pseudovirus was mediated by V1V2 region-dependent antibodies present in CAP256 serum.
FIG. 4.
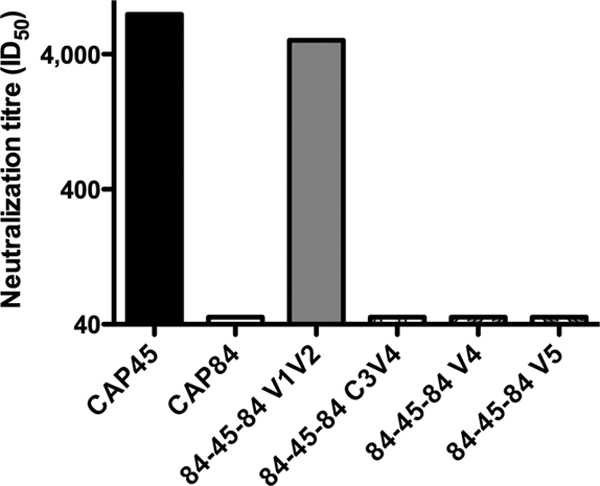
CAP256 plasma neutralizes CAP45 via a V1V2 region-dependent antibody. Neutralization of chimeric viruses constructed of CAP45 (sensitive) and CAP84 (resistant) is shown. Only the chimera containing the V1V2 region of CAP45 was sensitive to neutralization.
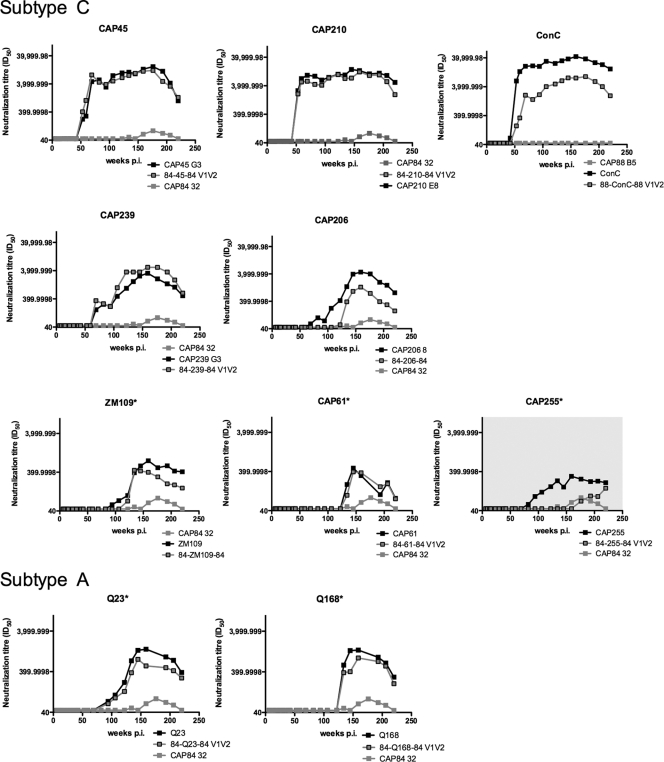
To assess whether CAP256 cross-neutralization of other viruses was also mediated by antibodies dependent on the V1V2 region and whether this remained the case over the duration of infection, we constructed nine additional V1V2 region chimeras, transferring the V1V2 region from sensitive heterologous viruses into a resistant backbone (either CAP88 or CAP84). In addition to CAP45 shown above, we included two subtype C viruses neutralized at extremely high titers by 1 year p.i. (CAP210 and ConC), as well as five subtype C viruses neutralized at much lower titers at 2 to 3 years p.i. (CAP239, CAP206, ZM109, CAP61, and CAP255) (Fig. 2). In order to assess whether subtype A viruses were also neutralized via the V1V2 region, we constructed chimeras using two subtype A viruses that were neutralized by plasma from 2 years p.i. Although we constructed chimeras for sensitive subtype B viruses (6535 and WITO), they were nonfunctional and could not be used in neutralization assays.
All chimeras were tested along with both parental viruses against longitudinal plasma spanning 4 years of infection. In 9 of 10 heterologous viruses (including CAP45), antibodies dependent on the V1V2 region were shown to mediate neutralization over the entire time span. This was reflected by acquisition of sensitivity of the resistant backbone (CAP84 or CAP88) into which the V1V2 region of each heterologous virus was introduced (Fig. 5). This confirmed that for the majority of the heterologous viruses, neutralization was mediated by cross-reactive V1V2 region-dependent antibodies in CAP256 plasma.
FIG. 5.
V1V2 region-dependent antibodies mediate cross-neutralization of subtype C and A viruses. Chimeric viruses containing the V1V2 region of sensitive subtype C or A viruses were tested against longitudinal plasma samples spanning 4 years of infection. Sensitive parental viruses are shown by black lines, gray lines denote resistant (backbone) viruses, and gray squares with black outlines are V1V2 region chimeras. Asterisks indicate graphs with differing scales to illustrate lower titers.
For one virus, CAP255, transfer of the V1V2 region into an insensitive backbone failed to transfer neutralization sensitivity (Fig. 5, gray shading). This suggested that CAP255 was neutralized not by anti-V1V2 region antibodies in CAP256 plasma but rather by different specificities targeting a region outside the V1V2 loops. The neutralization of CAP255 by CAP256 serum was of low titer even at 3 years of infection, similar to the profile of many of the subtype B viruses (Fig. 2). Interestingly, gp120 adsorption of CAP256 serum did not affect the neutralization of CAP255 (data not shown), suggesting that the antibody specificity mediating neutralization of this virus may also target a quaternary epitope within the trimeric form of the envelope.
The kinetics of the neutralization curves suggested that cross-neutralization was mediated in three waves, with the first appearing at 1 year p.i. and targeting the V1V2 region of CAP45, CAP210, and ConC (Fig. 2 and 5) and the second wave targeting the V1V2 region of subtype C viruses CAP239 and CAP206 and subtype A virus Q23. A third low-titer anti-V1V2 region response against ZM109 and CAP61, as well as subtype A virus Q168, emerged some months later. V1V2 region-dependent antibodies, as measured by the titers of antibodies against chimeric constructs, generally accounted for close to 100% of the parental neutralizing activity, suggesting that heterologous neutralization was restricted to antibodies recognizing this region. The consistent involvement of the V1V2 region in three waves of cross-neutralizing activity suggested the possibility that either multiple anti-V1V2 region antibodies developed in CAP256 over time or affinity maturation of a single nAb resulted in an expanded cross-neutralizing antibody response.
Mapping of the precise residues in the V1V2 region targeted by CAP256 antibodies.
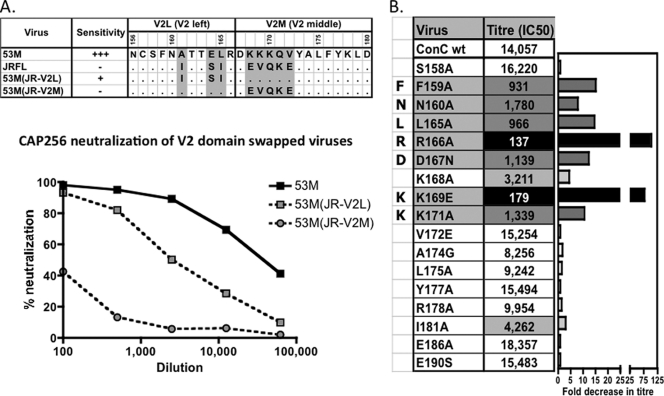
Since the V1 loop is considerably more variable that the V2 loop (48), we hypothesized that the BCN antibodies in CAP256 were most likely directed at V2. We therefore made use of subdomain V2 loop chimeras (V2-left or V2-middle) consisting of a resistant virus (JR-FL) and a highly sensitive virus (ZM53M) (Fig. 6 A). When the V2-L region from JR-FL (N156 to R166, spanning the N160 glycan site) was transferred into the ZM53M virus, there was a decrease in neutralization sensitivity. When the V2-M region (D167 to D180) was transferred, the chimera became almost completely resistant (Fig. 6A). This finding demonstrated that determinants of CAP256 sensitivity were located in both V2-L and V2-M, with residues 168 to 172 in V2-M making major contributions to the epitope.
FIG. 6.
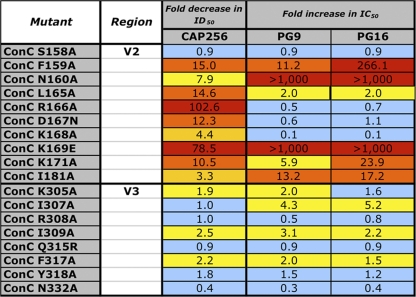
Identification of the FN/LRD-K-K motif in V2 as a major target of CAP256 plasma neutralizing activity. (A) V2-L and V2-M domain swap chimeras consisting of the ZM53M (sensitive) and JRFL (resistant) viruses. Shown at the top are residues in the sequences of the V2L and V2M regions of ZM53M and JRFL and the chimeras. The graph at the bottom shows the neutralization of the ZM53M and chimeric viruses containing the V2L or V2M domain from the JRFL virus, confirming the importance of both regions, but especially the V2M region, in forming the CAP256 epitope. (B) Site-directed mutagenesis of residues in V2 was used to map critical residues in the ConC background. Black highlighting denotes >75-fold titer reductions, dark gray denotes mutations resulting in >8-fold titer reductions, and light gray denotes >3-fold titer reductions. The FN/LRD-K-K motif represents residues affecting the CAP256 titers by more than 8-fold. Two other residues (168 and 181, not included in the motif) had a lesser effect. wt, wild type.
The precise residues involved in CAP256 neutralization were mapped more precisely by alanine scanning of the V2 region in ConC and ZM53M pseudoviruses. Plasma from visit 18 (82 weeks p.i.) was tested against ConC V1V2 region point mutants (selected data are shown in Fig. 6B), and plasma from visit 19 (94 weeks p.i.) was tested against ZM53M mutants (data not shown). In the ConC backbone, the most dramatic abrogation of CAP256 plasma neutralization was achieved through mutations at positions 166 and 169. For R166A, there was a >100-fold (>98%) titer reduction, from 1:14,057 to 1:137, while for K169E, there was a 79-fold titer reduction to 1:179 (Fig. 6B). In addition, there was an 8-fold (>90%) or greater reduction in the titers of antibodies against the ConC F159A (to 1:931), N160A (1:1780), L165A (1:966), D167N (1:1139), and K171A (1:1339) mutants. Overall, this antibody (or antibodies) appears to be dependent on F159, N160, L165, R166, D167, K169, and K171 (which is from here on referred to as the FN/LRD-K-K motif). Similar results were obtained for the ZM53M mutants, although in this backbone the dependence on N160 was not observed, and in fact there was a slight increase in titer (1:23,000) compared to that of wild-type ZM53M (1:14,800) (data not shown).
The discrepancy between the chimeras (Fig. 6A), which suggested that the V2-L region contributed only slightly to the CAP256 epitope, and the alanine scanning data (Fig. 6B), which showed the importance of, among others, the R166 residue (in the V2-L region), is likely due to the fact that both JR-FL and ZM53M contain an arginine residue at position 166. Therefore, swapping the V2-L region does not result in a dramatic change in titer in this case.
Changes in the fine specificity of the BCN anti-V2 antibody over time.
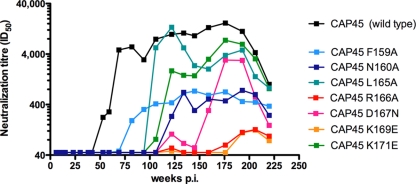
The mapping of the FN/LRD-K-K motif shown in Fig. 6 was performed using a single time point (visit 18, 82 weeks p.i.). Given that titers measured against CAP45 showed three distinct peaks at 82 weeks (titer of 1:5,555), 134 weeks (1:10,414), and 176 weeks (16,562) p.i. but remained exclusively targeted at the V1V2 region, with the titers against the chimera consistently mirroring those of the parental virus (Fig. 5), we queried whether there were changes in the fine specificity of this BCN antibody (antibodies) over time. Longitudinal plasmas from enrolment up to 4 years of infection were tested against CAP45 viruses containing mutations at all of the residues that make up the FN/LRD-K-K motif (F159A, N160A, L165A, R166A, D167N, K169E, and K171E). From the point at which breadth developed (approximately 1 year p.i.) through to 4 years p.i., the anti-V2 QNE antibody was consistently highly dependent on the R166 and K169 residues, with titers dropping more than 40-fold, from greater than 1:4,000 to between <1:45 (no neutralization) and 1:100 (Fig. 7).
FIG. 7.
Changes in the fine specificity of the V2-dependent nAb present in CAP256 plasma. Longitudinal assessment of the neutralization sensitivity of CAP45 (parent virus) compared to that of viruses with the F159A, N160A, L165A, R166A, D167N, K169E, and K171E mutations to CAP256 plasma from 1 to 4 years p.i.
In contrast, the antibody was variably dependent on the other residues over time. The L165A mutation abrogated neutralization during the early stages of infection, with the peak at 82 weeks reduced from a titer of 1:5,555 to <1:45. However, from 100 weeks onward, the dependence of the sera on the L165 residue was reduced, with titers rising to become similar to those of antibodies against the parental virus and then declining slightly by the third peak (to a titer of 1:3,771 at 176 weeks p.i.). Sensitivity to the K171E mutation was also limited to the earlier time points (<100 weeks)—this mutation entirely abrogated neutralization during the first peak but had a less dramatic effect on the second peak of neutralization (with a titer of 1:1,500 at 134 weeks p.i.) and even less of an effect on later neutralization (titer of 1:7,529 at 193 weeks p.i.). The D167N mutation prevented neutralization at both of the earlier peaks, but by the third peak at 176 weeks p.i., titers rose to 1:3,000, reflecting decreased dependence on the D167 residue. The N160A mutation had a significant effect on neutralization throughout the 4-year period, with neutralization abrogated during the first 2 years and thereafter rising slightly to titers of approximately 1:500, suggesting consistent involvement of this residue in the formation of the epitope. The effect of the F159A mutation on neutralization was also relatively consistent over time. Overall, the differing effect of each mutation reinforced the central role of the R166 and K169 residues in this epitope but also indicated that there were changes over time in the fine specificity of the anti-V2 QNE antibody.
The changes in the fine specificity of the CAP256 serum over time described above may explain the incremental increase in breadth (Fig. 2), with fewer heterologous viruses neutralized at year 1 and more neutralized at later time points. In particular, four of the six subtype C viruses that were neutralization resistant at year 2 but had titers of >1:100 at year 3 had polymorphisms at K171 (K171N/R/Q). The reduced dependence of the later sera on K171 may explain their increase in neutralization sensitivity.
The FN/LRD-K-K motif was associated with but not sufficient for high-titer neutralization in subtype C.
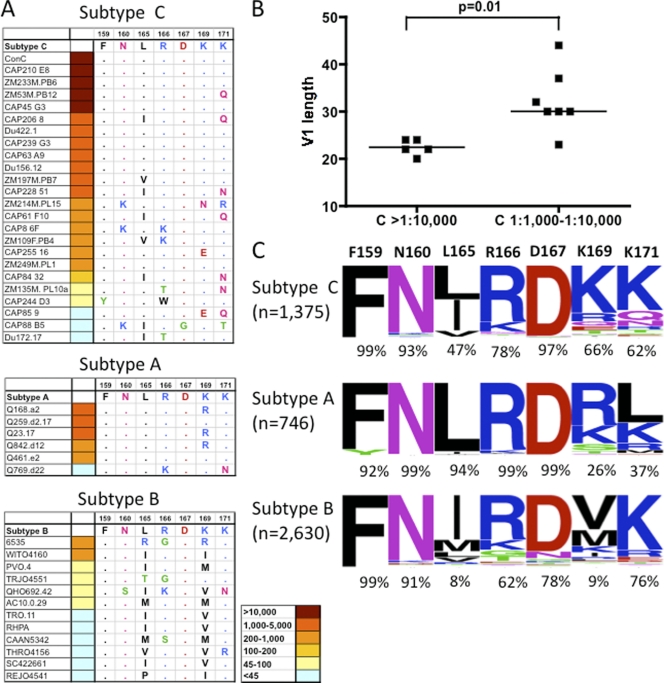
We analyzed the sequences of the heterologous viruses described in Fig. 2, focusing specifically on the FN/LRD-K-K motif to define the relationship, if any, between this motif and the neutralization titer. Among the subtype C viruses, ConC, CAP210, ZM53M, CAP45, and ZM233 all became sensitive to neutralization by 1 year and were all characterized by extreme sensitivity to CAP256 serum, with titers exceeding 1:13,000 at year 3. Analysis of the sequences showed that all but one of these viruses had an intact FN/LRD-K-K motif (Fig. 8 A). The exception, ZM53M, had an FN/LRD-K-Q motif, suggesting that 171Q does not compromise the epitope in this virus.
FIG. 8.
The FN/LRD-K-K motif and neutralization sensitivity of subtype C, A, and B viruses. (A) Analysis of the FN/LRD-K-K motif in subtype C, A, and B viruses (ranked by neutralization sensitivity at 3 years p.i.). Amino acids in blue are positively charged, and those in red are negatively charged. A point denotes conservation compared to the FNLDRKK motif. (B) Comparison of the V1 loop lengths of subtype C viruses neutralized at titers exceeding 1:10,000 at 3 years p.i. compared to those neutralized at 1:1,000 to 1:10,000 at the same time point. (C) Analysis of the frequencies of each residue within the FN/LRD-K-K motif, comparing envelope sequences of subtype C (n = 1,375), subtype A (n = 746), and subtype B (n = 2,630). Sequences were downloaded from the Los Alamos sequence database.
It was, however, also clear that an FN/LRD-K-K motif was not sufficient to confer extreme sensitivity to the CAP256 plasma, as four of the seven viruses that became sensitive by year 2, with moderate peak titers of 1:1,000 to 1:3,500, also contained a complete FN/LRD-K-K motif (Du422, CAP239, CAP63, and Du156, Fig. 2 and 8A). The remaining three viruses in this category had an L165I, an L165V, and/or a K171N/Q mutation, suggesting that these changes may contribute to the moderate neutralization sensitivity observed. Thus, the FN/LRD-K-K motif was necessary but not sufficient for high-level neutralization. In contrast, of the six subtype C viruses in Fig. 2 which were either resistant or only neutralized poorly (peak titers of less than 1:200), all contained a minimum of two mutations within the FN/LRD-K-K motif, with CAP88 containing polymorphisms at four of the seven residues in this motif.
There was a very striking distinction between the observed antibody titers against the five most sensitive subtype C viruses (with peak titers always above 1:13,000) and the next tier of sensitive subtype C viruses, none of which was neutralized at titers of >1:3,500. We hypothesized that although the essential residues that make up the epitope or acceptable substitutions (e.g., at L165 and K171) might be required for sensitivity, exposure of the epitope may modulate the titers at which these viruses were neutralized. In support of this hypothesis, we observed a statistically significant difference in the length of the V1 loop when comparing extremely sensitive viruses (ID50, >1:10,000; median V1 loop length of 22 amino acids) with moderately sensitive viruses (ID50, 1:1,000 to 1:10,000; median V1 loop length of 30 amino acids) (Fig. 8B, P = 0.01). This suggested that shorter V1 loops exposed the epitope in V2 to a greater extent or affected its conformation, resulting in increased neutralization titers.
The role of the FN/LRD-K-K motif in subtype A and B viruses.
Similarly, among the five sensitive subtype A viruses, two (Q259 and Q461) contained the intact FN/LRD-K-K motif while the remaining three had a single conservative change which maintained the positive charge at this residue (K169R). The single resistant subtype A virus contained two deviations from the FN/LRD-K-K motif, though interestingly, both of these changes (R166K, a conservative change between positively charged residues, and K171N) were present individually in the sensitive (but low-titer) subtype C viruses described above (Fig. 8A). All of the 12 subtype B viruses, none of which were neutralized at titers exceeding 1:1,000, had deviations away from the FN/LRD-K-K motif, primarily but not exclusively at the L165 and K169 residues, the latter being especially crucial in the ConC mapping described above.
While this variation within the FN/LRD-K-K motif may contribute to the observed subtype bias in CAP256 neutralization, the overall reduced titers even in subtype A viruses that contained the complete motif suggested that other factors influenced the sensitivity of these viruses to this plasma. One possible correlate is suggested by the result obtained for subtype C viruses, where viruses with longer V1 loops had lower titers than those with shorter loops. The increased length of the V1 loops of sensitive subtype A viruses (median V1 loop length of 26, P = 0.03), compared with those of the extremely sensitive (titers >1:10,000) subtype C viruses (median V1 loop length of 22 amino acids), might contribute to the reduced titers observed in all subtype A viruses. This association was, however, not significant (P = 0.13) when all of the sensitive subtype C viruses (regardless of titer) were compared with the sensitive subtype A viruses.
How common is the FN/LRD-K-K motif among viruses of subtypes A, B, and C?
We performed an analysis of the frequency of the residues comprising the FN/LRD-K-K motif within the V2 region from 746 subtype A, 2,630 subtype B, and 1,375 subtype C envelope sequences downloaded from the Los Alamos National Laboratory data bank (http://www.hiv.lanl.gov). Frequencies of the residues at these positions were plotted using http://weblogo.berkeley.edu (Fig. 8C). This analysis highlights the conserved nature of the F159 and N160 residues across all three subtypes (>91% across subtypes A, B, and C). The L165 residue was very conserved in subtype A viruses (94%) but substantially less conserved in subtype C viruses (47%) and rare in subtype B viruses (8%), with I165 also well represented in the latter two subtypes. However, the occasional presence of I165 in sensitive subtype C viruses (Fig. 8A) suggested that this residue may not have a great impact on neutralization sensitivity. The R166 residue was present in 99% of the subtype A viruses, compared to 78% and 62% of the subtype C and B sequences, respectively. Interestingly, in the sole resistant subtype A virus in our panel (Q769), there was a highly unusual K166 polymorphism. The D167 residue was well conserved across the three subtypes but particularly so in subtypes C and A (97% and 99%, respectively). The residue which varied the most between the subtypes was K169, which was present in two-thirds of the subtype C viruses but only a quarter of the subtype A viruses and less than 10% of the subtype B viruses. Though the K169 residue is relatively rare in subtype A viruses, the most common residue in this subtype, R169, was a conserved change maintaining a positive charge. In contrast, residue 169 in subtype B viruses was rarely positively charged. The extreme dependence of neutralization on residue 169 (as shown by the ConC mapping, Fig. 6B) is likely an important factor in the subtype preference observed in neutralization across subtypes A, B, and C. The K171 residue also varied considerably between subtypes (62% of subtype C viruses, compared with only 37% of subtype A viruses), but as polymorphisms at this residue occur in neutralization-sensitive viruses (e.g., Q171 is present in the highly sensitive subtype C virus ZM53M [neutralized at a peak titer exceeding 1:10,000]) and is a fairly common polymorphism in subtype C viruses (in 14% of the sequences examined), the significance of this intersubtype variation is less clear.
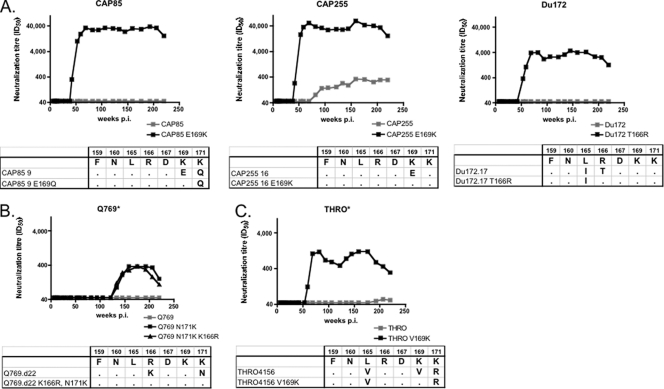
Introducing the FN/LRD-K-K motif into resistant subtype A, B, and C viruses.
We were interested in determining whether “repairing” the FN/LRD-K-K motif in resistant viruses could confer sensitivity. We focused first on the rare resistant subtype C viruses CAP85 and Du172, as well as CAP255, which was weakly neutralized via an undefined (but not anti-V1V2) specificity (Fig. 5, gray panel). Each of these three subtype C viruses had between one and three substitutions in the FN/LRD-K-K motif. For CAP85, introduction of the E169K mutation had a dramatic effect on neutralization sensitivity, with ID50 titers changing from complete neutralization resistance (<1:45) to greater than 1:40,000, similar to that seen with the ConC virus (Fig. 9 A). Interestingly, this was despite the presence of 171Q, which, as in the ZM53M virus, appeared to be tolerated by these antibodies. Similarly, conversion to the sensitive phenotype was also seen for CAP255, where the E169K mutation conferred extreme neutralization sensitivity. The introduction of a T166R mutation into a third completely resistant subtype C virus, Du172, made it highly sensitive, with peak titers of around 1:4,000, despite the presence of L165I, which was also seen in other sensitive subtype C viruses.
FIG. 9.
Restoring the FN/LRD-K-K motif in neutralization-resistant viruses confers neutralization sensitivity. Resistant subtype C (A), subtype B (B), and subtype A (C) viruses were mutated to restore the FN/LRD-K-K motif to various degrees. Parental viruses (resistant) are shown in gray. Mutant viruses are shown in black. Asterisks indicate graphs with differing scales to illustrate lower titers. Sequence changes from the FN/LRD-K-K motif and mutations introduced are shown below the graphs.
Similarly, the single subtype A virus which was resistant to neutralization, Q769, could be made neutralization sensitive (but at low levels) by an N171K mutation, with no further sensitivity conferred by repair of a second residue, K166R (a conservative polymorphism) (Fig. 9B). A resistant subtype B virus, THRO, which contained three substitutions in the FN/LRD-K-K motif, was rendered weakly sensitive by the V169K mutation, though in this case, we did not go on to repair the 165 and 171 residues, both of which had conservative changes from the FN/LRD-K-K motif (Fig. 9C).
Overall, in all three subtypes, we were able to confer sensitivity, sometimes extreme sensitivity, simply by making a single mutation within the FN/LRD-K-K motif in the V2 region. However, as with the neutralization-sensitive viruses, the titers varied considerably, even in the context of an intact FN/LRD-K-K motif, with titers ranging from approximately 1:400 to 1:40,000, confirming the role of other factors in modulating sensitivity. This variation in titers could not be ascribed to the length of the V1 loops in these cases.
The epitope recognized by CAP256 is distinct from but overlaps that of PG9/PG16.
We were interested in determining whether the epitope defined by CAP256 plasma overlapped that of the PG9 and PG16 MAbs. We therefore tested a subset of mutants either reported to affect neutralization by PG9/PG16 (43) or shown to affect the neutralization of ConC by CAP256 plasma. These mutants included the residues forming the FN/LRD-K-K motif plus residues 168 and 181, which had a lesser effect (Fig. 6). Like CAP256 plasma, both PG9 and PG16 were heavily dependent on K169 (this mutation was not tested by Walker et al. [43]) and somewhat dependent on K171 (Fig. 10). The effect of a mutation at residue 181 on CAP256 serum neutralization was also similar to that observed for PG9/PG16. Therefore, some similarities between the epitopes recognized by CAP256, PG9, and PG16 clearly exist.
FIG. 10.
Effects of V2 and V3 mutations on CAP256 plasma neutralization compared with that of MAbs PG9 and PG16.
Interestingly, a mutation abrogating the potential glycosylation site in V2 (N160A) had a profound affect on PG9 and PG16 (with a >1,000-fold increase in the IC50) but caused a more moderate 8-fold reduction in the ID50 in CAP256 (and had no effect on the neutralization of ZM53M [data not shown]). Unlike PG9 and PG16, where a L165A or K166A mutation had little effect, mutating these residues reduced the CAP256 ID50 more than 14- and 100-fold, respectively. The adjacent D167N residue also impacted CAP256 neutralization but not PG9/PG16, and a K168A mutation reduced CAP256 neutralization but in fact enhanced the neutralization of ConC by PG9/16.
Given the fact that PG9 and PG16 target a quaternary epitope in V2, like CAP256 plasma antibodies, but were also dependent on residues in V3, we examined the effect of ConC V3 mutations on CAP256 neutralization. Both PG9/16 and CAP256, in the context of the ConC backbone, were slightly affected by mutations in the V3 loop, with mutations at residues I305, I309, and F317 having a mild effect on neutralization sensitivity (1.5- to 3.1-fold reductions in IC50/ID50). The I307A mutation affected both PG9/PG16 slightly but had no discernible effect on CAP256 neutralization. The minor effect of V3 mutations on PG9/PG16 neutralization of ConC is contrary to a previous report (43), where mutations in V3 profoundly affected PG16 (though this effect was reduced for PG9), suggesting that the V3 dependence may be lessened in the ConC background. This inconsistency might be related to the fact that, like Du422, ConC in the monomeric form binds PG9 to some degree (data not shown). However, the discrepancies in the mapping of critical PG9/PG6 residues in JR-CSF (43) and ConC suggest the necessity for prudence in extrapolating such data between viruses. In summary, we observed some features in common between CAP256 and PG9/16 neutralization, but there were also significant differences, suggesting that although the epitopes recognized by these antibodies overlapped, they were not identical.
DISCUSSION
This study aimed to characterize the BCN activity in CAP256, an HIV-1 subtype C-infected (and subsequently superinfected) participant enrolled in the CAPRISA Acute Infection cohort. CAP256 developed cross-neutralizing activity by 1 year of infection, which increased in both titer and breadth over 3 years (with the ID50 against some subtype C viruses exceeding 1:10,000) before beginning to wane. The specificity conferring breadth targeted a quaternary (trimer-specific) epitope that was highly dependent on residues 159 to 171 in the V2 loop and overlapped the epitopes recognized by MAbs PG9 and PG16 (43). This adds to the growing body of data showing that antibodies recognizing a quaternary epitope involving V2 have considerable cross-neutralizing capacity and that this epitope may represent a useful vaccine target.
The question of whether BCN activity is due to a single very broad antibody (43) or to the accumulation of a limited number of moderately broad specificities (15, 36, 44) is still controversial, and both scenarios likely exist. In the case of CAP256, it seems that the vast majority of cross-neutralizing activity can be attributed to antibodies targeting a single region, which simplified the mapping of this specificity in a polyclonal sample. However, the observations that the V2-dependent response in CAP256 neutralized increasing numbers of viruses over time and that the fine specificity of the neutralizing activity of this plasma changed over time suggested two possible pathways to the evolution of breadth in this individual. Breadth could be associated with affinity maturation of a single V2-dependent antibody that evolved over time to become less dependent on residues aside from R166 and K169, thus acquiring the ability to neutralize an increasing number of heterologous viruses. This situation would be similar to that seen in the subtype A-infected individual from whom somatic variants PG9 and PG16 were obtained, both antibodies having been derived by affinity maturation from a single B cell clone but displaying various potencies and epitope specificities. Alternatively, the BCN antibody response may be composed of multiple V2-dependent antibodies with overlapping but different specificities. This would require the stimulation of multiple B cell lineages targeting the same region, which could be considered an exposed immunodominant epitope. This phenomenon has been reported in a Zambian subtype C-infected individual where two unrelated groups of autologous MAbs were isolated, both of which targeted the V2 region (24). The fact that in CAP256 a single mutation of either K169E or R166A completely abolished neutralization perhaps argues for the affinity maturation hypothesis rather than multiple unrelated antibodies that coincidentally depend on identical residues. However, a single mutation also conferred escape from both unrelated MAbs isolated from the Zambian individual (24). A more complete understanding of the pathway to evolution of breadth in CAP256 will depend on the future isolation of MAbs at multiple time points from this individual.
The factors that favor the development of breadth are not entirely clear. There appears to be an association between greater viral loads and BCN activity (30, 35), likely reflecting the need for ongoing antigenic stimulation of B cells. This is perhaps also reflected in the association of breadth with the duration of infection (35), which may be related to the high level of affinity maturation observed in many broadly reactive MAbs (43, 46, 47). CAP256 was referred for ARV treatment at 4.5 years after infection and had generally high viral loads (>100,000 copies/ml) over the duration of infection, which may have contributed to the development of BCN antibodies. Another factor that has been implicated in the development of breadth is HIV-1 sequence diversity, with individuals with greater envelope diversity early in infection developing greater breadth during chronic infection (30). This observation is supported by recent data showing that dual infection results in broader and more potent nAb responses (31). CAP256 was superinfected in the window between the initial infection and the development of autologous nAb responses, and recombination between the two infecting strains resulted in substantial viral variation over the first year of infection (data not shown). Therefore, a high level of viral diversity early in infection, in addition to the high viral loads, may also have driven the development of breadth in CAP256. The decline in titers observed by 4 years of infection, shortly before the initiation of ARV therapy, may reflect the lower CD4 counts and absence of CD4 help.
Mapping of the determinants of CAP256 neutralizing activity identified seven residues in V2 that contributed to this epitope, namely, the FN/LRD-K-K motif (ranging from F159 to K171). The presence of this motif was associated with neutralization sensitivity, and increasing numbers of polymorphisms at these residues were associated with neutralization resistance. Deviations from the FN/LRD-K-K motif were particularly pronounced among subtype B viruses, especially at residues L165 and K169. The role of this motif in conferring sensitivity was also confirmed by the finding that some resistant viruses (of subtypes A, B, and C) could be rendered sensitive (sometimes extremely so, with titers exceeding 1:10,000) by a single mutation in the FN/LRD-K-K motif. However, it was also clear that the existence of this motif was not sufficient to confer high-level sensitivity, with viruses containing the intact motif but bearing a relatively long V1 loop being much less sensitive than those with a shorter V1 loop. The effect of the V1 loop in modulating neutralization sensitivity may be mediated through shielding of the neutralization epitope; however, it is also possible that the V1 loop length affects the conformation of distal regions of the envelope, thereby exerting an indirect influence on neutralization titers. Furthermore, it is possible that additional conserved residues, which were not identified here, contribute to the epitope. The observation that CAP256 neutralization sensitivity is determined by the presence of crucial residues within the V2 epitope but also by the length of the adjacent V1 loop was not unexpected. Indeed, this is similar to 4E10, where the sensitivity of viruses containing the intact epitope in the MPER may be modulated by changes elsewhere in the envelope (13). The indirect modulation of sensitivity by other regions of the envelope likely provides a means of viral escape from antibodies targeting conserved regions of the envelope (40), as well as affecting the sensitivity of heterologous viruses to such specificities.
Unlike the V1 loop, which is likely to be structurally disordered and is very variable in length, the V2 loop contains a number of subdomains that are very conserved across all subtypes and shows a greater constraint on the minimum length tolerated (48). Thus, the ability of antibodies recognizing a relatively conserved region of V2 to cross-neutralize multiple subtypes is not unexpected. However, the fact that CAP256 neutralizing activity was especially potent against heterologous subtype C viruses, less potent but as broadly effective against subtype A viruses, but only weakly effective against subtype B viruses suggested that the epitope involved is subtype specific. The precise determinants of the subtype specificity observed in CAP256 plasma are difficult to pinpoint. Neutralization by CAP256 is heavily dependent on K169, which is rarely present in subtype B viruses, perhaps explaining the weak activity of this plasma against subtype B viruses. However, PG9/16 MAbs were also dependent on K169 but, unlike CAP256, cross-neutralized all subtypes equivalently. CAP256 was dependent on a wider range of residues in V2 than PG9/16 (e.g., R166, D167, and K168, none of which affected PG9/PG16), suggesting that the “footprint” of these MAbs may be more focused than that of CAP256, which may recognize a broader epitope that is less well maintained across multiple subtypes. However, R166, D167, and K168 are very well conserved across subtypes A, B, and C. The existence of subtype-specific epitopes is borne out by existing MAbs—2G12 and 2F5 fail to neutralize most subtype C viruses because of genetic variability in the epitope, and IgG1b12 neutralizes two-thirds of subtype B and C viruses but only a third of subtype A viruses (2). Preferential neutralization by polyclonal plasma of subtype-matched heterologous viruses has also been shown by us (32) and others (1, 3, 8, 38, 41). Indeed, within the CAPRISA cohort, all of whose members were infected with subtype C viruses, we note significantly better neutralization of subtype C than of subtype B viruses (data not shown). Although this has led to suggestions that vaccines should target locally prevalent subtypes (e.g., a subtype C vaccine in sub-Saharan Africa), the existence of antibodies targeting all subtypes (e.g., PG9/PG16, VRC01) perhaps obviates this need. At any rate, a greater understanding of the determinants of subtype-specific epitopes within the V2 region is required, especially if HIV-1 vaccine immunogen design becomes more focused on this region.
The increasing interest in antibodies targeting QNEs similar to PG9 and PG16 has led to the use of N160 mutants to screen for such specificities (7, 44). However, this approach may be affected by the various effects of this mutation within different viruses. The effect of an N160A mutation in ConC on CAP256 neutralization sensitivity was moderate (with an 8-fold reduction in sensitivity). In contrast, within the context of the ZM53M envelope, an N160A mutation did not abrogate neutralization; in fact, neutralization was slightly enhanced. The context dependence of this mutation is not specific to CAP256, as similar observations were made by Walker et al. (44), who made an N160A mutation in five viruses and saw various effects on neutralization by individual plasmas, ranging from abrogation to enhancement. Furthermore, CAP256 was affected by mutations at both N160 and L165. The effect of these mutations was considered to represent separate specificities in a recent analysis of polyclonal sera, with those specificities sensitive to N160 mutation considered to be PG9/PG16 like, and distinct from L165A-sensitive antibodies (44). However, assuming that a single specificity mediates breadth in this case, CAP256 spans both of these categories, suggesting caution in interpreting such data as being due to unrelated antibodies.
The development of BCN activity in CAP256, targeting a subtype-specific epitope existing within the trimeric form of the envelope, adds to a growing body of data suggesting that QNEs may represent fairly common neutralization targets in HIV-1-infected individuals. The extremely high titers observed in CAP256 also highlight the potential potency of such specificities. This finding confirms the existence of potential vaccine targets in this region of the envelope that should be further investigated in immunogenicity studies.
Acknowledgments
We thank the participants in the CAPRISA Acute Infection cohort and the clinical and laboratory staff at CAPRISA for providing specimens. We are grateful to Mary Phoswa and Sarah Cohen for sample and data management. We thank Dennis Burton (The Scripps Research Institute, La Jolla, CA) for providing MAbs PG9 and PG16.
This work was funded by CAPRISA, CHAVI, and the South African HIV/AIDS Research and Innovation Platform of the Department of Science and Technology and by NIH grants AI088610 and AI078410 to A.P. CAPRISA was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services (grant U19 AI51794). P.M. is a Wellcome Trust Intermediate Fellow in Public Health and Tropical Medicine (grant 089933/Z/09/Z).
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Binley, J. M., et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binley, J. M., et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, B. K., et al. 2008. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375:529-538. [DOI] [PubMed] [Google Scholar]
- 4.Bunnik, E. M., L. Pisas, A. C. van Nuenen, and H. Schuitemaker. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, D. R., et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 6.Dhillon, A. K., et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doores, K. J., and D. R. Burton. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510-10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Euler, Z., et al. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 201:1045-1053. [DOI] [PubMed] [Google Scholar]
- 9.Gao, F., et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorny, M. K., et al. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, E. S., et al. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 83:11265-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray, E. S., et al. 2008. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J. Virol. 82:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, E. S., et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, E. S., et al. 2009. Antibody specificities associated with neutralization breadth in plasma from HIV-1 subtype C infected blood donors. J. Virol. 83:8925-8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haynes, B. F., and D. C. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honnen, W. J., et al. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81:1424-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, S. L., and L. Stamatatos. 2007. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr. HIV Res. 5:507-513. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson Hedestam, G. B., et al. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 20.Li, B., et al. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch, R. M., et al. 2011. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J. Virol. 85:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, P. L., et al. 2008. The C3-V4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, P. L., et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancera, M., et al. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098-8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pejchal, R., et al. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483-11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phogat, S., and R. Wyatt. 2007. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr. Pharm. Des. 13:213-227. [DOI] [PubMed] [Google Scholar]
- 30.Piantadosi, A., et al. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell, R. L., T. Kinge, and P. N. Nyambi. 2010. Infection by discordant strains of HIV-1 markedly enhances the neutralizing antibody response against heterologous virus. J. Virol. 84:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rademeyer, C., et al. 2007. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368:172-181. [DOI] [PubMed] [Google Scholar]
- 33.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson, J. E., et al. 2010. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J. Virol. 84:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sather, D. N., et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheid, J. F., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 37.Seaman, M. S., et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simek, M. D., et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatatos, L., L. Morris, D. R. Burton, and J. R. Mascola. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866-870. [DOI] [PubMed] [Google Scholar]
- 40.van Gils, M. J., et al. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84:3576-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Gils, M. J., D. Edo-Matas, B. Schweighardt, T. Wrin, and H. Schuitemaker. 2010. High prevalence of neutralizing activity against multiple unrelated human immunodeficiency virus type 1 (HIV-1) subtype B variants in sera from HIV-1 subtype B-infected individuals: evidence for subtype-specific rather than strain-specific neutralizing activity. J. Gen. Virol. 91:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Loggerenberg, F., et al. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker, L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker, L. M., et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6(8):e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, X., et al. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolla-Pazner, S., and T. Cardozo. 2010. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]