Abstract
Aberrant DNA methylation (DNAm) was first linked to cancer over 25 yr ago. Since then, many studies have associated hypermethylation of tumor suppressor genes and hypomethylation of oncogenes to the tumorigenic process. However, most of these studies have been limited to the analysis of promoters and CpG islands (CGIs). Recently, new technologies for whole-genome DNAm (methylome) analysis have been developed, enabling unbiased analysis of cancer methylomes. By using MeDIP-seq, we report a sequencing-based comparative methylome analysis of malignant peripheral nerve sheath tumors (MPNSTs), benign neurofibromas, and normal Schwann cells. Analysis of these methylomes revealed a complex landscape of DNAm alterations. In contrast to what has been reported for other tumor types, no significant global hypomethylation was observed in MPNSTs using methylome analysis by MeDIP-seq. However, a highly significant (P < 10−100) directional difference in DNAm was found in satellite repeats, suggesting these repeats to be the main target for hypomethylation in MPNSTs. Comparative analysis of the MPNST and Schwann cell methylomes identified 101,466 cancer-associated differentially methylated regions (cDMRs). Analysis showed these cDMRs to be significantly enriched for two satellite repeat types (SATR1 and ARLα) and suggests an association between aberrant DNAm of these sequences and transition from healthy cells to malignant disease. Significant enrichment of hypermethylated cDMRs in CGI shores (P < 10−60), non–CGI-associated promoters (P < 10−4) and hypomethylated cDMRs in SINE repeats (P < 10−100) was also identified. Integration of DNAm and gene expression data showed that the expression pattern of genes associated with CGI shore cDMRs was able to discriminate between disease phenotypes. This study establishes MeDIP-seq as an effective method to analyze cancer methylomes.
The development of human cancer is driven by changes in the genetic and epigenetic landscape of the cell. It is now clear that epigenetic changes, such as the hypermethylation of tumor suppressor genes and the hypomethylation of oncogenes, are hallmarks of cancer and play a key role in the development and maintenance of the malignant phenotype (Issa 2004; Laird 2005; Baylin and Ohm 2006). Epigenetic alterations also appear to be some of the earliest abnormalities occurring in the tumorigenic process and have the potential to predispose stem/progenitor cells to subsequent genetic alterations involved in tumor development and progression (Feinberg et al. 2006) and, therefore, may represent good markers for early disease diagnosis. The role of aberrant DNAm in the development and progression of human cancer has been studied in several tumors such as breast, prostate, lung, and colorectal cancers (Santourlidis et al. 1999; Ordway et al. 2007; Barault et al. 2008; Vaissiere et al. 2009). However, until recently the majority of studies have focused on those regions where DNA methylation (DNAm) was assumed to have the greatest functional significance in the regulation of gene expression, such as promoters and CpG islands (CGIs). This has led to the identification of many genes where differential DNAm in CGIs results in aberrant gene expression and association with disease phenotype. However, as only 7% of CpGs reside within CGIs, it is highly likely that many potentially informative sites have not yet been analyzed (Rollins et al. 2006). This was recently confirmed by the identification of CGI shores as key DNAm gene regulatory sites (Doi et al. 2009; Irizarry et al. 2009), and suggests a truly comprehensive map of the DNAm landscape in human malignancies is yet to be defined.
Recently, next-generation sequencing technologies have emerged as powerful tools to allow whole-genome profiling of epigenetic modifications, including DNAm (Beck and Rakyan 2008; Laird 2010). MeDIP-seq, for instance, combines methylated DNA immunoprecipitation (MeDIP) with next-generation sequencing, provides an efficient approach for whole-genome DNAm (methylome) analysis, and has been used to generate the methylome of human spermatozoa (Down et al. 2008). Similarly, whole-genome bisulfite sequencing (BS-seq) has recently been used to generate the first single-base resolution profiles of DNAm in human embryonic stem cells and differentiated cells (Lister et al. 2009; Laurent et al. 2010).
Neurofibromas (NFs) are benign tumors that originate from Schwann cells (SCs) in a peripheral nerve sheath. Neurofibromatosis type 1 (NF1) is predominantly an autosomal dominant neurocutaneous disorder affecting one in 3000 individuals world-wide and is caused by germline mutations of the NF1 gene (Levy et al. 2004). However, it also occurs sporadically. Approximately 5%–10% of patients with NF1 develop malignant peripheral nerve sheath tumors (MPNSTs) (Evans et al. 2002). Although the incidence of MPNSTs is relatively low, only 43% of patients with this diagnosis survive beyond 5 yr (Evans et al. 2002; Zhou et al. 2003). We still do not understand the epigenetic or genetic alterations driving the development of MPNSTs, or the molecular markers that are associated with the development and progression of this disease. So far, few studies have assessed the DNAm state of MPNSTs, and most of those have been restricted to the NF1 gene itself (Harder et al. 2004) or selected CGIs and promoters (Gonzalez-Gomez et al. 2003). Nevertheless, these studies have identified differential DNAm of several genes thought to be involved in MPNST development and progression, including CDKN2A and MSH2 (Kawaguchi et al. 2006; Titze et al. 2010). To provide a more comprehensive catalog of DNAm differences in MPNST, we performed unbiased methylome analysis of benign and malignant NF samples as well as controls.
Results
Methylome analysis was conducted on three pools of genomic DNA extracted from 10 MPNST, 10 NF, and six normal non-neoplastic SC samples. SC samples were derived from biopsies and expanded in culture. By using MeDIP-seq (Down et al. 2008), we generated on average 65 million paired-end reads per cohort pool, of which 72% could be uniquely aligned to the human genome (build NCBI36) (Supplemental Table 1). Absolute DNAm values were inferred for 100-bp windows using the Batman algorithm (Down et al. 2008). To correct for potential bias in DNAm differences due to changes in copy number, sequence read counts were normalized for copy number variation (CNV) prior to Batman processing. CNV profiles were generated using the Affymetrix Human SNP 6.0 microarray (Supplemental Fig. 1; Materials and Methods). Due to the study design, which in this case involved sample pooling, it is difficult to estimate the contribution made by individual patients to specific CNVs. However, any CNV observed will have to be present in a significant proportion of the samples in order to be identified in the first place. The fact that we were able to identify known CNVs associated with MPNSTs (e.g., loss of NF1 and CDKN2A) confirmed our ability to detect common CNVs. After normalization of the DNAm data for CNVs, there was no significant difference in the number of DMRs found within regions of CNV gain or loss compared with the control regions. Based on this methylome analysis, we covered on average 67% (range, 65%–70%) of the ∼26.7 million CpG sites in the haploid autosomal human genome reference sequence for each of the three methylomes (sex chromosomes were removed from the analysis because of the pooled study design) (Supplemental Fig. 2). Furthermore, saturation analysis indicates we have sufficient reads to give reproducible genome-wide methylation profiles (Supplemental Fig. 3; Chavez et al. 2010). We validated our DNAm data for each cohort pool (R2 = 0.78 for MPNSTs, R2 = 0.80 for NFs, R2 = 0.77 for SCs) using the Infinium Human Methylation 27K BeadArray (Supplemental Fig. 4; Bibikova et al. 2009). Although slightly lower, these results are consistent with previously reported correlations between BeadArray and BS-seq (R2 = 0.82) and between MeDIP-seq and BS-seq (R2 = 0.82) (Down et al. 2008; Bibikova et al. 2009; Smith et al. 2009; Gu et al. 2010). In addition, we further validated our MeDIP-seq data by confirming the DNAm status of three selected cancer-associated differentially methylated regions (cDMRs; R2 = 0.97) using targeted pyrosequencing (Supplemental Fig. 5) and using BS-seq, which had a concordance of 77% with Batman methylation scores across 64,943 CpG sites.
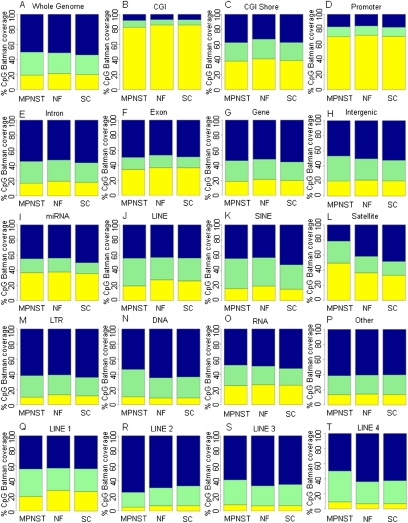
We first performed a comprehensive global analysis of the three methylomes to identify major directional changes in DNAm. This does not include balanced and undirectional changes in DNAm, which will be described in the DMR section. Batman DNAm scores for the individual CpG sites were categorized into three DNAm states: low DNAm (DNAm score, <40%), intermediate (DNAm, 40%–60%), and high (DNAm, >60%) DNAm. As shown in Figure 1, there was only a 0.7% change in global DNAm in MPNSTs compared with NF and SC controls. This finding is in contrast to our current understanding based on other cancer types, that cancer genomes undergo global hypomethylation with an average reduction in DNAm of between 10% and 20% between cancer and normal (Feinberg and Vogelstein 1983; Gama-Sosa et al. 1983; Ehrlich 2002). We then analyzed CpG DNAm in a further 16 distinct genomic features, including CGI and other regulatory regions, exons, introns, and the repeat families annotated in the Ensembl database (Fig. 1). This analysis revealed that the main targets of differential DNAm in MPNSTs were satellite repeats (Fig. 1), with a significant (P < 10−100) increase in the percentage of CpG sites with low DNAm within satellite repeats between SCs (33%) and NFs (39%) and MPNSTs (49%). Compared to this, the level of global DNAm across the most commonly studied and usually unmethylated CGIs (Baylin and Ohm 2006) remained quite similar for all three phenotypes (MPNSTs, 12%; NFs, 10%; SCs, 11%) (Fig. 1B). Little change in the global DNAm of other regulatory features such as CGI shores (Fig. 1C) and promoters (Fig. 1D) as well as genic features (Fig. 1E–H) was observed.
Figure 1.
Global DNA methylation profiles. (A–T) Global patterns of methylation in unique genomic features from the Ensembl genome database (Ensembl 52). Shown are the color-coded methylation states of CpGs for the MPNST, NF, and SC samples (x-axis). The mean Batman methylation states of CpGs within a 1-kb window were calculated and categorized into low methylation (yellow, 0%–40%), intermediate methylation (green, 41%–60%), and high methylation (blue, 61%–100%) for each genomic feature. MeDIP-seq/Batman CpG coverage (y-axis) shows the proportion of CpGs covered with Batman methylation scores at low, intermediate, or high methylation levels. Coordinates for genomic features were taken from the Ensembl genome database, with the exception of CGI shores, which were defined as 2 kb on either side of the CGI, and promoters defined as 1 kb upstream of and 0.5 kb downstream from the TSS.
One of the most commonly cited features of cancer epigenomes concerns directional hypomethylation of repeats (Ehrlich 2002; Wilson et al. 2007). Historically, repeat sequences proved to be refractory to many analysis approaches (particularly array-based approaches), but this was not a problem for the MeDIP-seq approach used here, which provided excellent coverage of all major repeat types. In contrast to previous studies in other cancers (Dante et al. 1992; Jurgens et al. 1996; Santourlidis et al. 1999), we did not observe global hypomethylation of LINE repeats (Fig. 1J). In fact, we saw a decrease (8%) in the proportion of low DNAm (<40%) at CpGs between MPNSTs and SCs. Interestingly, analysis of the different subtypes of the LINE repeat family revealed different patterns of global DNAm profiles, with L2 and L1 repeats showing an increase in the proportion of highly methylated (>60% DNAm) CpGs, whereas the L3 and L4 LINE repeats showed a decrease in the proportion of highly methylated (>60% DNAm) CpGs in MPNSTs (Fig. 1Q–T). SINE repeat elements (Fig. 1K) showed a small 6% decrease in the number of highly methylated CpGs between MPNSTs and SCs (Table 1). However, the starkest difference in global DNAm was observed in satellite repeats (Fig. 1L), with a large change in the proportion of methylated CpGs seen during transition from the normal to benign to malignant states. Although the pooling strategy employed in this study somewhat limited the sensitivity at which rare DNAm changes could be detected, we have already showed above that we were able to detect common changes that hold the best potential for future diagnostic and prognostic markers.
Table 1.
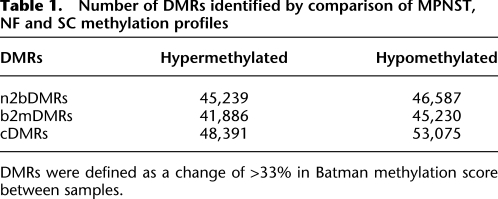
Number of DMRs identified by comparison of MPNST, NF and SC methylation profiles
DMRs were defined as a change of >33% in Batman methylation score between samples.
Next, we carried out pairwise comparisons between the three methylomes to identify DMRs associated with progression from non-neoplastic SCs to benign NFs (n2bDMRs), progression from benign NFs to malignant MPNSTs (b2mDMRs), cDMRs between SCs and MPNSTs and tissue-specific DMRs (tDMRs) between MPNSTs, and previously determined tDMRs between germline and somatic tissues (Down et al. 2008; Irizarry et al. 2009). Since DNAm at neighboring CpG sites is highly correlated up to 1000 bp (Eckhardt et al. 2006), DMRs were defined as the average Batman DNAm score over 1-kb windows with a minimum difference of ≥33% (equivalent to the 95th percentile of DNAm scores) in Batman DNAm score between samples (Down et al. 2008). Each 1-kb window was subsequently mapped back to its nearest corresponding genomic feature. By using this approach, we identified 101,466 unique cDMRs with a false discovery rate (FDR) of 3.7%. These cDMRs can further be subdivided into 48,391 hypermethylated and 53,075 hypomethylated cDMRs (Supplemental Fig. 2; Table 1). Interestingly, the lowest number of cDMRs was identified in the NF/MPNST comparison (b2mDMRs), suggesting the benign methylome to be more closely related to the malignant than the normal methylome (Supplemental Fig. 2). Changing this threshold from the 95th to the 90th and 99th percentiles of DMR scores (difference in Batman DNAm score of ≥43% and ≥27%, respectively) showed that the numbers of DMRs associated with different genomic features changed proportionally with the change in threshold, suggesting that the noise is evenly distributed across different genomic features.
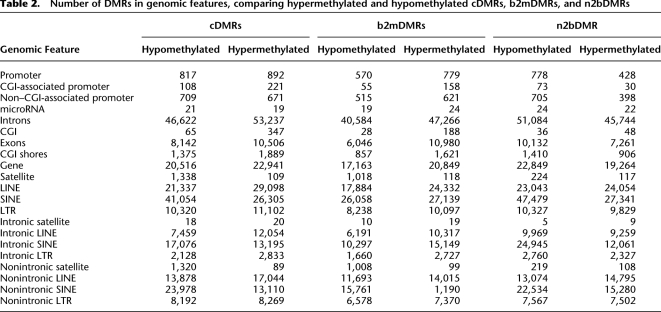
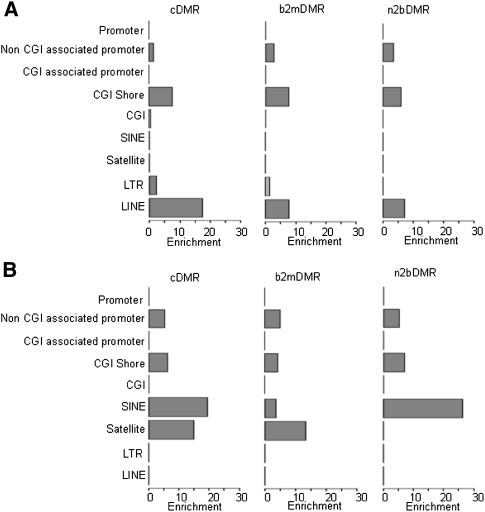
We subsequently mapped all DMRs to their nearest genomic feature (Table 2) and performed an enrichment analysis on the number of DMRs in unique genomic features. Analysis of these DMRs showed significant (P > 10−3; range 10−3–10−60) enrichment of hypermethylated cDMRs (Fig. 2) in all three sample types (defined by cDMRs n2bDMRs and b2mDMRs) in non–CGI-associated promoters, CGI shores, LTRs, and LINEs, but not in CGIs. Conversely, hypomethylated DMRs were enriched in non–CGI-associated promoters, CGI shores, SINEs, and satellites, but, again, not in CGIs (Fig. 2). Although the observed number of 412 CGI-associated cDMRs (1.4% of total) was statistically not significant, this does not exclude them from having functional relevance. Of all the DMRs, only CGI shore-associated DMRs showed both highly significant gain and loss of DNAm, emphasizing the special role of DNAm at this genomic feature. The majority of the CGI-associated cDMRs were hypermethylated (367, or 89%). Interestingly only 11% (44 hypermethylated and three hypomethylated) of cDMRs were within 3 kb of the nearest annotated gene. Analysis of gene promoter regions identified a similar number of hyper- and hypomethylated cDMRs (892 hypermethylated, 817 hypomethylated) (Table 2; Supplemental Fig. 6). When promoters were subdivided according to association or nonassociation with CGIs, we observed a significant enrichment of both hypermethylated (P = 9.9 × 10−4) and hypomethylated cDMRs in non–CGI-associated promoters compared with no enrichment of CGI-associated promoters (Fig. 2). No enrichment of promoter-associated DMRs in bivalent chromatin domains was observed. Similarly no enrichment of DMRs containing microRNAs was observed. These data are consistent with the recent finding that the majority of differential DNAm occurs outside CGIs (Irizarry et al. 2009). Consequently, many previous studies using CGI-biased profiling approaches have only assayed a fraction of the differential DNAm potential in the tumorigenic epigenome.
Table 2.
Number of DMRs in genomic features, comparing hypermethylated and hypomethylated cDMRs, b2mDMRs, and n2bDMRs
Figure 2.
Relative DMR enrichment. Relative enrichment of DMRs in CGI, CGI shore, promoter, and repeat regions (SINE, satellite, LTR, and LINE), for hypermethylated (A) and hypomethylated (B) DMRs for the three sample comparisons (n2bDMRs, b2mDMRs, and cDMRs). Relative enrichment analysis was carried out on the number of DMRs in each genomic feature compared to the expected number. Features with significant relative enrichment (P < 0.001) are highlight in red. The x-axis shows the relative enrichment score; the y-axis, the relevant genomic features.
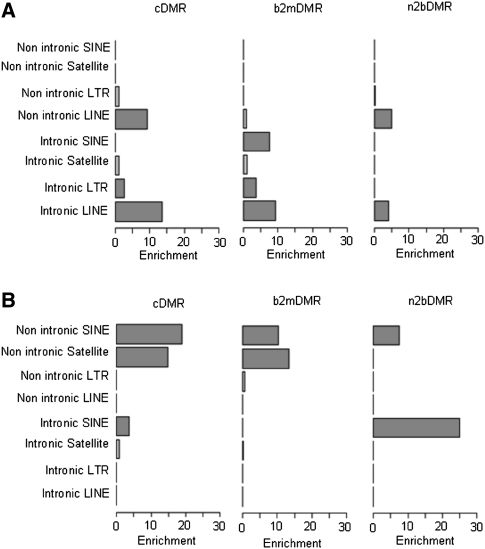
As expected, the majority of cDMRs mapped to repeat elements (Table 2). The propensity for hypermethylation of LINE repeat elements identified in the global analysis persisted (Fig. 3), with the enrichment of hypermethylated cDMRs (P < 10−100), n2bDMRs (P = 6.47 × 10−32), and b2mDMRs (P = 4.33 × 10−37) in both intronic and nonintronic LINE repeats (Fig. 3). Intronic and nonintronic LINE cDMRs showed the largest level of enrichment and n2bDMRs the lowest, raising the possibility that hypermethylation of LINE repeats increases with disease progression. Contrary to LINE repeats, the other major class of repeats, SINEs, showed significant enrichment of hypomethylated DMRs [cDMRs (P < 10−100), n2bDMRs (P < 10−100), and b2mDMRs (P = 2.48 × 10−8)] (Fig. 3).
Figure 3.
Relative DMR enrichment of intronic and non-intronic repeats. Shown are the relative enrichment of unique repeats (SINE, satellite, LINE, and LTR) located within introns and outside introns for hypermethylated (A) and hypomethylated (B) DMRs. The x-axis shows the level of relative enrichment; the y-axis, the relevant repeat type. Features highlighted in red show significant enrichment (P < 0.001).
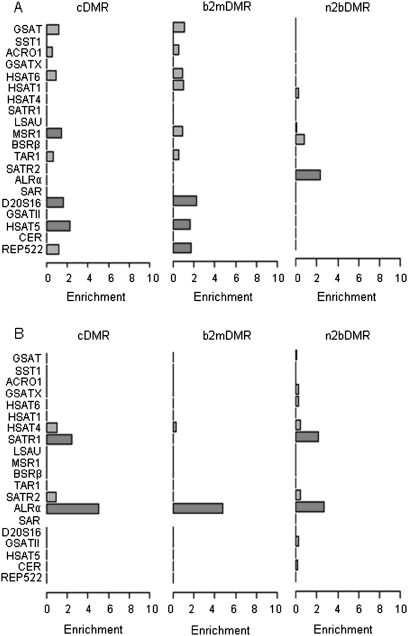
Satellite repeats showed significant enrichment of hypomethylated DMRs, particularly those repeats in nonintronic regions (cDMRs, P < 10−100; b2mDMRs, P < 10−100) (Figs. 3, 4). Of the 1447 satellite repeat cDMRs, 1384 (107 hypermethylated, 1241 hypomethylated) mapped to regions outside of CNV. Only 13 of the 1241 hypomethylated satellite cDMRs mapped to regions of genomic loss. These data suggest that hypomethylation of satellite repeats observed in the MPNST epigenome is not due to changes in the underlying genomic copy number of these loci. It is also possible that hypomethylation events, particularly in repeat regions, may be under-called as a result of the removal of multi-mapping reads prior to methylome analysis. However, as part of our methylome analysis, similar proportions of multi-mapping reads were removed from both the normal and malignant methylomes (20.43% and 23.23%, respectively), suggesting that we did not under-call hypomethylation events. Further analysis of these DMRs according to satellite repeat subtypes showed significant enrichment of hypomethylated cDMRs in the ARLα (P = 8.7 × 10−65) and SATR1 (P = 3.7 × 10−9) satellite repeats. Interestingly, we observed a progressive increase in the enrichment of the hypomethylated ALRα DMRs during the progression from non-neoplastic to malignant disease, whereas DMRs in SATR1 repeats appeared enriched to a similar level in n2bDMRs and cDMRs with no enrichment in b2mDMRs, suggesting that the hypomethylation of SATR1 repeats may be an early event in the tumorigenic process. These data suggest that specific subtypes of satellite repeats undergo preferential hypomethylation during tumorigenic development.
Figure 4.
DMR enrichment of satellite repeat types. Relative enrichment of 19 satellite repeat types for hypermethylated (A) and hypomethylated (B) DMRs. The x-axis shows the level of enrichment; the y-axis, the relevant genomic features. Features highlighted in red show significant enrichment (P < 0.001).
The DMR analysis revealed 3690 genes potentially involved in MPNST development and progression (Supplemental Table 2): for example, the hypermethylation of the promoter region of MEST, a target of both loss of imprinting and hypermethylation in multiple tumor types, including breast cancer and glioblastoma (Pedersen et al. 2002; Martinez et al. 2009). Intriguingly, this study also identified the hypermethylation of the promoter region of the Wilms tumor gene, WT1, a tumor suppressor gene associated with the development of several tumor types (Pelletier et al. 1991; Chen et al. 2003). Consistent with previous studies of DNAm in MPNSTs, we also found no difference in DNAm at the NF1 promoter (Luijten et al. 2000; Harder et al. 2004; Fishbein et al. 2005), although we observed the heterozygous loss of the NF1 locus in the MPNST samples (Supplemental Fig. 1). It has recently been suggested that DNAm may impinge on the expression of microRNAs (Lujambio et al. 2007). Although we did not observe any enrichment of aberrantly methylated microRNAs, several interesting candidate microRNAs were shown to be differentially methylated. For example, miR-124 mapped to a hypermethylated cDMR. This microRNA has recently been shown to be a predictive DNAm biomarker for high-grade disease in cervical cancer (Lujambio et al. 2007; Wilting et al. 2010). In addition, we also observed hypomethylation of miR-30, which has been shown to be up-regulated in a variety of both hematological and solid malignancies (Volinia et al. 2010).
Recently, it has been shown that tDMRs frequently coincide with cDMRs (Irizarry et al. 2009). Comparison with these previously determined DMRs showed that 15% of the tDMRs and 14% of cDMRs were also sites of differential DNAm in MPNSTs, including the hypomethylated oncogenes YES1, MATK, E2F3, EGFR, and AFF1 and the hypermethylated tumor suppressor genes TSC2, RUNX1, FOXD3, and RASSF1 among others.
To assess if the genes associated with cDMRs were enriched for certain pathways or networks, we performed gene ontology (GO) analysis (multiple test corrected P < 0.01). This analysis showed hypermethylated promoter-associated cDMRs to be enriched for genes involved in the regulation of transcription factor activity, DNA binding, and cell surface receptor signaling and hypomethylated cDMRs to be enriched for genes involved in differentiation and development, particularly brain and neural devolvement. Hypermethylated CGI shore cDMRs showed enrichment for genes involved in cell adhesion and regulation of transcription, along with genes involved in neurogenesis. The latter was also observed for CGI shore n2bDMRs, suggesting that genes in this ontology may be involved in the early development of MPNSTs.
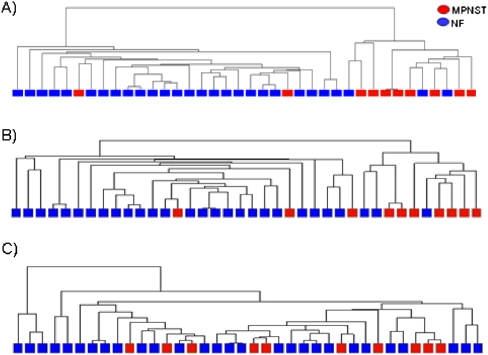
Finally, we explored the relationship between differential DNAm and gene expression by integrating our DNAm data with published gene expression data from NFs and MPNSTs (Henderson et al. 2005; Miller et al. 2009). We next asked whether gene expression correlated sufficiently with aberrant DNAm to discriminate between benign and malignant disease phenotypes. We performed partition clustering of the MPNST and NF samples using the expression of genes associated with b2mDMRs in CGIs, CGI shores, or promoters. The expression patterns of genes associated with CGI shore hypermethylated and hypomethylated b2mDMRs were able to discriminate between the MPNST and NF phenotypes, significantly better (P = 0.0001) than expected by chance (Fig. 5). We also observed significant discrimination between the MPNST and NF samples based on the expression of genes associated with hypermethylated non–CGI-associated promoter b2mDMRs (P = 0.0003). No association was seen between gene expression and CGI-associated promoter b2mDMRs (hypermethylation, P = 0.1057; hypomethylation, P = 0.129) or hypomethylated promoter-associated b2mDMRs (P = 0.797).
Figure 5.
Clustering of MPNST and NF samples using the expression of genes associated with b2mDMRs. (A) The expression level of 1056 genes that contain a hypermethylated b2mDMR in associated CGI shores were used to cluster the MPNST and NF samples. (B) Clustering of the MPNST and NF samples based on the expression of 667 genes associated with the non–CGI-associated promoter b2mDMRs. The two major branches of the dendrogram correspond to a MPNST cluster and NF cluster, respectively. (B) Clustering of the MPNST and NF samples based on the expression of genes associated with hypermethylated non–CGI-associated promoter b2mDMRs. Columns represent individual samples (MPNST, red; NF, blue). (C) Clustering of the MPNST and NF samples based on the expression of genes associated with the hypermethylated CGI-associated promoter b2mDMRs.
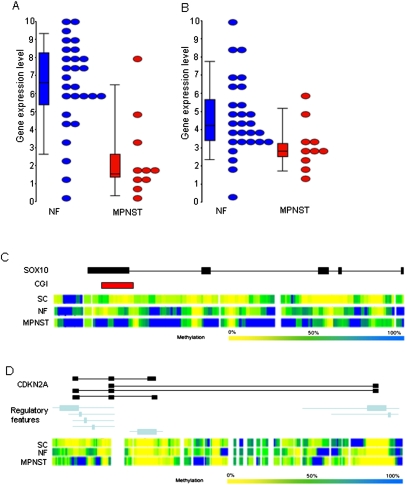
Integration of gene expression data also allowed the identification of several candidate genes involved in the development and progression of MPNST. For example, we observed hypermethylation and a concomitant reduction in gene expression of the tumor suppressor gene SOX10 (P = 0.00054) (Fig. 6C; Miller et al. 2006). We also observed reduced expression and hypermethylation of CDKN2A (P = 0.00004), implying that the suppression of the CDKN2A locus in these tumors may be driven by increased DNAm (Fig. 6D) as well as genomic loss (Supplemental Fig. 1) in MPNSTs (Nielsen et al. 1999).
Figure 6.
Example of genes displaying differential gene expression and methylation. (Top) Gene expression in NFs (blue) and MPNSTs (red) for SOX10 (A) and CDKN2A (B). (Bottom) Genomic organization of the gene (black) with genomic regulatory features (blue) and CGIs (red) and the Batman methylation profiles for the three disease states (MPNSTs, NFs, and SCs) for SOX10 (C) and CDKN2A (D), respectively.
Discussion
Many studies have shown that epigenetic changes play an important role in cancer development (Ehrlich 2002). However, our knowledge of how changes in DNAm impinge on the neoplastic genome is largely still limited to its effect on CGIs. In order to elucidate the impact of aberrant DNAm on cancer development, we have carried out a whole-genome comparative methylome analysis of MPNSTs, benign NFs, and normal non-neoplastic SCs. This analysis reveals a complex pattern of epigenetic changes during disease progression, the majority of which occur outside of regions previously assumed to be the predominant regions involved in the epigenetic regulation of cancer.
Based on our methylome analysis, we identified satellite repeats to be the predominant target of aberrant DNAm in MPNSTs. Although aberrant repeat DNAm has been observed before (Weisenberger et al. 2005; Wilson et al. 2007), it has previously been challenging to assess the DNAm state of individual repeats at the whole-genome level due to the technical and physical limitations of microarray- and PCR-based assays. Digressing from the current understanding of repeat DNAm in human malignancies, our data indicate that the statistically most significant change in repeat DNAm is seen in satellite repeats rather than LINE and SINE repeats. Furthermore, our analysis revealed two unique satellite subtypes to be the main targets of hypomethylation, ARLα and SATR1, which are predominantly located in nonintronic regions of the genome.
A similar increase in hypomethylation was also observed in benign neoplasms compared with non-neoplastic SCs. Analysis of the DNAm of satellite repeats located in nonintronic compared with intronic regions suggested that changes in global satellite DNAm occur predominately in nonintronic regions. These data suggest that aberrant DNAm of satellites may be an early and essential step in the tumorigenic process (Saito et al. 2001; Wong et al. 2001). The distinct DNAm patterns associated with different types of satellite repeats suggests that either regional or sequence-specific determinants influence the propensity of satellite repeats to undergo aberrant DNAm. It is therefore interesting to note that specific satellite repeat sequences appear to be targeted by specific members of the DNMT methyltransferase family (Robertson et al. 2004). However, as genomic instability does not appear to be a common feature in MPNSTs (Serra et al. 1997; Kobayashi et al. 2006), aberrant satellite DNAm may result in altered expression of nearby genes due to changes in chromatin accessibility by transcription factors (Cadieux et al. 2006). It will be interesting to investigate further whether the distinct pattern of satellite repeat DNAm observed in MPNSTs also occurs in other tumor types, whether it is under sequence-specific control, and how it affects the expression of nearby genes.
In addition to satellite repeats, the other major target for aberrant DNAm in MPNSTs is CGI shores, confirming the involvement of CGI shores in a second lineage of human cancers in addition to colorectal cancer (Irizarry et al. 2009). Previous studies have suggested that these regions surrounding CGIs play a more significant role in the regulation of gene expression than do CGIs themselves (Doi et al. 2009; Irizarry et al. 2009). The balanced number of hypermethylated and hypomethylated cDMRs in CGI shores may further imply they are functionally implicated in MPNST development, allowing both the activation of oncogenes and the suppression of tumor suppressor genes. In support of such a possible functional role for CGI shores, integration of the gene expression data showed that the aberrant DNAm of CGI shores had a stronger association with gene expression than did aberrant DNAm of CGIs. Although the independent expression data correlate well with our pooled methylation data, the conclusions drawn from the integration of the two data sets must be viewed in the context that both data sets were derived from different individuals and other explanations may exist. Furthermore, the high degree of overlap between DMRs from other cancer types (Irizarry et al. 2009) suggests that these regions may constitute core sites of aberrant DNAm involved in the development and progression of human malignancies, a concept consistent with the epigenetic progenitor model of cancer (Feinberg et al. 2006).
In summary, we have performed a comparative methylome analysis of normal SCs along with benign and malignant nerve sheath tumors. Our data show the suitability of MeDIP-seq to investigate cancer methylomes and to identify novel sites of differential DNAm, which constitute potential biomarkers for disease development and progression in human malignancies. However, further work is necessary to assess the biological function and clinical utility of such epigenetic biomarkers in the development and progression of MPNSTs.
Methods
Clinical samples
This study complies with Central Office for Research Ethics Committees standards (REC 07/Q0506/8). The material, snap-frozen tissue, was obtained from the histopathology department of The Royal National Orthopaedic Hospital. The clinical data were retrieved from the clinical notes. The cohort studied consisted of NFs with NF1 from 10 patients and MPNSTs from 10 patients.
Primary human SCs were isolated from 10- to 15-cm lengths of human sciatic/brachial nerve from amputation specimens from patients without NF1 or nerve sheath tumors. The protocol was adapted from that described by Muir and Manthorpe (1992). Briefly, after removal of the perineurium, the nerve was cut into ∼2-mm transverse sections with a scalpel and placed in cell culture plates with hSC medium, containing DMEM/F12 supplemented with 2 μM of Forskolin (Calbiochem), recombinant GGF (20 ng/mL; R&D Systems), 3% FCS (Invitrogen), kanamycin monosulfate (80 μg/mL; Sigma), and gentamicin (40 μg/mL; GIBCO). The plates were incubated at 37°C and 5% CO2 and fed every 2–3 d for 2–3 wk. Then the nerve fragments were dissociated overnight at 37°C with a mixture of 300 U/mL collagenase Type XI (Sigma) and 1.25 U/mL of Dispase (Roche) containing the antibiotics kanamycin/gentamicin. The single cell suspension was then plated into 1× Poly-L-Lysin/Laminin-coated tissue culture plates (Sigma) and incubated at 37°C 5% CO2 for another few days before CD271 (low-affinity nerve growth factor receptor, LNGFR)-positive cells were selected by MACS according to the manufacturer's instructions (Miltenyi Biotec Ltd.). After selection, the nucleic acid was extracted.
MeDIP-seq
Five micrograms of pooled DNA from each sample cohort was sonicated to between 50 and 350 bp. Sonicated DNA was then subjected to Illumina's paired-end library preparation and MeDIP enrichment as previously described by Down et al. (2008). Next-generation sequencing (50-bp paired-end reads) was performed on pooled libraries (size-selected to be between 150 and 200 bp for MPNSTs and between 100 and 150 bp for NFs and SCs) for each pooled sample type. Failure of the MPNST 100- to 150-bp library resulted in the larger library (150–200 bp) being sequenced for MPNSTs. To assess if different library sizes effected enrichment, we assessed the relative representation of fragments containing the same number of CpG sites. This showed the enrichment of fragments with the same number CpGs to be similar across samples (Supplemental Fig. 5). Reads were mapped to the human genome reference sequence (Build 36) using the alignment software MAQ (http://maq.sf.net/) (Li et al. 2008). Reads with a MAQ score of <10 were removed from the subsequent downstream analysis. Absolute DNAm states were inferred from raw reads using the Batman algorithm as previously described by Down et al. (2008). The MeDIP-seq data have been deposited in GEO under accession no. GSE21714.
BS-seq
BS-seq was performed as previously described (Lister et al. 2009). One microgram of pooled DNA from MPNST was sonicated to between 50 and 350 bp. Sonicated DNA was then subjected to Illumina's paired-end library preparation, with the exception that 5-methyl-cytosine–containing paired-end adapters were used. Adapter ligated fragments were bisulfite converted using the EZ DNA methylation kit (Zymo Research) and subsequently size-selected by Agarose gel electrophoresis. Size-selected libraries between 250 and 350 bp were then subjected to 18 rounds of amplification prior to sequencing on the Illumina GAIIx. We mapped 75-bp-long raw reads using the Bismarck software (http://www.bioinformatics.bbsrc.ac.uk/projects/bismark/). For mapping quality assurances, the first 10 bases were removed from each read prior to mapping. DNAm states were inferred from raw reads using the SeqMonk package (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk/). Concordance with the Batman methylation score was assessed for those CpG sites covered by at least 10 reads.
Pyrosequencing
One microgram each of pooled DNA from the MPNST, NF, and SC samples was bisulfite-treated using a EZ DNA methylation kit (Zymo Research). Bisulfite-treated DNA was used for generating PCR-amplified templates for pyrosequencing using target specific primers (Supplemental Table 3). Ten microliters of the biotinylated PCR product was sequenced according to the manufacturer's recommendation. Pyrosequencing was carried out on the PSQ HS 96 System and PyroMark MD System using Pyro Gold Reagent kits (Biotage). Methylation was quantified using Pyro Q-CpG Software that calculates the ratio of converted Cs (Ts) to unconverted Cs at each CpG and expresses this as a percentage of DNAm. Average DNAm across the target region was determined for each sample and compared to the Batman and Infinium DNAm scores.
CNV profiling
Pooled DNA from each of the sample cohorts were interrogated for CNVs using the SNP 6.0 Arrays (Affymetrix). Arrays were processed in accordance with the manufacturer's instructions. Changes in CNVs for each sample were inferred using a hidden Markov model algorithm (Zhao et al. 2004). Regions of gain were called using a cut-off of 2.3 copies, and regions of loss were called using a cut-off of 1.7 copies; regions were only called altered if a minimum of 10 loci were above (or below) the threshold. Raw MeDIP-seq read counts were normalized to correct for any bias introduced by changes in genomic copy number by dividing the sequence read count number by the change in copy number. Sex chromosomes were removed from the analysis because of the pooled study design.
Global DNAm analysis
Changes in global DNAm were assessed by binning CpG DNAm levels into three states: low DNAm (<40% DNAm), intermediate DNAm (40%–60% DNAm), and high DNAm (>60% DNAm). The significance of differences in global DNAm was assessed using the Fisher test in Bioconductor (Gentleman et al. 2004).
DMR identification
To detect regions of differential methylation between disease phenotypes and non-neoplastic SC controls, we screened genomic windows of 1 kb. For each window, the Batman methylation scores (100 bp) were averaged and compared between sample cohorts. Using the distribution of differences between sample methylation scores, we defined a conservative threshold for calling DMRs which was based on the 95th percentile of the difference in methylation score. This resulted in a difference in methylation score of ≥33% between samples being considered as DMRs.
To assess the FDR in the DMR calling, we randomized the Batman DNAm scores for each sample and repeated the DMR analysis (10,000 permutations). This analysis gave an FDR of 3.7% across the genome.
To investigate if DMRs were enriched among certain structural genomic features we used the epitools R-package to compute odds ratios for any given structural genomic feature (e.g., CGI) against all other feature types. Significance of odds ratio values was calculated using median-unbiased estimation (mid-p). Since feature types differed greatly in terms of the total number of 1-kb windows that mapped to each of them, thus resulting in large differences in the expected variance for each genomic feature type, we computed and displayed an enrichment score, which measured the number of 1.96 sigma deviations from the null value of 1. Specifically, the relative enrichment score for hypermethylated (or hypomethylated) DMRs to be of a given feature type f is given by
where OR is the odds ratio for feature type f, and D is the estimated 95% confidence interval (−1.96 × sigma, +1.96 × sigma). Thus, the variance is scaled out allowing for a more suitable graphical representation of the significance of OR values.
GO analysis
GO analysis was performed to find enriched categories using the GOstats package in Bioconductor (Gentleman et al. 2004). P-values were multiple test corrected in order to reduce false-positive rates. GO terms with adjusted P-values of <0.01 were considered significant.
DNAm profiling using Infinium human 27K BeadArrays
One microgram of pooled DNA from MPNST, NF and non-neoplastic SCs was bisulfite converted using the EZ DNA methylation kit (Zymo Research). The samples were then hybridized to 27K BeadArrays according to the manufacturer's recommendation. The BeadStudio software (Illumina) was used to infer methylation scores from image intensities. Two technical replicates of each sample were run and data averaged for subsequent analysis.
Gene expression analysis
Gene expression analysis was performed using publically available data (MPNSTs, n = 10; NFs, n = 27) (Henderson et al. 2005; Miller et al. 2009). Gene expression data were derived from raw intensities using RMA (Irizarry et al. 2003). As the two data sets were from two different Affymetrix gene expression platforms, U95 (Henderson et al. 2005) and U133Plus2 (Miller et al. 2009), only the common probe sets (22218) between the two platforms were used in the feature analyses. DMRs and gene expression data were associated by Ensembl ID number. Significant differential gene expression was assessed using the LImma package in Bioconductor (Gentleman et al. 2004), and P-values were corrected for multiple testing. Clustering of gene expression data by DMR was carried out using the Cluster package in BioConductor(Gentleman et al. 2004). Significance between expression and sample phenotype was determined by carrying out 10,000 permutations based on the same number of genes from each DMR category.
Acknowledgments
A.F. and S.B. were supported by the Wellcome Trust (084071), and S.B. was also supported by a Royal Society Wolfson Research Merit Award. N.P. was supported by Skeletal Action Cancer Trust, Stanmore, United Kingdom. G.A.W. was supported by an EU grant (Highthroughput Epigenetic Regulatory Organisation in Chromatin [HEROIC], LSHG-CT-2005-018883) under the Sixth Framework Programme to S.B. A.E.T. was supported by a Heller Research Fellowship. L.A.N. and A.C.L. were supported by a Cancer Research UK program. The experimental work was supported in part by the UCLH/UCL Comprehensive Biomedical Research Centre, Illumina, and the Wellcome Trust. We thank UCL Genomics for its excellent technical support. We also thank the patients who donated their samples for research and the staff of the London Sarcoma Service who were involved in the care of the patients.
Footnotes
[Supplemental material is available for this article. The MeDIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE21714.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109678.110.
References
- Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, et al. 2008. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 68: 8541–8546 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE 2006. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116 [DOI] [PubMed] [Google Scholar]
- Beck S, Rakyan VK 2008. The methylome: Approaches for global DNA methylation profiling. Trends Genet 24: 231–237 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL 2009. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1: 177–200 [DOI] [PubMed] [Google Scholar]
- Cadieux B, Ching TT, VandenBerg SR, Costello JF 2006. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res 66: 8469–8476 [DOI] [PubMed] [Google Scholar]
- Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J 2010. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res 20: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Chen HL, Hsiau TH, Hsiau AH, Shi H, Brock GJ, Wei SH, Caldwell CW, Yan PS, Huang TH 2003. Methylation target array for rapid analysis of CpG island hypermethylation in multiple tissue genomes. Am J Pathol 163: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dante R, Dante-Paire J, Rigal D, Roizes G 1992. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Res 12: 559–563 [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. 2008. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38: 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M 2002. DNA methylation in cancer: too much, but also too little. Oncogene 21: 5400–5413 [DOI] [PubMed] [Google Scholar]
- Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A 2002. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet 39: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S 2006. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Fishbein L, Eady B, Sanek N, Muir D, Wallace MR 2005. Analysis of somatic NF1 promoter methylation in plexiform neurofibromas and Schwann cells. Cancer Genet Cytogenet 157: 181–186 [DOI] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M 1983. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 11: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gomez P, Bello MJ, Arjona D, Alonso ME, Lomas J, De Campos JM, Kusak ME, Gutierrez M, Sarasa JL, Rey JA 2003. Aberrant CpG island methylation in neurofibromas and neurofibrosarcomas. Oncol Rep 10: 1519–1523 [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A 2010. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 7: 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder A, Rosche M, Reuss DE, Holtkamp N, Uhlmann K, Friedrich R, Mautner VF, von Deimling A 2004. Methylation analysis of the neurofibromatosis type 1 (NF1) promoter in peripheral nerve sheath tumours. Eur J Cancer 40: 2820–2828 [DOI] [PubMed] [Google Scholar]
- Henderson SR, Guiliano D, Presneau N, McLean S, Frow R, Vujovic S, Anderson J, Sebire N, Whelan J, Athanasou N, et al. 2005. A molecular map of mesenchymal tumors. Genome Biol 6: R76 doi: 10.1186/gb-2005-6-9-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP 2004. CpG island methylator phenotype in cancer. Nat Rev Cancer 4: 988–993 [DOI] [PubMed] [Google Scholar]
- Jurgens B, Schmitz-Drager BJ, Schulz WA 1996. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Res 56: 5698–5703 [PubMed] [Google Scholar]
- Kawaguchi K, Oda Y, Saito T, Yamamoto H, Takahira T, Kobayashi C, Tamiya S, Tateishi N, Iwamoto Y, Tsuneyoshi M 2006. DNA hypermethylation status of multiple genes in soft tissue sarcomas. Mod Pathol 19: 106–114 [DOI] [PubMed] [Google Scholar]
- Kobayashi C, Oda Y, Takahira T, Izumi T, Kawaguchi K, Yamamoto H, Tamiya S, Yamada T, Oda S, Tanaka K, et al. 2006. Chromosomal aberrations and microsatellite instability of malignant peripheral nerve sheath tumors: A study of 10 tumors from nine patients. Cancer Genet Cytogenet 165: 98–105 [DOI] [PubMed] [Google Scholar]
- Laird PW 2005. Cancer epigenetics. Hum Mol Genet 14: R65–R76 [DOI] [PubMed] [Google Scholar]
- Laird PW 2010. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11: 191–203 [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy P, Vidaud D, Leroy K, Laurendeau I, Wechsler J, Bolasco G, Parfait B, Wolkenstein P, Vidaud M, Bieche I 2004. Molecular profiling of malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1, based on large-scale real-time RT-PCR. Mol Cancer 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijten M, Redeker S, van Noesel MM, Troost D, Westerveld A, Hulsebos TJ 2000. Microsatellite instability and promoter methylation as possible causes of NF1 gene inactivation in neurofibromas. Eur J Hum Genet 8: 939–945 [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. 2007. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M 2009. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics 4: 255–264 [DOI] [PubMed] [Google Scholar]
- Miller SJ, Rangwala F, Williams J, Ackerman P, Kong S, Jegga AG, Kaiser S, Aronow BJ, Frahm S, Kluwe L, et al. 2006. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res 66: 2584–2591 [DOI] [PubMed] [Google Scholar]
- Miller SJ, Jessen WJ, Mehta T, Hardiman A, Sites E, Kaiser S, Jegga AG, Li H, Upadhyaya M, Giovannini M, et al. 2009. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med 1: 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir D, Manthorpe M 1992. Stromelysin generates a fibronectin fragment that inhibits Schwann cell proliferation. J Cell Biol 116: 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen GP, Stemmer-Rachamimov AO, Ino Y, Moller MB, Rosenberg AE, Louis DN 1999. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol 155: 1879–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, Budiman MA, Korshunova Y, Maloney RK, Bedell JA, Citek RW, Bacher B, Peterson S, Rohlfing T, Hall J, et al. 2007. Identification of novel high-frequency DNA methylation changes in breast cancer. PLoS ONE 2: e1314 doi: 10.1371/journal.pone.0001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IS, Dervan P, McGoldrick A, Harrison M, Ponchel F, Speirs V, Isaacs JD, Gorey T, McCann A 2002. Promoter switch: A novel mechanism causing biallelic PEG1/MEST expression in invasive breast cancer. Hum Mol Genet 11: 1449–1453 [DOI] [PubMed] [Google Scholar]
- Pelletier J, Bruening W, Li FP, Haber DA, Glaser T, Housman DE 1991. WT1 mutations contribute to abnormal genital system development and hereditary Wilms' tumour. Nature 353: 431–434 [DOI] [PubMed] [Google Scholar]
- Robertson AK, Geiman TM, Sankpal UT, Hager GL, Robertson KD 2004. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem Biophys Res Commun 322: 110–118 [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH 2006. Large-scale structure of genomic methylation patterns. Genome Res 16: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S 2001. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 33: 561–568 [DOI] [PubMed] [Google Scholar]
- Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA 1999. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate 39: 166–174 [DOI] [PubMed] [Google Scholar]
- Serra E, Puig S, Otero D, Gaona A, Kruyer H, Ars E, Estivill X, Lazaro C 1997. Confirmation of a double-hit model for the NF1 gene in benign neurofibromas. Am J Hum Genet 61: 512–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Gu H, Bock C, Gnirke A, Meissner A 2009. High-throughput bisulfite sequencing in mammalian genomes. Methods 48: 226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titze S, Peters H, Wahrisch S, Harder T, Guse K, Buske A, Tinschert S, Harder A 2010. Differential MSH2 promoter methylation in blood cells of neurofibromatosis type 1 (NF1) patients. Eur J Hum Genet 18: 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, et al. 2009. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 69: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu JR, et al. 2010. Reprogramming of miRNA networks in cancer and leukemia. Genome Res 20: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW 2005. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 33: 6823–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL 2007. DNA hypomethylation and human diseases. Biochim Biophys Acta 1775: 138–162 [DOI] [PubMed] [Google Scholar]
- Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD 2010. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 9: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ 2001. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol 159: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al. 2004. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 64: 3060–3071 [DOI] [PubMed] [Google Scholar]
- Zhou H, Coffin CM, Perkins SL, Tripp SR, Liew M, Viskochil DH 2003. Malignant peripheral nerve sheath tumor: a comparison of grade, immunophenotype, and cell cycle/growth activation marker expression in sporadic and neurofibromatosis 1-related lesions. Am J Surg Pathol 27: 1337–1345 [DOI] [PubMed] [Google Scholar]